Abstract
Synthetic molecules derived from natural sugars with a positively charged amino group or ammonium salt and two lipophilic chains have been shown to inhibit TLR4 activation in vitro and in vivo. In order to characterize the mechanism of action of this class of molecules, we investigated possible interactions with the extracellular components that bind and shuttle endotoxin (lipopolysaccharide, LPS) to TLR4, namely LBP, CD14, and MD-2. Molecules that inhibited TLR4 activation inhibited LBP·CD14-dependent transfer of endotoxin monomers derived from aggregates of tritiated lipooligosaccharide ([3H]LOS) from Neisseria meninigitidis to MD-2·TLR4, resulting in reduced formation of a [[3H]LOS·MD-2·TLR4ECD]2 (Mr ~190,000) complex. This effect was due to inhibition of the transfer of [3H]LOS from aggregates in solution to sCD14 with little or no effect on [3H]LOS shuttling from [3H]LOS·sCD14 to MD-2. These compounds also inhibited transfer of [3H]LOS monomer from full length CD14 to a truncated, polyhistidine tagged CD14. Dose-dependent inhibition of [3H]LOS transfer between the two forms of CD14 was observed with each of three different synthetic compounds that inhibited TLR4 activation but not by another structurally related analog that lacked TLR4 antagonistic activity. Saturation transfer difference (STD) NMR data showed direct binding to CD14 by the synthetic TLR4 antagonist mediated principally through the lipid chains of the synthetic compound. Taken together, our findings strongly suggest that these compounds inhibit TLR4 activation by endotoxin by competitively occupying CD14 and thereby reducing the delivery of activating endotoxin to MD-2·TLR4.
Innate immunity is the first line of defense against microbial infections. Defense responses are activated when microbial components are recognized by a variety of pathogen sensors, including Toll-like receptors (TLRs) that activate the host defense effector system by rapidly triggering pro-inflammatory processes (1). Among microbial components, lipopolysaccharides (LPS) and lipooligosaccharides (LOS) and their bioactive portions, the lipodisaccharide lipid A, commonly defined as endotoxins (E), are potent stimulants of immune responses, but small differences in LPS structure can have a great influence on host immune responses (2). Endotoxin is an amphiphilic molecule and under physiological conditions is an integral membrane constituent. After extraction and purification, endotoxin forms large aggregates whose supramolecular structure depends on the chemical structure of endotoxin and, in particular, the lipid A moiety (3–5). However, as for every amphiphilic system, monomers are also present in a dynamic equilibrium. The induction of inflammatory responses by endotoxin is achieved by the coordinate and sequential action of four principal endotoxin-binding proteins: the lipopolysaccharide binding protein (LBP), the cluster differentiation antigen CD14, the myeloid differentiation protein (MD-2) and Toll-like receptor 4 (TLR4) (6). LBP interacts with endotoxin-rich bacterial membranes and purified endotoxin aggregates, catalyzing extraction and transfer of E monomers to CD14 in the presence of serum albumin (7, 8). Monomeric E·CD14 complexes are the most efficient vehicle for transfer of E monomers to MD-2 and to MD-2·TLR4 heterodimer, explaining the importance of LBP and CD14 for LPS signaling at low concentrations of endotoxin (9, 10). CD14 also has an important role in TRIF-dependent intracellular signaling triggered after TLR4 activation by endotoxin (11). The transfer of LPS from CD14 to MD-2, coupled with binding of MD-2 to TLR4, is required for TLR4 activation (12–14). Activation includes the formation of a dimer of the ternary [TLR4·MD-2·E]2 complex (15). Receptor dimerization leads to the recruitment of adapter proteins to the intracellular domain of TLR4, initiating the intracellular signal cascade that culminates in translocation of transcription factors to the nucleus and the biosynthesis of cytokines. The very recent determination of the crystal structure of [TLR4·MD-2·LPS]2 complex (16), together with crystallographic data of MD-2 bound to TLR4 antagonists lipid IVa (17) and Eritoran (18), has revealed some fundamental structural aspects of the TLR4 dimerization process and the molecular basis of TLR4 agonism and antagonism. The majority of antisepsis agents designed to be TLR4 antagonists, such as Eritoran (19), are comprised of a β(1–6) N-acetylglucosamine disaccharide scaffold with two phosphates in 1 and 4′ positions and four lipid chains instead of the six present in lipid A (20–23). Comparison of the crystal structure of the [TLR4·MD-2·LPS]2 complex (16) and that of TLR4·MD-2·Eritoran (18) indicates that the size of the MD-2 pocket is the same whether hexaacylated agonists or tetraacylated antagonists are bound. The MD-2 cavity volume can readily accommodate the four lipid chains of antagonists and additional space for lipid binding is generated, at least in the case of hexaacylated E. coli LPS, by displacing the glucosamine backbone upward by about 5 Å (16). This shift of the anomeric phosphate and resulting rearrangement of the lipid A acyl chains may be essential for the interaction of activating LPS·MD-2 from one TLR4·MD-2·LPS ternary complex to TLR4 from a second ternary complex, leading to formation of the [TLR4·MD-2·LPS]2 dimer.
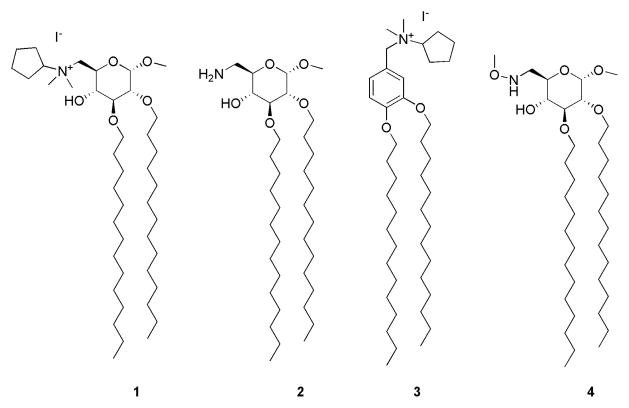
Many other compounds whose structures are not related to that of lipid A have also been described that interfere with TLR4 activation. These include the cyclohexene derivative named TAK242 (24, 25), now in clinical phase III trials, and both synthetic and natural (host) polycationic amphiphiles that, by binding LPS, sequester LPS from the CD14/MD-2/TLR4 pathway and protect animals against endotoxin-induced lethality (26–28). We recently developed a new class of inhibitory compounds, namely amphiphilic glycolipids 1, 2 and benzylammonium lipid 3 (Figure 1). We found that these compounds (1-3) inhibit LPS-induced TLR4 activation on HEK/TLR4 cells and LPS-induced septic shock in mice (29, 30). Compounds 1 and 2 are also able to inhibit other pathologies caused by TLR4 activation, such as inflammation and neuropathic pain (31). In contrast, glycolipid 4, which differs in structure from compounds 1 and 2 only by the presence of a neutral methoxyamino group instead of a charged amine, was inactive both in vitro and in vivo. The aim of the present work was to clarify the mode of action of these molecules with particular focus on the lead compound molecule 1, by analyzing possible interactions with the extracellular components that bind and shuttle endotoxin to TLR4, namely LBP, CD14, and MD-2 (free and TLR4-bound).
Figure 1.
Structure of methyl 6-deoxy- 6-N-dimethyl-N-cyclopentylamonium-2,3-di-O-tetradecyl-α-D-glucopyranoside (1) methyl 6-deoxy-6-amino-2,3-di-O-tetradecyl-α-D-glucopyranoside (2), N-(3,4-bis-tetradecyloxy-benzyl)-N-cyclopentyl-N,N-dimethylammonium (3), and methyl 6-deoxy-6-methoxyamino-2,3-di-O-tetradecyl-α-D-glucopyranoside (4).
EXPERIMENTAL PROCEDURES
Materials
[3H]LOS (25,000cpm/pmol) was metabolically labeled and isolated from an acetate auxotroph of Neisseria meningitidis serogroup B as described (32). LBP and sCD14 were gifts from Xoma (Berkley, CA) and Amgen Corp. (Thousand Oaks, CA), respectively. Human Serum Albumin (HSA) was obtained as an endotoxin-free, 25% stock solution (Baxter Health Care, Glendale, CA). Chromatography matrices (Sephacryl HR S200 and S300) were purchased from GE Healthcare and the silica-based metal chelation matrix, HisLink, is from Promega. ESF921 medium for High Five insect cells was purchased from Expressions Systems. Molecules 1-4 (Fig. 1) were prepared, purified and characterized as previously described (29, 30). In the assays described below, stocks of molecules 1-4 (ca. 15 mM) were freshly prepared in 50% DMSO/50% ethanol (vol/vol) and then further diluted with PBS/0.1% HSA to the desired concentration.
Preparation of recombinant proteins
Preparative amounts of sMD-2 and a truncated form of CD14 were generated from infections of High Five (Invitrogen) insect cells with baculovirus containing the cDNA for human MD-2 inserted into pBAC3 (His6-MD-2) or for human tCD14 (amino acids 1-156, tCD14-His6) inserted into pBAC11 as described previously (15). Conditioned medium containing MD-2 was stored at −80°C until needed. tCD14-His6 was purified using Ni FF Sepharose resin (GE Healthcare) on an Explorer100 FPLC (GEHealthcare) with an imidazole gradient. Purified tCD14-His6 was stored at 4°C (15). Conditioned medium containing MD-2 associated with TLR4 ectodomain (TLR4ECD), MD-2/TLR4ECD, was produced by transient transfection in HEK293T cells as described previously (15). Expression vectors containing cDNA of interest for production of FLAG-TLR4ECD, amino acids 24-631, (pFLAG-CMV-TLR4) and MD-2-FLAG-His (pEF-BOS) have been previously described and characterized (15). Media containing secreted proteins were concentrated 10–20-fold using Millipore Centricon-10 before use. Conditioned medium containing secreted MD-2/TLR4ECD proteins maintained activity to react with [3H]LOS·sCD14 for at least 6 months when stored at 4°C.
Size exclusion chromatography
Sephacryl S300 and S200 columns were used for resolution and identification of different [3H]LOS·protein products (i.e., ([3H]LOS·MD-2·TLR4ECD)2 (Mr ~190,000), [3H]LOS·sCD14 (Mr ~60,000), and ([3H]LOS·MD-2 (Mr ~25,000). For determination of apparent Mr, these columns were calibrated with the following Mr standards: blue dextran (2 × 106, V0), thyroglobulin (650,000), ferritin (440,000), catalase (232,000), IgG (158,000), HSA (66,000), ovalbumin (44,500), myoglobin (17,500), vitamin B12 (1,200, Vi).
In experiments in which the formation of [3H]LOS·MD-2 and [3H]LOS·CD14 complexes was monitored, chromatographic analyses were carried out, using a Sephacryl HR S200 column (1.6 × 30 cm) pre-equilibrated in PBS, pH 7.4, 0.1% HSA and eluted in the same buffer at a flow-rate of 0.5 mL/min at room temperature using AKTA Purifier or Explorer 100 fast protein liquid chromatography (GE Healthcare). Fractions of 1 mL were collected. In experiments in which the generation of [LOS·MD-2·TLR4ECD]2 was also monitored, the reaction mixtures were applied to a Sephacryl HR S300 column (1.6 × 70 cm) and chromatography was carried out as described above.
Radioactivity ([3H]LOS) in collected fractions was analyzed by liquid scintillation spectroscopy (Beckman LS liquid scintillation counter). Total recovery of [3H]LOS was ≥ 70% in all experiments; experimental results in chromatograms are reported as % cpm of total radiolabeled LOS recovered.
Production and purification of [3H]LOSagg, [3H]LOS·sCD14 and [3H]LOS·MD-2 complexes
[3H]LOSagg and [3H]LOS·sCD14 complex were prepared as previously described (8). Briefly, [3H]LOSagg (Mr > 20 × 106) were obtained after hot phenol extraction of [3H]LOS from metabolically labeled bacteria, followed by ethanol precipitation of [3H]LOSagg, and ultracentrifugation. Monomeric [3H]LOS·CD14 complexes (Mr ~ 60,000) were prepared by treatment of [3H]LOSagg for 30 min at 37°C with substoichiometric LBP (molar ratio 200:1 LOS:LBP) and 1–1.5 × molar excess sCD14 followed by gel exclusion chromatography (Sephacryl S200, 1.6 × 70 cm column) in PBS, pH 7.4, 0.1 % HSA to isolate monomeric [3H]LOS·sCD14 complex. [3H]LOS·MD-2 (Mr ~25,000) was generated by treatment of [3H]LOS·sCD14 (30 min at 37°C) with High Five insect cell medium containing His6-MD-2 followed by isolation of [3H]LOS·MD-2 by S200 chromatography. Radiochemical purity of [3H]LOS·sCD14 and 3H]LOS·MD-2 was confirmed by S200 chromatography (12).
Effect of synthetic molecules on formation of ([3H]LOS·MD-2·TLR4ECD)2 from [3H]LOSagg
In these experiments, sCD14 (0.8 nM, final concentration) were pre-incubated with LBP (4 pM, final concentration) ± synthetic molecules (0–10 μM) for 30 min at 37 °C in 0.875 ml of PBS, pH 7.4, 0.1% HSA. After this pre-incubation, [3H]LOSagg was added (up to 5 μl, yielding final LOS concentration of 0.8 nM) and the mixture was incubated an additional 30 min at 37° C. This was followed by addition of conditioned HEK293T cell medium containing preformed MD-2·TLR4ECD heterodimer (15) (0.125 ml, ca. 0.2 nM, final concentration, of reactive MD-2·TLR4ECD heterodimer) and an additional incubation for 15 min at 37 °C. Analysis of the reaction products was determined by gel size exclusion chromatography as described above.
Effect of the synthetic molecules on LOS transfer from [3H]LOS·sCD14 to either His-tagged tCD14 or sMD-2
Conditioned medium containing either His-tagged protein (corresponding to a final concentration in the incubation mixture of ca. 1.2 nM) was pre-incubated with sCD14 (0.8 nM) ± synthetic molecules in PBS, 1% HSA, 30 min at 37 °C. sCD14 was added to the preincubation to facilitate interaction of the synthetic compound with either His-tagged protein. LBP was not needed. This pre-incubation was followed by incubation for 30 min at 37 °C with [3H]LOS·sCD14 (0.8 nM) to allow transfer of [3H]LOS to available His-tagged tCD14 or MD-2. Products of the reactions were analyzed by size exclusion chromatography (see above) and/or by co-capture to HISLINK resin (see below).
Evaluation of transfer of [3H]LOS to His6-tagged proteins by co-capture to metal chelating resin
tCD14-His6 or His6-sMD-2 (1.6 nM, final concentration) was pre-incubated in 0.3 ml PBS, 1% HSA ± LBP (4 pM) for 30′ at 37°C ± different concentrations of synthetic compounds. [3H]LOSagg (0.8 nM) (with tCD14) or [3H]LOS·sCD14 (0.8 nM) (with tCD14 or with MD-2) was added followed by another incubation for 30 min at 37 °C. The reaction mixture was then incubated with HISLINK resin (20 μl) for 15 min at 25 °C, allowing His-tagged tCD14 and sMD-2 to be adsorbed onto the beads. The resin was spun down for 2 min at 2000× g, the supernatant was removed and the resin washed 2× with PBS, 1% HSA using the same procedure. The [3H]LOS absorbed onto the beads or recovered in the supernatant was quantified by liquid scintillation spectroscopy.
Endotoxin displacement assay
tCD14-His6 (10 μl, final concentration of 1.6 nM) was preincubated in PBS, pH 7.4, 0.1% HSA in the presence of LBP (8 pM) and [3H]LOSagg (1.6 nM) for 30 min at 37°C. These samples were then incubated an additional 30 min at 37° C ± synthetic compounds (10 μM), to allow displacement of bound [3H]LOS by the synthetic compound. Accumulation of [3H]LOS·His6-tCD14 was monitored by size exclusion chromatography and by capture by HISLINK resin as described above.
Assay of glycolipid molecule 1-sCD14 interaction by saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectroscopy
A freshly prepared stock (1.5 mM) of molecule 1 (in 1:1 DMSO-d6:methanol-d4, v/v) was diluted to 19 μM in a buffer containing 10 mM sodium phosphate (pH 6.8) in 100% 2H2O (sample A). An aliquot of sample A (0.5 ml) was mixed with 2 μl of a 120 μM stock solution of sCD14 (in the same sodium phosphate buffer) to give a final concentration of sCD14 of 0.5 μM (sample B). This sample was used to collect the STD NMR data. The control sample (sample C) contained 0.5 μM sCD14 alone in the same buffer (10 mM sodium phosphate (pH 6.8) in 100% 2H2O).
STD NMR data were collected at 25 °C on a Bruker Avance II 800 MHz NMR spectrometer equipped with a sensitive cryoprobe. STD data were collected as previously described (33–36). Since no aromatic resonances are present for molecule 1, the sCD14 protein can be selectively saturated by irradiating (on-resonance) at 7.6 and 8.0 ppm on the protein resonances with a train of Gauss-shaped pulses with a total length of saturation time of 1.0 s. The off-resonance radiation was performed at a chemical shift of 13.0 ppm where no resonances were observed. The on-resonance and off-resonance spectra were collected in an interleaved fashion. The total number of scan collected was 10K. The spectra were processed with the topspin v2.1 software on the Bruker spectrometer.
RESULTS AND DISCUSSION
Molecule 1 inhibits LBP/sCD14-dependent extraction and transfer of [3H]LOS monomers from [3H]LOS aggregates to CD14 and to MD-2·TLR4
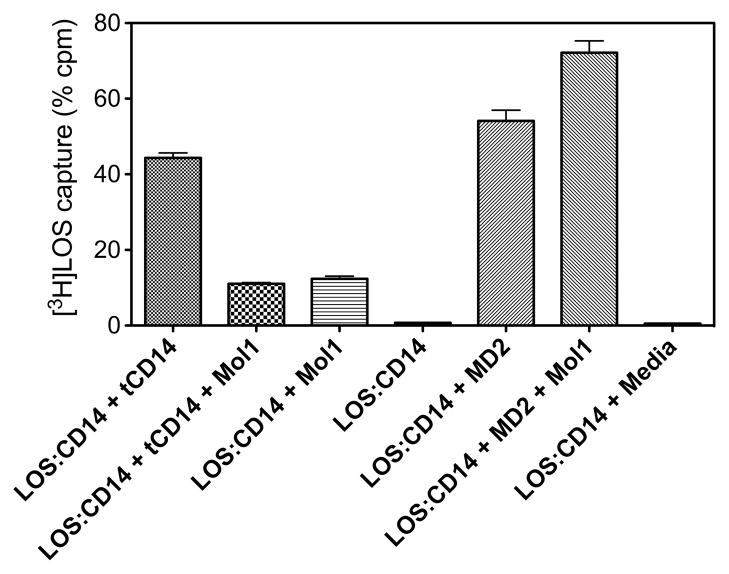
Molecule 1 was selected as a lead compound for biochemical characterization. We have previously shown that this compound inhibits LPS- and lipid A-stimulated cytokine production in cellular and animal models (29,30). To better define the mechanism of the inhibitory action of molecule 1 on LPS (lipid A)-triggered TLR4 activation, we tested the ability of this compound to inhibit LBP/CD14-dependent transfer of endotoxin ([3H]LOS) monomers from [3H]LOS aggregates to MD-2/TLR4, resulting in reduced formation of a [[3H]LOS·MD-2·TLR4ECD]2 (Mr ~190,000) complex (15).
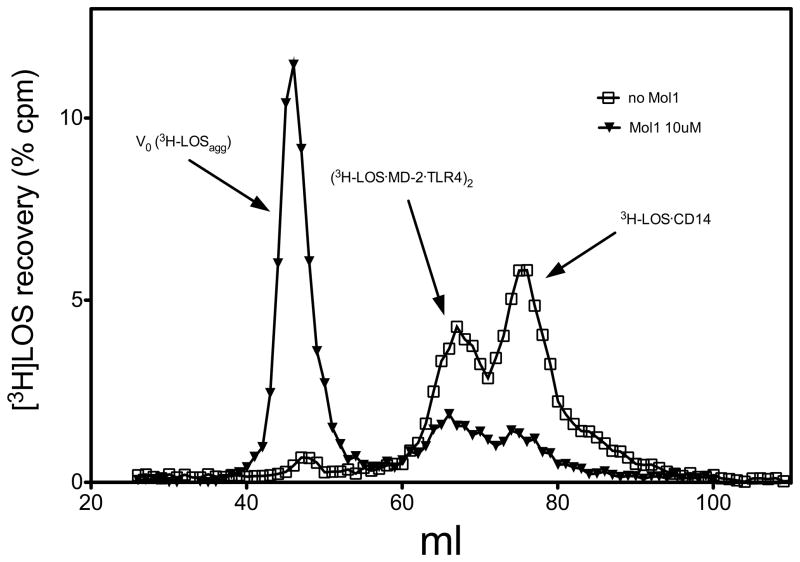
For this purpose, sCD14 was pre-incubated with LBP in the presence or absence of molecule 1 (10 μM), and then incubated with [3H]LOSagg followed by conditioned medium containing preformed MD-2·TLR4ECD heterodimer. Size-exclusion chromatography of the reaction mixture (Figure 2) showed that, in the absence of molecule 1, virtually all [3H]LOS aggregates (LOSagg) were converted to later eluting species (i.e., smaller [3H]LOS-containing complexes) corresponding to [3H]LOS·sCD14 (Mr~60,000) and [[3H]LOS·MD-2·TLR4ECD]2 (Mr~190,000). The identity of these complexes was confirmed by co-capture of [3H]LOS with immobilized monoclonal antibodies to CD14 (8) or to epitope tags in His6-MD-2 or in Flag-TLR4ECD (15) and by co-elution with purified, defined [3H]LOS·protein complexes. The generation of [3H]LOS·sCD14 reflects extraction and transfer of [3H]LOS monomers from [3H]LOSagg to sCD14 by the combined action of LBP and sCD14. Formation of [[3H]LOS·MD-2·TLR4ECD]2 reflects transfer of [3H]LOS monomers from [3H]LOS·sCD14 to MD-2·TLR4ECD (15). Thus, the chromatographic profile of [3H]LOSagg incubated first with LBP and sCD14 and then with conditioned medium containing sMD-2·TLR4ECD suggests that, under these experimental conditions, there is nearly complete extraction and transfer of [3H]LOS monomers from [3H]LOSagg to [3H]LOS·sCD14 followed by transfer of [3H]LOS monomers from about half of the [3H]LOS·sCD14 formed to MD-2·TLR4ECD. In contrast, in the presence of 10 μM molecule 1, accumulation of both [3H]LOS·sCD14 and [[3H]LOS·MD-2. ·TLR4ECD]2 was markedly reduced and most [3H]LOS was recovered in the void volume presumably as large [3H]LOS aggregates (Figure 2). The inhibition of accumulation of [3H]LOS·sCD14 suggested a primary effect of molecule 1 on LBP/sCD14-dependent extraction and transfer of [3H]LOS monomers from [3H]LOSagg to [3H]LOS·sCD14.
Figure 2.
Molecule 1 inhibits conversion of [3H]LOS·agg to [3H]LOS·sCD14 and [[3H]LOS·MD-2·TLR4]2. sCD14 (0.8 nM) was pre-incubated ± molecule 1 (10 μM) for 30 min at 37 °C in the presence of LBP (4 pM). Subsequently, [3H]LOSagg (0.8 nM) was added and this mixture was incubated for 30 min at 37° C. After the second incubation, conditioned HEK293T cell medium containing preformed reactive MD-2·TLR4ECD heterodimer (ca. 0.2 nM, final concentration) was added and incubated again for 15 min at 37 °C. This reaction mixture was applied to Sephacryl S200 as described under Experimental Procedures to measure conversion of [3H]LOSagg (void volume) to [3H]LOS·sCD14 (Mr ~60,000) and [[3H]LOS·MD-2·TLR4]2.(Mr ~190,000). The chromatographic profiles shown are representative of ≥4 experiments. Overall recoveries of [3H]LOS were ≥ 70%.
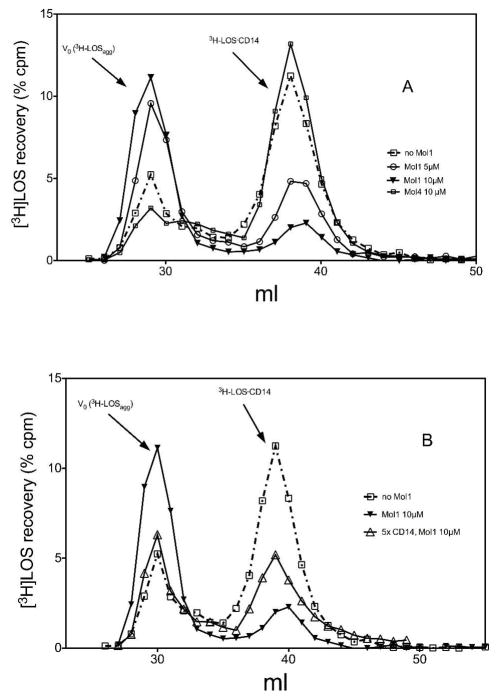
To test this hypothesis more directly, we examined the effect of increasing concentration of molecule 1 on LBP/sCD14-dependent conversion of [3H]LOSagg to [3H]LOS·sCD14. As shown in Figure 3A, molecule 1 caused a dose-dependent reduction in conversion of [3H]LOSagg to [3H]LOS·sCD14, with decreased accumulation of [3H]LOS·sCD14 accompanied by increased recovery of [3H]LOS as high Mr LOS aggregates (Figure 3A). In contrast, molecule 4, a compound with a chemical structure similar to molecule 1 (Fig. 1) but not active as an inhibitor of LPS-triggered TLR4 activation in vitro and in vivo (29,30), had little or no effect on LBP/sCD14-dependent conversion of [3H]LOSagg to [3H]LOS·sCD14 (Figure 3A). Thus, the ability of molecule 1, but not molecule 4, to inhibit LPS-triggered TLR4 signaling was paralleled by its ability to inhibit LBP/sCD14-dependent formation of monomeric E·CD14 complex.
Figure 3.
(A) Dose-dependent inhibition by molecule 1 but not by molecule 4 of LBP/sCD14-dependent extraction and transfer of [3H]LOS monomers from [3H]LOS aggregates to sCD14. sCD14 (0.8 nM) was pre-incubated with LBP (4 pM) and varying concentrations of the indicated synthetic molecules (0, 5, 10 μM) for 30 min at 37 °C in PBS, pH 7.4, 0.1% HSA. [3H]LOSagg (0.8 nM) was then added to the reaction mixture followed by an incubation for 30 min at 37° C and reaction products were analyzed using Sephacryl S200 chromatography. (B) Inhibition by molecule 1 of transfer of [3H]LOS from LOS aggregates to sCD14 is reduced by increasing sCD14 concentration. Incubations and analysis were as described in (A) except for the addition of 5x higher sCD14 concentration (4 nM). The chromatographic profiles shown are representative of ≥3 experiments. Overall recoveries of [3H]LOS were ≥ 70%.
One possible mechanism by which molecule 1 inhibits formation of E·CD14 could be by competing with E (e.g., [3H]LOS) for binding to CD14. In that case, increasing CD14 concentration should reduce the inhibitory effect of molecule 1. To test this hypothesis, we examined the effect of increasing the sCD14 concentration five-fold on the ability of molecule 1 to inhibit conversion of [3H]LOSagg to [3H]LOS·sCD14. A less inhibitory effect of molecule 1 was observed under these conditions (Figure 3B), consistent with a direct competition between molecule 1 and LOS for CD14.
Molecule 1 inhibits binding of [3H]LOS to CD14 but not to MD-2
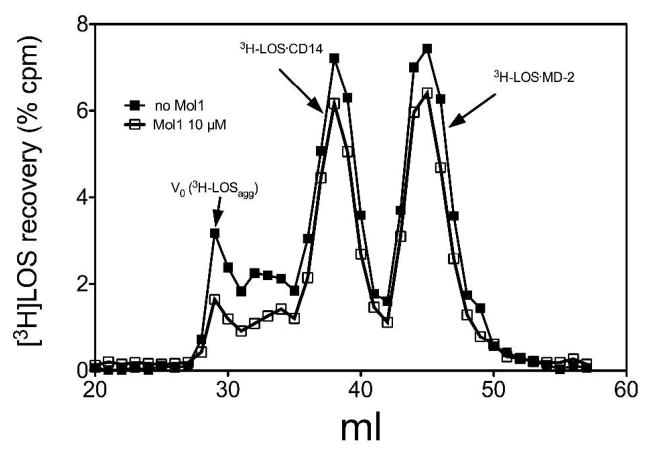
To test if molecule 1 can also inhibit transfer of [3H]LOS monomers from CD14 to MD-2, we examined the effect of molecule 1 on this reaction. For this purpose, conditioned medium containing sMD-2 was pre-incubated +/− molecule 1 (10μM) in the presence of LBP and sCD14 to allow formation of CD14·molecule 1 complex possibly needed for transfer of molecule 1 to MD-2. This pre-incubated mixture was then incubated with [3H]LOS·sCD14 to measure the degree of transfer of LOS from sCD14 to MD-2 that had been pre-incubated ± molecule 1 to assess the extent to which MD-2 is occupied by molecule 1, inhibiting transfer of [3H]LOS from CD14 to MD-2. Figure 4 shows that, in contrast to its effects on conversion of [3H]LOSagg to [3H]LOS·sCD14, molecule 1 had little or no effect on transfer of [3H]LOS monomers from [3H]LOS·sCD14 to MD-2, indicating that little or no MD-2 is occupied by molecule 1.
Figure 4.
Molecule 1 does not inhibit transfer of [3H]LOS monomers from CD14 to MD-2. sMD-2 (1.2 nM) containing medium was pre-incubated with sCD14 (0.8 nM) ± molecule 1 for 30 min at 37 °C to give molecule 1 an opportunity to interact with MD-2. This pre-incubation mixture was then incubated for an additional 30 min at 37° C with [3H]LOS·sCD14 (0.8 nM) to allow for transfer of [3H]LOS to unoccupied MD-2. The reaction mixture products were analyzed by Sephacryl S200 chromatography. Chromatograms shown are representative of ≥3 independent experiments. Overall recoveries [3H]LOS were > 70%.
The above experiments demonstrate an ability of molecule 1 to inhibit extraction and transfer of [3H]LOS monomers from [3H]LOS aggregates to CD14 without affecting subsequent transfer of E monomers from [3H]LOS·sCD14 to MD-2. This targeted inhibitory effect could be explained by a selective effect on extraction of E monomers from [3H]LOS aggregates that is needed for generation of [3H]LOS·sCD14 or a selective effect on net transfer of [3H]LOS monomers to sCD14 vs. sMD-2. To test the latter possibility more directly, we took advantage of the ability of monomeric E·sCD14 complexes to serve as a donor of E monomers to either CD14 or MD-2. For this purpose, we used [3H]LOS·sCD14 (full-length sCD14, no His tag) as [3H]LOS donor and either His6-tagged truncated sCD14 (tCD14; residues 1-156; 15) or His6-tagged sMD-2 as [3H]LOS acceptors. Thus, [3H]LOS transfer from [3H]LOS·sCD14 to tCD14-His6 or to His6-sMD-2 could be readily measured by assay of co-capture of [3H]LOS to HISLINK resin as described in Methods.
As expected, transfer of [3H]LOS from [3H]LOS ·sCD14 to tCD14-His6 and to His6-sMD-2 was similar (Fig. 5.). Molecule 1 markedly inhibited transfer of [3H]LOS from full-length sCD14 (no His tag) to tCD14-His6 but did not inhibit transfer of [3H]LOS to His6-sMD-2 (Fig. 5). Note that the presence of molecule 1 increased by about 10% the nonspecific capture of [3H]LOS after incubation of LOS·sCD14 ± MD-2 (Figure 5). This may be explained by the ability of molecule 1 to promote displacement of [3H]LOS from LOS·sCD14 (see below), followed by non-specific adsorption of some of the displaced monomeric [3H]LOS to the HISLINK resin. These results demonstrate that molecule 1 inhibits transfer (binding) of [3H]LOS monomer to CD14 but not to sMD-2.
Figure 5.
Molecule 1 selectively inhibits transfer of [3H]LOS monomer to tCD14 but not to MD-2. His-tagged tCD14 or sMD2 (1.6 nM) was pre-incubated for 30′ at 37°C with 10 uM molecule 1, followed by addition of [3H]LOS·sCD14 (0.8 nM) and further incubation for 30 min at 37 °C. Transfer of [3H]LOS to His-tagged tCD14 and MD-2 was assayed by co-capture of [3H]LOS to the HISLINK resin as described under Experimental Procedures. Capture of [3H]LOS·sCD14 (no His-tag) before and after incubation in medium without His-tagged protein was < 4% and subtracted from each of the experimental samples shown. Data shown are representative of 3 experiments, each in duplicate, and are expressed as mean ± SEM.
Molecule 1 promotes displacement of [3H]LOS from [3H]LOS·CD14
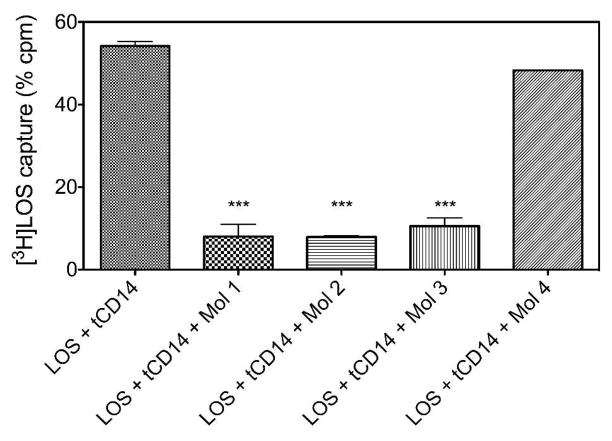
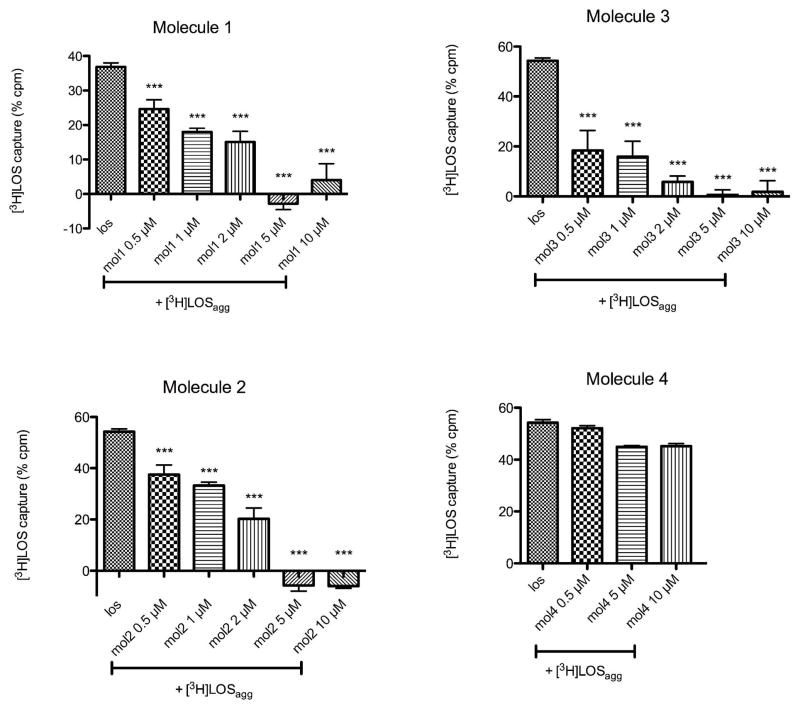
Molecule 1 could inhibit net binding of [3H]LOS to CD14 by inhibiting transfer of [3H]LOS to CD14 and/or by promoting displacement of bound [3H]LOS from CD14. To test the latter possibility more directly, we incubated [3H]LOSagg with tCD14 in the presence of LBP at 37 °C for 30 min in PBS/0.1% albumin, thus generating [3H]LOS·tCD14-His6. This mixture containing [3H]LOS·tCD14-His6 complex was then incubated ± 10 μM molecule 1 at 37 °C for 30 min. Loss of [3H]LOS from tCD14-His6 was observed by both gel sieving chromatography (data not shown) and co-capture on the HISLINK resin (Figure 6). Both assays showed a marked diminution of [3H]LOS bound to tCD14-His6 induced by incubation with molecule 1. Closely similar effects were seen with molecules 2 and 3, but not with molecule 4 (Figure 6), paralleling the ability of molecules 1-3, but not 4, at 1–10 μM concentrations, to inhibit LPS-triggered TLR4 signaling in vitro and in vivo (29,30). We took advantage of this simpler and faster co-capture assay to quantify the inhibition of LOS·CD14 complex formation by our synthetic compounds (Figure 7). Molecules 1-3 showed a dose-dependent inhibition of the transfer of [3H]LOS monomers from [3H]LOSagg to tCD14-His6, with a calculated IC50 of 0.8, 2, and 0.4 μM for molecules 1, 2, and 3, respectively. In contrast, molecule 4, even at 10 μM, produced little inhibition of the transfer of [3H]LOS monomers from [3H]LOSagg to tCD14-His6.
Figure 6.
Molecules 1-3, but not molecule 4, promote displacement of [3H]LOS from [3H]LOS·tCD14-His6. [3H]LOS··tCD14-His6 was formed by pre-incubation of tCD14-His6 with [3H]LOS·agg (0.8 nM) + LBP (4 pM), and then incubated an additional 30 min at 37° C ± 10 μM molecules 1-4, as indicated. The amount of remaining [3H]LOS·tCD14-His6 was assayed by co-capture of [3H]LOS To HISLINK resin as described under Experimental Procedures. Non-specific binding of [3H]LOSagg incubated in the absence of ·tCD14-His6 was < 9% and subtracted from each of the experimental samples shown. Results shown represent the mean of 3 experiments each in duplicates and data are expressed as mean ± SEM. ***indicates conditions in which incubation with the indicated synthetic molecule resulted in a statistically significant (p < 0.01 (ANOVA; Dunnett’s test)) reduction in co-capture of [3H]LOS (i.e., retention of [3H]LOS.tCD14-His6).
Figure 7.
Dose-dependent inhibition of the net transfer of [3H]LOS monomers from [3H]LOSagg to tCD14-His6 by molecules 1-4 as assayed by co-capture of [3H]LOS to HISLINK resin. tCD14-His6 (1.6 nM) was pre-incubated + LBP (4 pM) for 30′ at 37°C ± increasing concentrations of the indicated synthetic compounds. [3H]LOSagg (0.8 nM) was then added and samples were incubated for an additional 30 min at 37 °C. Results shown are the mean ± SEM of ≥ 3 independent experiments, each in duplicate. Non-specific binding, measured as [3H]LOS binding in absence of tCD14-His6, was subtracted from each sample. ***indicates conditions in which pre-incubation of tCD14-His6 with the added synthetic molecule resulted in a statistically significant (p < 0.01 (ANOVA; Dunnett’s test)) reduction in co-capture of [3H]LOS (i.e., reduced net formation of [3H]LOS.tCD14-His6).
Saturation Transfer Difference (STD) NMR shows direct binding between molecule 1 and sCD14
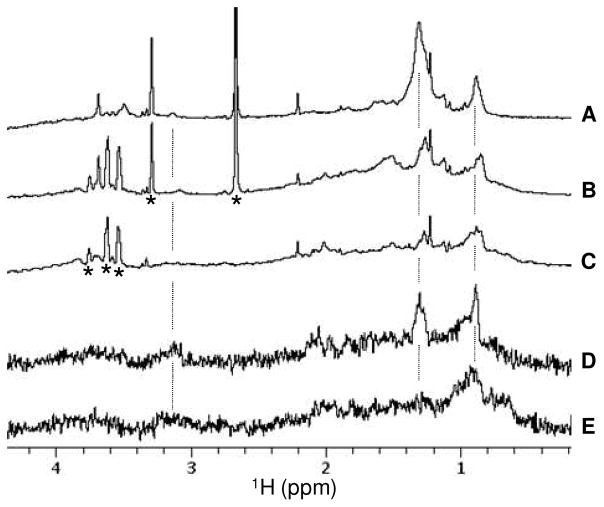
The ability of molecule 1 to inhibit net transfer/binding of LOS to sCD14 could be most easily explained by an ability of molecule 1 (and related molecules 2 and 3) to bind to CD14 and thus reduce the interaction of LOS with CD14. To test more directly the possible interaction of molecule 1 with sCD14, we made use of STD NMR (33–35) to probe intermolecular interactions between molecule 1 and sCD14 and to determine sites in molecule 1 that interact with sCD14. STD NMR experiments performed on the sample that contained both molecule 1 and sCD14 clearly showed saturation transfer from sCD14 to molecule 1, most strongly at 0.88 and 1.30 ppm and weakly at 3.13 ppm when the protein resonances were saturated by RF irradiation (Figure 8D). Identical STD NMR experiments performed on the control sample that contained only sCD14 showed no resonances at these chemical shift positions except for the broad peaks of background signals (Figure 8E). Excellent cancellation in the STD spectra of the strong solvent peaks present in the samples (Figures 8B–E) confirmed the good quality of the STD data. The STD peaks detected at 0.88 and 1.30 ppm are derived from the lipid chain –CH3 and –CH2 groups, respectively (Figure 8A). The weak STD peak at 3.13 ppm is likely derived from the lipid chain –OCH2 group near the sugar moiety. These STD data strongly suggest that molecule 1 binds to CD14 and that the lipid chains have a major role in its interaction with CD14.
Figure 8.
Saturation transfer difference (STD) NMR analysis of interaction between molecule 1 and sCD14. (A–C) 1D 1H NMR spectra of 19 μM compound 1 (sample A), 19 μM compound 1 plus 0.5 μM sCD14 (sample B), and 0.5 μM sCD14 (sample C), respectively. (D) STD NMR spectrum obtained on sample B. (E) STD NMR spectrum collected on the control sample C. The stars indicate 1H peaks derived from solvents (DMSO at 2.66 ppm, methanol at 3.29 ppm) and/or buffer components (e.g. glycerol from sCD14 initial stock at 3.5–3.8ppm).
CONCLUSION
Previous experimental observations on molecule 1 and similar glycolipids or benzylammonium lipids suggested that the anti-endotoxic activity of these molecules is due to an interference with the TLR4 pathway (29, 30). The lipid A-stimulated cytokine production by dendritic cells and macrophages is inhibited by compound 1 and an antagonistic effect on TLR4-dependent but not on TLR2- or TLR9-dependent cell activation was observed in selectively transfected HEK293 cells (29). The main aim of this work was to better understand the mechanism of action of cationic glycolipid 1, as a representative prototype of this class of compounds. For this purpose, we tested the effect of molecule 1 on the sequential extraction and transfer of [3H]LOS monomers from [3H]LOSagg to LBP, CD14, and MD-2(·TLR4ECD), reactions that are key to efficient delivery of activating endotoxin to MD-2·TLR4 (12, 15). Our findings demonstrate that molecule 1, and structurally related molecules 2 and 3, affect this multistep pathway in a relatively selective manner, acting mainly to inhibit transfer to and/or stable occupation of CD14 by lipooligosaccharide ([3H]LOS) monomers. Similar inhibitory effects of molecule 1 were seen when the transfer of [3H]LOS from either [3H]LOS aggregates or from monomeric [3H]LOS·sCD14 was examined (Figures 3, 5, and 6), indicating that an additional effect of molecule 1 on interactions between LBP and E-rich interfaces was unlikely and not necessary for the inhibitory action of molecule 1. Remarkably, shuttling of E monomers from CD14 to MD-2 is unaffected by molecule 1. The apparently selective targeting of CD14 by these compounds bearing two lipid chains is reminiscent of the previously described ability of CD14, but not MD-2, to interact stably with other compounds with three lipid chains such as phospholipids (36) and lipopeptides (37, 38). The targeting of CD14 by molecules 1-3 is consistent with their ability to inhibit CD14-dependent TLR4 activation by endotoxin (lipid A/LPS) but not CD14-independent cell activation by TLR2 and TLR9 agonists (29). Moreover, as predicted from our biochemical studies, molecule 1 does not block TLR4 activation when endotoxin (LOS) is presented as a pre-formed LOS·sCD14 complex that needs only to transfer LOS to MD-2 to induce TLR4 activation (data not shown).
Whether or not the selective inhibition of LOS binding to CD14, but not to MD-2, reflects a higher affinity of these dialkylated compounds for CD14 vs. MD-2 or rather a higher affinity and more stable association of LOS with MD-2 than with CD14 cannot be judged. The inability of 1 to inhibit transfer of [3H]LOS to MD-2 from [3H]LOS·CD14 (Figures 4, 5) underscores the efficiency of that transfer reaction and strongly suggests that 1 and related compounds must have time to occupy CD14 and thereby exclude E from CD14 to exert their maximum inhibitory effects. Although the STD NMR data did not provide the ligand binding site(s) on CD14 for the compound, these NMR data together with the fact that 1 inhibits the formation of LOS·sCD14 complex is most compatible with interaction of 1 with the hydrophobic cavity of CD14 believed to provide the binding site for endotoxins (lipid A) and a variety of other molecules, including triacylated lipids (37, 39).
Compounds 2 and 3 but not 4 were active in inhibiting endotoxin displacement from the complex with CD14 and compound 4 was also inactive in inhibiting CD14-mediated endotoxin presentation to MD-2·TLR4. The basic nitrogen present in compound 2 and the quaternary ammonium of 1 and 3 are replaced in 4 by the less basic methyl hydroxylamine. As a consequence, while sugar moieties of compounds 1, 2 and 3 are cationic (compound 2 is protonated at neutral pH), compound 4 is mostly neutral in the conditions used for biological assays. The absence of a cationic group could affect dramatically the biological activity of 4, influencing the aggregation state in solution as well the affinity for CD14. Work is in progress to determine the thermodynamic parameters of binding and affinity (included stoichiometry of binding) between compounds 1-3 and CD14 and to further clarify the structural basis of the apparently lower reactivity of molecule 4 with CD14. The experiments reported here suggest that the affinity of compound 1 for CD14 is probably ca. 1000-fold lower than that of LBP-treated E aggregates or monomeric E·CD14 for CD14. The functional properties described suggest that molecules 1-3 could be considered lead compounds in the development of new anti-endotoxic agents selectively targeting CD14. The broader role of CD14 (vs. MD-2) in other biological recognition/response pathways may further expand the potential applications of these compounds.
Acknowledgments
We thank Xoma Corp. (Berkeley, CA) for recombinant LBP, Amgen Corp. (Thousand Oaks, CA) for sCD14 and Baxter Health Care (Glendale, CA) for endotoxin-free Human Serum Albumin (HSA). We are also grateful to DeSheng Zhang for preparation of radiolabeled LOS, to Gaetana Damore for the synthesis of some molecules.
ABBREVIATIONS
- CD14
cluster differentiation antigen
- tCD14
truncated CD14
- sCD14
soluble CD14
- CMV
- DMSO
dimethylsulfoxide
- E
endotoxin
- FPLC
fast protein liquid chromatography
- HEK cells
Human Embryonic Kidney cells
- HSA
human serum albumin
- LPS
lipopolysaccharide
- LBP
Lipid Binding Protein
- LOS
lipooligosaccharide
- MD-2
myeloid differentiation protein
- Mr
apparent molecular weight
- pBAC3
bacterial artificial chromosome (BAC) vector
- STD
Saturation Transfer Difference
- TLR4
Toll-like receptor 4
- TLR4ECD
TRL4 ectodomain
Footnotes
This work was supported by grants from the National Institute of Allergy and Infectious Disease (AI05732) and the Veterans’ Administration.
References
- 1.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 2.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol. 2006;7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 3.Mueller M, Lindner B, Dedrick R, Schromm AB, Seydel U. Endotoxin: Physical requirements for cell activation. J of Endotoxin Res. 2005;11:299–303. doi: 10.1179/096805105X46574. [DOI] [PubMed] [Google Scholar]
- 4.Seydel U, Hawkins L, Schromm AB, Heine H, Scheel O, Koch MH, Brandenburg K. The generalized endotoxic principle. Eur J Immunol. 2003;33:1586–1592. doi: 10.1002/eji.200323649. [DOI] [PubMed] [Google Scholar]
- 5.Gutsmann T, Schromm AB, Brandenburg K. The physicochemistry of endotoxins in relation to bioactivity. Int J Microbiol. 2007;297:341–352. doi: 10.1016/j.ijmm.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Jerala R. Structural biology of the LPS recognition. Int J Med Microbiol. 2007;297:353–363. doi: 10.1016/j.ijmm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 8.Gioannini TL, Zhang DS, Teghanemt A, Weiss JP. Essential Role for Albumin in the Interaction of Endotoxin with Lipopolysaccharide-binding Protein and sCD14 and Resultant Cell Activation. J Biol Chem. 2002;277:47818–47825. doi: 10.1074/jbc.M206404200. [DOI] [PubMed] [Google Scholar]
- 9.Gangloff SC, Hijiya N, Haziot A, Goyert SM. Clin Infect Dis. 1999;2(8):491–496. doi: 10.1086/515176. [DOI] [PubMed] [Google Scholar]
- 10.Lynn WA, Liu Y, Golenbock DT. Infect Immun. 1993;61:4452–4461. doi: 10.1128/iai.61.10.4452-4461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. Nat Immunol. 2005;6:565–569. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 12.Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci USA. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teghanemt A, Widstrom RL, Gioannini TL, Weiss JP. Isolation of Monomeric and Dimeric Secreted MD-2: endotoxins-CD14 and toll-like receptor 4 ectodomain selectively react with the monomeric form of secreted MD-2. J Biol Chem. 2008;283:21881–21889. doi: 10.1074/jbc.M800672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teghanemt A, Re F, Prohinar P, Widstrom R, Gioannini TL, Weiss JP. Novel Roles in Human MD-2 of Phenylalanines 121 and 126 and Tyrosine 131 in Activation of Toll-like Receptor 4 by Endotoxin. J Biol Chem. 2008;283:1257–1266. doi: 10.1074/jbc.M705994200. [DOI] [PubMed] [Google Scholar]
- 15.Prohinar P, Re F, Widstrom R, Zhang D, Teghanemt A, Gibson BW, Weiss JP. Specific High Affinity Interactions of Monomeric Endotoxin-Protein Complexes with Toll-like Receptor 4 Ectodomain. J Biol Chem. 2007;282:1010–1017. doi: 10.1074/jbc.M609400200. [DOI] [PubMed] [Google Scholar]
- 16.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature Letters. 2009:1–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 17.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structure of Human MD-2 and its complex with antiendotoxic Lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 18.Kim HM, Park BS, Kim JI, Lee SE, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee J-O. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Mullarkey M, Rose JR, Bristol J, Kawata T, Kimura A, Kobayashi S, Przetak M, Chow J, Gusovsky F, Christ WJ, Rossignol DP. Inhibition of endotoxin response by E5564, a novel Toll-like receptor 4-directed endotoxin antagonist. J Pharmacol and Exp Therapeutics. 2003;304:1093–1102. doi: 10.1124/jpet.102.044487. [DOI] [PubMed] [Google Scholar]
- 20.Netea MG, van Deuren M, Kullberg BJ, Cavaillon JM, Van der Meer JW. Does the shape of lipid A determine the interaction of LPS with Toll-like receptors? Trends in Immunol. 2002;23:135–139. doi: 10.1016/s1471-4906(01)02169-x. [DOI] [PubMed] [Google Scholar]
- 21.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow G, Ulmer AJ, Zahringer U, Seydel U, Di Padova F, Schreier M, Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 22.Seydel U, Oikawa M, Fukase K, Kusumoto S, Brandenburg K. Intrinsic conformation of lipid A is responsible for agonistic and antagonistic activity. Eur J Biochem. 2000;267:3032–3039. doi: 10.1046/j.1432-1033.2000.01326.x. [DOI] [PubMed] [Google Scholar]
- 23.Seydel U, Schromm AB, Blunck R, Brandenburg K. Chemical structure, molecular conformation, and bioactivity of endotoxins. Chem Immunol. 2000;74:5–24. doi: 10.1159/000058754. [DOI] [PubMed] [Google Scholar]
- 24.Yamada M, Ichikawa T, Ii M, Sunamoto M, Itoh K, Tamura N, Kitazaki T. Discovery of Novel and Potent Small-Molecule Inhibitors of NO and Cytokine Production as Antisepsis Agents: Synthesis and Biological Activity of Alkyl 6-(N-Substituted sulfamoyl)cyclohex-1-ene-1-carboxylate. J Med Chem. 2005;48:7457–7467. doi: 10.1021/jm050623t. [DOI] [PubMed] [Google Scholar]
- 25.Takashima K, Matsunaga N, Yoshimatsu M, Hazeki K, Kaisho T, Uekata M, Hazeki O, Akira S, Iizawa Y, Ii M. Analysis of binding site for the novel small-molecule TLR4 signal transduction inhibitor TAK-242 and its therapeutic effect on mouse sepsis model. British J Pharmacol. 2009;157:1250–1262. doi: 10.1111/j.1476-5381.2009.00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller KA, Kumar EVK, Wood SJ, Cromer JR, Datta A, David SA. Lipopolysaccharide Sequestrants: Structural Correlates of Activity and Toxicity in Novel Acylhomospermines. J Med Chem. 2005;48:2589–2599. doi: 10.1021/jm049449j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns MR, Jenkins SA, Kimbrell MR, Balakrishna R, Nguyen TB, Abbo BG, David SA. Polycationic Sulfonamides for the Sequestration of Endotoxin. J Med Chem. 2007;50:877–888. doi: 10.1021/jm061198m. [DOI] [PubMed] [Google Scholar]
- 28.David SA. Towards a rational development of anti-endotoxin agents: novel approaches to sequestration of bacterial endotoxins with small molecules. J Mol Recognition. 2001;14:370–387. doi: 10.1002/jmr.549. [DOI] [PubMed] [Google Scholar]
- 29.Peri F, Granucci F, Costa B, Zanoni I, Marinzi C, Nicotra F. Inhibition of lipid A stimulated activation of human dendritic cells and macrophages by amino and hydroxylamino monosaccharides. Angew Chem Int Ed. 2007;46:3308–3312. doi: 10.1002/anie.200604932. [DOI] [PubMed] [Google Scholar]
- 30.Piazza M, Rossini C, Della Fiorentina S, Pozzi C, Comelli F, Bettoni S, Fusi P, Costa B, Peri F. Glycolipids and Benzylammonium Lipids as Novel Antisepsis Agents: Synthesis and Biological Characterization. J Med Chem. 2009;52:1209–1213. doi: 10.1021/jm801333m. [DOI] [PubMed] [Google Scholar]
- 31.Bettoni I, Comelli F, Rossini C, Granucci F, Giagnoni G, Peri F, Costa B. Glial TLR4 Receptor as New Target to Treat Neuropatic Pain: Efficacy of a New Receptor Antagonist in A Model Peripheral Nerve Injury in Mice. Glia. 2008;56:1312–1319. doi: 10.1002/glia.20699. [DOI] [PubMed] [Google Scholar]
- 32.Giardina PC, Gioannini T, Buscher BA, Zaleski A, Zhang DS, Stoll L, Teghanemt A, Apicella MA, Weiss J. Construction of acetate auxotrophs of Neisseria meningitidis to study host-meningococcal endotoxin interactions. J Biol Chem. 2001;276:5883–5891. doi: 10.1074/jbc.M009273200. [DOI] [PubMed] [Google Scholar]
- 33.Meinecke R, Meyer B. Determination of the Binding Specificity of an Integral Membrane Protein by Saturation Transfer Difference NMR: RGD Peptide Ligands Binding to Integrin aIIbβ3. J Med Chem. 2001;44:3059–3065. doi: 10.1021/jm0109154. [DOI] [PubMed] [Google Scholar]
- 34.Streiff JH, Juranic NO, Macura SI, Warner DO, Jones KA, Perkins WJ. Transfer Difference Nuclear Magnetic Resonance Spectroscopy As a Method for Screening Proteins for Anesthetic Binding. Mol Pharmacol. 2004;66:929–935. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 35.Mayer M, Meyer B. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 36.Post D, Zhang De S, Eastvold J, Teghanemt A, Gibson BW, Weiss JP. Biochemical and Functional Characterization of Membrane Blebs Purified from Neisseria meningitidis Serogroup B. J Biol Chem. 2005;280:38383–38394. doi: 10.1074/jbc.M508063200. [DOI] [PubMed] [Google Scholar]
- 37.Nakata T, Yasuda M, Fujita M, Kataoka H, Kiura K, Sano H, Shibata K. CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cell Microbiol. 2006;8:1899–1909. doi: 10.1111/j.1462-5822.2006.00756.x. [DOI] [PubMed] [Google Scholar]
- 38.Manukyan M, Triantafilou K, Mackie A, Nilsen N, Espevik T, Wiesmuller KH, Ulmer AJ, Heine H. Binding of lipopeptide to CD14 induces physical proximity of CD14, TLR2 and TLR1. Eur J Immunol. 2005;35:911–921. doi: 10.1002/eji.200425336. [DOI] [PubMed] [Google Scholar]
- 39.Kim JI, Lee CL, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling. J Biol Chem. 2005;25:11347–11351. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]