Abstract
S. cerevisiae mutants lacking VPS4 missort several vacuolar proteins to the extracellular space, including carboxypeptidase (CPY), vacuolar protease A (PrA), and vacuolar protease B (PrB). In addition, certain soluble secretory proteins, such as invertase and acid phosphatase, are missorted from the pre-vacuolar compartment (PVC) to the general secretory pathway prior to exocytosis. Although little is known about sorting of proteins via the PVC in C. albicans, we have previously demonstrated that the C. albicans vps4Δ null mutant missorts PrA and CPY extracellularly, but fails to secrete the aspartyl proteases Sap2p and Sap4-6p. To further define the role of C. albicans VPS4 in the trafficking of pre-vacuolar proteins, we have used 2 dimensional gel electrophoresis (2-DE) and mass spectrometry techniques to study soluble proteins in the supernatants of planktonic cultures obtained from the C. albicans vps4Δ mutant compared to control strain DAY185. Results indicated that lack of VPS4 results in a decrease of canonically secreted proteins whilst having a limited effect on non-canonically secreted extracellular proteins. Four canonically secreted proteins (Cht3p, Pra1p, Mp65p and Sun41p) were identified as reduced in the supernatants from the mutant strain. We also indentified two other major consequences of lack of VPS4, likely associated with secretion defects: altered branching and biofilm formation.
Keywords: Candida albicans, protein secretion, proteomics, secreted aspartyl proteases, vacuole, VPS4
INTRODUCTION
Candida albicans is a versatile commensal organism which possesses a number of attributes which enhances its ability to survive in diverse environments and enables it to transition from harmless commensal to invasive pathogen. Recently, several molecular studies have investigated the role of the vacuole and vacuolar protein sorting (VPS) genes in secretion, filamentation, and pathogenesis [1, 2, 3]. C. albicans vps34Δ mutants, which are vacuolar class “E” mutants lacking the phostphatidylinositol 3-kinase Vps34p, are sensitive to temperature and osmotic stresses, defective in filamentation and adhesion, and are avirulent in a mouse model of disseminated candidiasis [1]. C. albicans mutants lacking the class “E” endocytic genes VPS28 or VPS32 are defective in growth at high alkaline pH, and are hypovirulent in a mouse model of disseminated candidiasis [2]. C. albicans vps11 Δ mutants, which are vacuolar class “C” mutants defective in multiple protein sorting pathways to the PVC and vacuole, are also sensitive to temperature and osmotic stresses and are defective in filamentation [3]. Furthermore, the vps11Δ mutant is unable to kill macrophages efficiently in vitro [4].
S. cerevisiae vps4 mutants accumulate vacuolar, endocytic, and late-Golgi marker proteins in an aberrant multilamellar compartment, which is thought to represent an abnormal pre-vacuolar compartment (PVC) [5]. Vps4p function is required for efficient transport out of the PVC whether this traffic is retrograde to the Golgi or antegrade to the vacuole. Vps4p functions as a key component which interacts with the ESCRT-III complex required for multivesicular body formation and endocytosis (reviewed in [6]). Studies of a C. albicans vps4Δ null mutant have revealed impaired endocytosis, and minor defects in filamentation on M199 solid media but not in serum [7]. We have previously shown that a C. albicans vps4Δ mutant missorts CPY and vacuolar protease A (Apr1p) to the extracellular space [8]. Conversely, the vps4Δ mutant does not produce extracellular Sap2p [8] or Sap4-6p [9].
In the present series of experiments, we sought to further define the role VPS4 in the sorting of pre-vacuolar secretory proteins in C. albicans, and describe two new phenotypes of the vps4Δ mutant, altered branching and biofilm formation, likely associated with defects in secretory proteins.
MATERIALS and METHODS
C. albicans strains and media
C. albicans strains used in this study were BWP17 (ura3Δ∷imm434/ura3Δ∷imm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG) and control strain DAY185 (both from A. Mitchell, Columbia University). The C. albicans vps4Δ mutant and VPS4 reintegrant strains used in this study were generated by the integration of empty pGEM-HIS1 (from A. Mitchell) or pGEM-HIS1 bearing a single wild-type copy of VPS4, respectively, into the mutant strains previously published [8] to generate prototrophic, isogenic null (ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/HIS1 vps4Δ∷ dpl200-URA3- dpl200/vps4Δ∷ARG4 and reintegrant (ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/HIS1-VPS4 vps4Δ∷ dpl200-URA3- dpl200/vps4Δ∷ARG4) strains.
Isolation of secreted proteins from in vitro cultures
For generation of protein extracts, C. albicans was propagated in RPMI-1640 supplemented with L-glutamine and buffered with 165 mM MOPS to pH 7.0. Batches of media (200 mL) were inoculated to a starting density of approximately 1 × 105 cells/ml and incubated overnight in an orbital shaker at 30 or 37 °C. Secreted proteins from planktonic cultures were obtained as previously described by our group and others [10, 11] with slight modifications. Briefly, at the end of the incubation time, the liquid culture supernatants were obtained by centrifugation. The cells were pelleted by centrifugation at 3500 g for 5 min, and the remaining supernatant was filter–sterilized and processed through a Millipore Ultrafree-15 centrifugal filter device (Millipore, Billerica, MA) for desalting and concentration.
2-DE
IPG strips (11 cm, pH 4–7) (BioRad, Hercules CA) were rehydrated for 16 h at 20°C in 200 μL of rehydration/sample buffer containing C. albicans supernatant extracts (approximately 250 μg expressed as total protein). IEF was carried out using the PROTEAN IEF (BioRad, Hercules CA) under the following conditions: Step 1, 250V for 20 min; Step 2, ramped to 8000 V over 2.5 h; and Step 3, 8000 V for a total of 30 000 V/h. Strips were then placed into equilibration buffer and disulfide groups were subsequently blocked with iodacetamide. Equilibrated IPG strips were then placed and fixed using hot agarose on the top of SDS-PAGE 12.5% polyacrylamide Criterion Precast Gels (BioRad, Hercules CA) and separation of proteins in the second dimension carried out under reducing conditions. After electrophoresis, protein spots were visualized by staining with SYPRO Ruby gel stain (BioRad, Hercules CA) following manufacturer’s instructions. Gel comparison analysis was performed by PDQuest (BioRad, Hercules CA). To assure maximal coverage, initial experiments were also performed with pH 3–10 IPG strips in the first dimension and 4–20% acrylamide gradient gels in the second dimension. These experiments revealed that the vast majority of proteins appeared at a pI between 5 and 6, and molecular mass between 15 and 100 kDa. For differential expression analysis, protein spots were cataloged that showed consistent and reproducible changes in protein abundance as determined by PDQuest using a pairwise t-test analysis of multiple experiments (three biological replicates) with 95 percent confidence limits. Gels were normalized via total density in valid spots after the exclusion of very abundant spots with large visible changes.
MS protein identification
Identification of gel spots after 2-DE was accomplished by mass spectrometry. Gels were stained with SYPRO Ruby (Molecular Probes, Invitrogen, Carlsbad CA) and imaged using an FX Pro Plus fluorescent imager (Bio-Rad, Hercules CA). A Proteome Works spot cutter (Bio-Rad, Hercules CA) was used to robotically excise protein spots of interest which were digested in situ with trypsin (Promega, Madison WI) in 40 mM NH4HCO3 at 37 °C for 4 hr according to standard protocols based on the initial work of Mann and coworkers [12]. The digests were analyzed by mass spectrometry without further purification. Capillary HPLC-electrospray ionization tandem mass spectra (HPLC-ESI-MS/MS) were acquired on a Thermo Fisher (Waltham MA) LTQ linear ion trap mass spectrometer fitted with a New Objective (Woburn, MA) PicoView 550 nanospray interface. On-line HPLC separation of the digests was accomplished with an Eksigent (Dublin CA) NanoLC micro HPLC column, PicoFrit™ (New Objective; Woburn, MA 75 μm i.d.) packed to 10 cm with C18 adsorbent (Vydac; Grace, Deerfield IL, 218MS 5 μm, 300 Å); mobile phase A, 0.5% acetic acid (HAc)/0.005% trifluoroacetic acid (TFA); mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA; gradient 2 to 42% B in 30 min; flow rate, 0.4 μl/min. MS conditions were: ESI voltage, 2.9 kV; isolation window for MS/MS, 3; relative collision energy, 35%; scan strategy, survey scan followed by acquisition of data dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan above a set threshold. The un-interpreted CID spectra were searched against the NCBInr databases (NCBInr_20060418) by means of Mascot (Matrix Science Inc, Boston MA, version 2.1.0). Methionine oxidation was considered as a variable modification for all searches. Cross-correlation of the Mascot results with X! Tandem and determination of protein identity probabilities was accomplished by Scaffold™ (Proteome Software Inc, Portland OR, version 2.02.03).
Enzyme-linked immunosorbent assay (ELISA)
For ELISA, wells of a microtiter plate (Immulon 2 High Binding, Nunc, Thermo Fisher Scientific, Waltham MA) were coated with equal volumes of the parallel culture protein extracts in coating buffer overnight at 4°C. The plate was then washed twice in PBS containing 0.05% Tween-20 (PBST), and blocked for 30min using 1% BSA in PBS. The samples were then incubated with the two primary antibodies (anti-enolase [13] and anti-mp58 [14]) in PBST 1% BSA for 1 hr at 37°C and then washed twice with PBST. Subsequently the plate was incubated for 1 hour with a 1:2000 dilution of secondary antibody (peroxidase conjugated goat anti-mouse IgG, Bio-Rad) before washing twice with PBST and once with PBS. Then the plate was then developed with 100 μl of o-phenylenediamine substrate per well and incubated in the dark for 10 min with gentle agitation. Color development was stopped by the addition of 50 μl of 1 M H2SO4 per well and read on a BenchMark microplate reader (Bio-Rad, Hercules CA) at 492nm.
Analysis of filamentation
Filamentation was assayed at 30 and 37 °C in RPMI-1640 (Invitrogen, Carlsbad CA) supplemented with L-glutamine (US Biological, Swampscott MA) and buffered with 165mM MOPS to pH 7.0. Liquid medium was inoculated with cells from overnight cultures to achieve a starting density of 5 × 106 cells ml−1, followed by incubation with shaking at 200 rpm, and visualization by light microscopy.
Analysis of fungal biofilms
Formation of C. albicans biofilms and the XTT-reduction assay of biofilm metabolic activity were performed as described [15]. Scanning electron microscopy was performed on biofilm samples formed on a coverslip (Thermanox, Nunc, Thermo Fisher Scientific, Waltham MA) after 24 h incubation of a 0.5 ml inoculum containing 1 × 106 cells ml−1 according to previously described methods [16]. Antifungal susceptibility testing of cells within the biofilms against fluconazole, amphotericin B, and caspofungin was performed as previously described [15]. In brief, measurement of metabolic activities of the sessile cells growing within the biofilm is based on the reduction of 2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide (XTT), which yields a formazan colored product that is measured using spectrophotometry Biofilms of the C. albicans strains were formed by growing overnight cultures at 30°C in YPD. Cells were harvested and washed in sterile phosphate-buffered saline (PBS), pH 7.4, and re-suspended in RPMI-1640 supplemented with glutamine and buffered with MOPS. Biofilms were formed by pipetting standardized cell suspensions (100 ml of 1.0 × 106 cells/ml) into selected wells of pre-sterilized, polystyrene, flat-bottomed, 96-well microtiter plates (Corning Incorporated, Corning, NY, USA) and incubated for 48 h at 37 °C. The biofilms were then washed thoroughly with sterile PBS, followed by addition of antifungal agents in serially double diluted concentrations and incubated again for 48 h at 37°C. After thorough washing with PBS, XTT reduction was measured in 4 replicates for each condition in a standard ELISA plate reader. The degree of inhibition was expressed as a percentage of the treatment wells relative to the untreated control wells.
Fluconazole was obtained from Pfizer, Inc (New York, NY), and amphotericin B (Bristol-Myers Squibb, Princeton, NJ), and caspofungin (Merck, Rahway, NJ) were purchased from the hospital pharmacy.
Statistical analyses
Proteomic statistical comparisons were obtained using a pairwise t-test, with P < 0.05 considered significant. All other statistically significant differences were determined by analysis of variance, with P < 0.05 considered to be significant.
RESULTS and DISCUSSION
Analysis of soluble secreted proteins in the C. albicans vps4Δ mutant
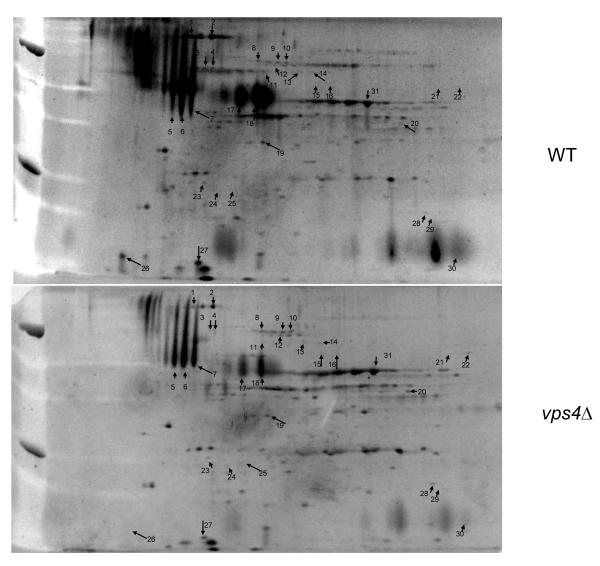
Analysis of the secreted proteins from the vps4Δ mutant in comparison to control strain DAY185 in planktonic form indicated major changes in protein complement (Fig. 1, Table 1). Most notably, a decrease in all of the identified traditionally secreted proteins (RER-Golgi-Membrane) was demonstrated (p<0.05). Pra1p, Mp65p, Cht3p, and Sun41p, one of which is predicted to be a cell wall protein (encoded by SUN41), are reduced in the extracellular supernatant of the vps4Δ mutant. Of note, whilst the traditionally secreted proteins were diminished, they remained present in the supernatant obtained from the vps4Δ mutant, with the exception of Sun41p. In contrast, there is little difference in the presence or abundance of proteins known to be secreted by a non-canonical pathway (i.e. glycolytic enzymes, heat stress proteins, etc.). A list of many of the proteins in this fraction along with all changes of significance after normalization and analysis of replicates are shown in Table 1.
Fig. 1. 2DE analysis of culture supernatants from the C. albicans vps4Δ null mutant compared with control strain DAY185.
Cultures were inoculated with a starting density of approximately 1 × 105 cells/ml grown overnight at 37 °C in RPMI-1640 supplemented with L-glutamine and buffered with 165 mM MOPS to pH 7.0. Cells were then pelleted, and the culture supernatants separated by 2DE using a pI range of 4-7. Differences in protein spots are indicated in Table 1. HPLC-ESI-MS/MS Mass spectrometry data is included in supplementary information.
Table 1. Identified Secreted Proteins: WT vs. vps4Δ mutant.
Proteins were identified by searching the un-interpreted CID spectra against the NCBInr database (NCBInr_20060418) using Mascot (Matrix Science Inc, Boston MA, version 2.1.0). Cross-correlation of the Mascot results with X! Tandem and determination of protein identity probabilities was accomplished by Scaffold™ (Proteome Software Inc, Portland OR, version 2.02.03).
| Spot # | Protein Information | Gene # | Gene Name | Downregulated in Δvps4 |
ACC |
|---|---|---|---|---|---|
| 1 | similar to heat shock protease protein; Hsp5; Hsp20 region | 19.8442 | 46442969 | ||
| 2 | secreted chitinase | 19.7586 | CHT3 | X | 46441161 |
| 3 | involved in translocation of nascent polypetides across the ER membrane and in nuclear fusion during mating |
19.9564 | KAR2 | 46441414 | |
| 4 | involved in translocation of nascent polypetides across the ER membrane and in nuclear fusion during mating |
19.9564 | KAR2 | 46441414 | |
| 5 | Mp65p, potential glycosyl hydrolase [16] | 19.1779 | MP65 | X | EAK95313 |
| 6 | Mp65p, potential glycosyl hydrolase [16] | 19.1779 | MP65 | X | EAK95313 |
| 7 | Mp65p, potential glycosyl hydrolase [16] | 19.1779 | MP65 | X | EAK95313 |
| 8 | Hsp70p aka Ssa4p aka Ssa1p | 19.4980 | HSP70 | 68467506 | |
| 9 | Ssb1 in SSB subfamily of HSP70 | 19.13724 | SSB1 | 68479520 | |
| 9 | Co-chaperonin interacts with Cdc37p and Crk1p, chaperonin binding motif | 19.10702 | STI1 | 46436331 | |
| 9 | mitochondrial import protein, DNAK, HSP70 and ftsA regions | 19.9452 | SSC1 | 68485837 | |
| 10 | Ssb1p in SSB subfamily of HSP70 | 19.13724 | SSB1 | 68479520 | |
| 11 | Hsp60p, mitochondrial chaperonin, GroEL type 1 chaperonin | 19.717 | HSP60 | 68486010 | |
| 12 | Ssb1p in SSB subfamily of HSP70 | 19.13724 | SSB1 | 68479520 | |
| 12 | Hsp70p aka Ssa4p aka Ssa1p | 19.4980 | HSP70 | 68467506 | |
| 12 | mitochondrial import protein, DNAK, HSP70 and ftsA regions | 19.9452 | SSC1 | 68485837 | |
| 13 | pyruvate decarboxylase | 19.10395 | PDC11 | 68480872 | |
| 14 | pyruvate decarboxylase | 19.10395 | PDC11 | 68480872 | |
| 14 | Co-chaperonin interacts with Cdc37p and Crk1p, chaperonin binding motif | 19.10702 | STI1 | 46436331 | |
| 14 | mitochondrial import protein, DNAK, HSP70 and ftsA regions | 19.9452 | SSC1 | 68485837 | |
| 15 | dihydrolipoamide acetyl transferase (mitochondrial) | 19.6561 | LAT1 | 76573599 | |
| 16 | dihydrolipoamide acetyl transferase (mitochondrial) | 19.6561 | LAT1 | 76573599 | |
| 17 | pH regulated protein (formerly mp58p) [16] | 19.3111 | PRA1 | X | EAK96466 |
| 18 | pH regulated protein (formerly mp58p) [16] | 19.3111 | PRA1 | X | EAK96466 |
| 19 | RNA recognition motif. Predicted ORF in Assemblies 19, 20 & 21; induced in core caspofungin response; increased expression observed in an ssr1 homozygous null mutant; induced by nitric oxide in yhb1 mutant |
19.3932 | 46443783 | ||
| inorganic pyrophophatase | 19.11072 | IPP1 | 46436055 | ||
| 20 | Unidentified | X | |||
| 21 | putative mitochondrial matrix dihydrolipoamide. Putative dihydrolipoamide dehydrogenase; soluble protein in hyphae; antigenic during human oral infection and murine systemic infection; macrophage-induced protein |
19.6127 | LPD1 | 68487571 | |
| 22 | putative mitochondrial matrix dihydrolipoamide. Putative dihydrolipoamide dehydrogenase; soluble protein in hyphae; antigenic during human oral infection and murine systemic infection; macrophage-induced protein |
19.6127 | LPD1 | 68487571 | |
| 23 | Unidentified | X | |||
| 24 | involved in translocation of nascent polypeptides across the ER membrane and in nuclear fusion during mating |
19.9564 | KAR2 | 46441414 | |
| 25 | similar to heat shock protease protein; Hsp5; Hsp20 region | 19.8442 | 46442969 | ||
| 26 | similar to S. cerevisiae Sun4p (septation role) hyphal induced; caspofungin repressed; Efg1p, Cph1p regulated; predicted signal sequence, O-glycosylation, potential Kex2p substrate; 9 5′ E-boxes; 5′- UTR intron |
19.11124 | SUN41 | X | 68489582 |
| 27 | Predicted ORF in Assemblies 19, 20 and 21; regulated by Tsa1p, Tsa1Bp under H2O2 stress conditions [16] |
19.10993 | LDG8 | 46432072 | |
| 28 | putative flavodoxin WrbA, fungal specific, similar to S. cerevisiae Pst2p | 19.5285 | PST3 | 46431555 | |
| 29 | ATP synthase subunit D (normally mitochondrial). Putative subunit of the F1F0-ATPase complex; shows colony morphology-related gene regulation by Ssn6p; macrophage-downregulated protein abundance |
19.2785 | ATP7 | 68488854 | |
| 30 | glyceraldehyde-3-dehydrogenase | 19.6814 | TDH1 | 2494636 | |
| 31 | Enolase [16] | 19.395 | ENO1 | 46433227 |
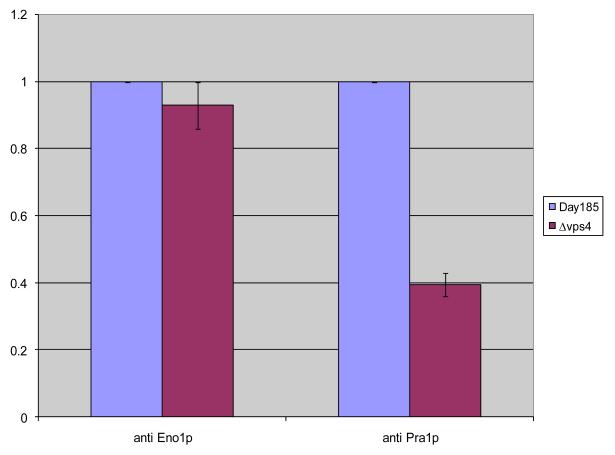
Overall these changes seemed to involve very dominant proteins and thus affected protein quantification and loading. Therefore, we employed an ELISA assay in order to discriminate whether all the canonically secreted proteins were decreased or the non-canonical proteins were increased in the vps4Δ supernatant. Using this approach, we determined protein levels within the supernatants of parallel cultures inoculated with identical number of cells into identical culture volumes at 37 °C in RPMI. The cultures were grown for 3-5 hours and the cell densities were checked to confirm that the growth rates had been the same. As shown in Fig. 2, results of the ELISA clearly indicated that quantities of enolase (a non-canonically secreted protein previously shown to be in these extracts [17]) were similar. However levels of Pra1p (a canonically secreted glycoprotein) were reduced in the supernatant compared to levels recovered from the wild type strain.
Fig. 2. The C. albicans vps4Δ mutant secretes reduced extracellular Pra1p, but not enolase.
ELISA was used to assay protein levels of supernatants from parallel cultures 3-5 hours following inoculation with 3 × 107 cells in RPMI at 37 °C. Quantities of enolase (a non-canonically secreted protein) were similar between control strain DAY185 and the vps4Δ mutant. In contrast, the vps4Δ mutant secreted reduced levels of Pra1p (a canonically secreted glycoprotein) compared to the DAY185 control strain. Data are expressed relative to DAY185.
Properties of biofilms formed by the C. albicans vps4Δ mutant
We hypothesized that secretion of extracellular proteins is required for normal biofilm formation, and consequently, that altered pre-vacuolar secretion could potentially lead to defective biofilm formation. Previously, we have examined the role of the late-Golgi trafficking gene VPS1 in secretion of secreted asparytl proteases and biofilm formation. Using a tetracycline-regulated C. albicans VPS1 mutant, in repressing conditions, this pre-vacuolar mutant was defective in filamentation, and secreted reduced Sap2p and lipolytic activity [18]. Furthermore, biofilm production and filamentation within the biofilm were markedly reduced.
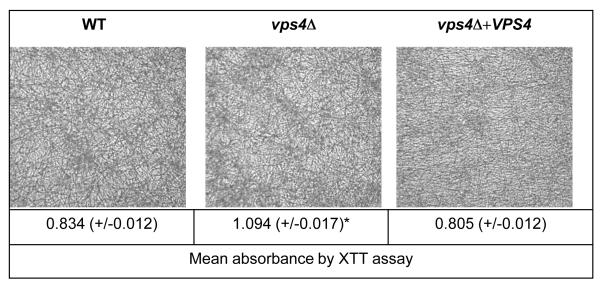
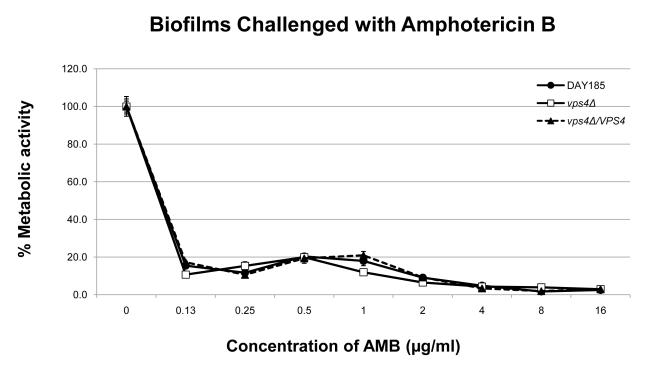
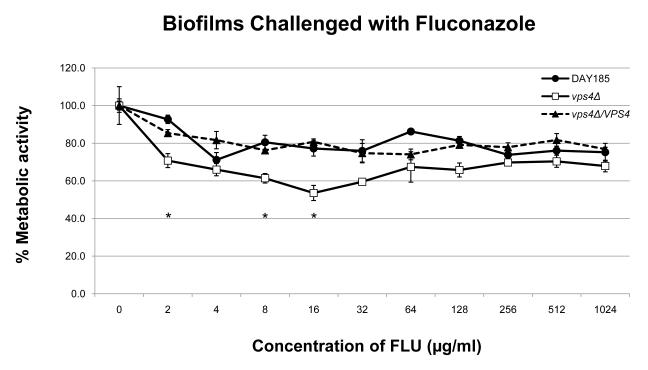
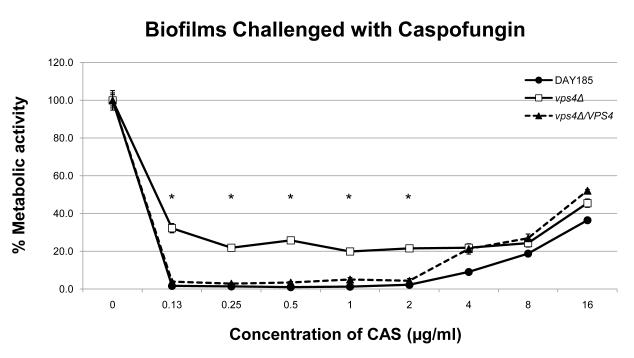
In order to define the role of VPS4 in biofilm formation, we examined biofilm formation by light microscopy and the XTT-reduction assay. Interestingly, the vps4Δ mutant showed increased biofilm formation compared to the wild-type and isogenic reintegrant strain (Fig. 3). Qualitatively, an increased density of the biofilm cellular component in the vps4Δ mutant was demonstrated by scanning electron microscopy (Fig. 4). Functionally, the biofilm formed by the vps4Δ mutant showed no statistically significant difference in response to amphotericin B, and a modest increase in susceptibility to fluconazole, although overall, fluconazole remained poorly active against the biofilm. Most notably, the biofilm formed by the vps4Δ mutant demonstrated greater resistance to caspofungin than prototrophic control strain DAY185 or its isogenic reintegrant strain (Fig. 5). As expected, an in vitro paradoxical effect was seen at high concentrations of caspofungin [19-21]. Taken together, these findings suggest that pre-vacuolar secretion plays a key role in biofilm formation. Specifically, absence of VPS4 leads to a denser biofilm which is functionally more resistant to capsofungin than biofilms formed by the wild-type or isogenic control strains.
Fig. 3. The C. albicans vps4Δ mutant shows increased biofilm formation.
Biofilms were generated in microtiter plates in RPMI-1640 supplemented with L-glutamine and buffered with 165 mM MOPS to pH 7.0 at 37°C, incubated for 24 h, washed, visualized by light microscopy, and analyzed by the XTT reduction assay. Although crude architecture was similar in the vps4Δ mutant, wild-type, and reconstituted controls, the vps4Δ mutant generated denser biofilms when assayed by XTT reduction. (*p<0.0001 by analysis of variance)
Fig. 4. Scanning electron microscopy of C. albicans vps4Δ mutant biofilms.
Biofilms were formed and visualized using scanning electron microscopy. Control strain DAY185 formed biofilms consisting of predominantly of mature hyphae. The vps4Δ mutant strain demonstrated markedly denser biofilms throughout. The figures shown depict representative structures at 2,000x magnification with a 10 μm bar indicated.
Fig. 5. Determination of antifungal susceptibilities within C. albicans vps4Δ mutant biofilms.
Antifungal susceptibilities within the fungal biofilms were assayed as previously described using the XTT-reduction assay. No statistically significant difference is seen in the antifungal susceptibilities in biofilms when challenged with amphotericin B. A modest increase in fluconazole susceptibility is seen in the vps4Δ mutant at several intermediate concentrations of fluconazole compared to DAY185 (* p<0.05). Most notably, the vps4Δ mutant demonstrates decreased susceptibility to the echinocandin caspofungin beginning at a concentration of 2.0 μg/ml of CAS compared to DAY185 (* p<0.05). As expected, an in vitro paradoxical effect is seen at high concentrations of CAS.
The C. albicans vps4Δ mutant filaments with characteristic altered branching
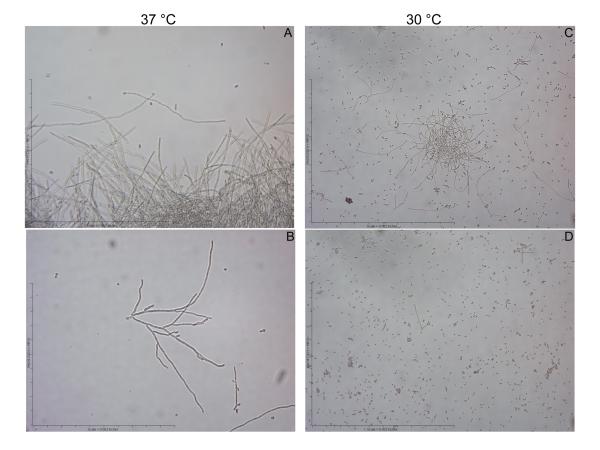
To further analyze the production of increased biomass during biofilm formation we examined the C. albicans vps4Δ null mutant in typical planktonic growth conditions. Here, the strain demonstrated a characteristic class “E” phenotype, which in vitro includes more frequent branching during filamentation. At 37 °C, control strain DAY185 filamented with typical long parallel-sided filaments with infrequent branching (Fig. 6A). In comparison, the vps4Δ mutant filamented with a frequently branching phenotype, as previously described for other “class E” vacuolar mutants (Fig. 6B). At 30 °C, DAY185 also produced a majority of filaments, whereas the vps4Δ mutant produced filaments rarely (~1%), but with the same characteristic branching phenotype (Fig. 6C, D). Thus it is possible that the increase in branching at 37 °C leads to increased biofilm formation in the vps4Δ mutant.
Fig. 6. The C. albicans vps4Δ mutant mutant exhibits a characteristic “class E” branching filamentous phenotype.
Filamentation was assayed at 30 and 37 °C in RPMI-1640 supplemented with L-glutamine and buffered with 165 mM MOPS to pH 7.0. Liquid medium was inoculated with cells from overnight cultures to achieve a starting density of 5 × 106 cells/ml, followed by incubation with shaking at 200 rpm, and visualization by light microscopy. DAY185 filaments robustly at 37 °C with long parallel-sided filaments and infrequent branching (A). In contrast the vps4Δ mutant filaments with a branching phenotype as previously described for other vacuolar “class E” mutants (B). At 30 °C, DAY185 filaments modestly (C), whereas the vps4Δ mutant filaments only rarely (<1%), but with the same branching phenotype (D).
There are some important considerations that need to be taken into account when trying to correlate the proteomics results with the observed phenotypes in the vps4Δ mutant strain. There is a marked difference in Sun41p levels in the vps4Δ strain compared to wild-type. Sun41p is intimately linked to septation, and normal C. albicans branching occurs at sites of previous septa (reviewed by [22]), and thus mislocalization of Sun41p may contribute to the altered branching phenotype of the vps4Δ mutant. Also of note, Velours et al [23] demonstrated that whilst Sun41p can be associated with the cell wall, it can also be localized to the mitochondria. More recently sun41Δ mutants have been described to be defective in biofilm formation and cell separation, and to show enhanced adhesion [24-26]. Conditional inactivation of all the C. albicans SUN family genes results in defects at the septum and atypical bud scar formation [24]. Regarding Cht3p, it is regulated by Ras1p, a signal transduction GTPase linked to hyphal induction and virulence, and under yeast form growth conditions both Cht3p [27] and Sun41p [24] are required for mother-daughter cell separation. Moreover, both Sun4p (the S. cerevisiae ortholog of Sun41p) and C. albicans Mp65p are substrates of Kex2p, a proprotein convertase linked to filamentation and virulence which is integral to the Golgi membrane. Kex2p normally cycles between the Golgi and multi-vesicular body (MVB). The vps4Δ mutation prevents proteins from leaving the MVB. An obvious correlation thus exists with Vps10p (the CPY receptor), which usually cycles between the Golgi and MVB carrying CPY; in the vps4Δ mutant Vps10p is restricted to the MVB, causing CPY accumulation in the Golgi which is later secreted outside the cell. Although we did not detect extracellular CPY in the proteomics experiments, this is likely due to culture conditions. Presumably the vps4Δ mutation results in reduction of Kex2p in the Golgi, affecting some proproteins. This is supported by the absence of secreted Sap2p [8] and may correlate with a similar phenotype in vps11Δ strains [3]. Newport and Agabian [28] showed a lesser decrease in a kex2Δ mutant, although this insertion removed an internal fragment of the gene containing one of four active site residues, rather than the whole gene.
Although the overall proteomic changes between the vps4Δ mutant and wild-type strain are very specific, there are clearly differences in extracellular secretion of several key proteins, and clear structural and functional changes in the biofilm formed by the vps4Δ pre-vacuolar secretory mutant. Overall, these findings suggest that pre-vacuolar secretion plays an important role in exocytosis in C. albicans. Furthermore, since secretion and biofilm formation are closely related, these results also suggest that pre-vacuolar secretion impacts C. albicans biofilm formation, although this may be due to an indirect effect. Further studies of the role of pre-vacuolar secretion and trafficking of secreted virulence-associated proteins and biofilm formation are currently in progress.
Supplementary Material
ACKNOWLEDGEMENTS
Sequence data for C. albicans was obtained from the Candida Genome Database at http://www.candidagenome.org/. Proteomic data reflect sequence Assembly 21 deposited in NCBInr (NCBInr_20060418). We would like to thank Anna Lazzell for technical assistance with the ELISA assays, and Stella M. Bernardo for assistance with data analysis.
This work was supported by a grant from the Department of Veterans’ Affairs (MERIT Award to SAL), the Biomedical Research Institute of New Mexico (SAL) and by NIH grant R21DE115079 from NIDCR (to JLL-R).
The MS analyses were conducted in the UTHSCSA Institutional Mass Spectrometry Shared Resource Facility funded in part by the San Antonio Cancer Institute (NCI CA54174), directed by Dr. Susan Weintraub.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Bruckmann A, Kunkel W, Hartl A, Wetzker R, Eck R. A phosphatidylinositol 3-kinase of Candida albicans influences adhesion, filamentous growth and virulence. Microbiology-Uk. 2000;146:2755–64. doi: 10.1099/00221287-146-11-2755. [DOI] [PubMed] [Google Scholar]
- [2].Cornet M, Bidard F, Schwarz P, Da Costa G, Blanchin-Roland S, Dromer F, et al. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect Immun. 2005;73:7977–87. doi: 10.1128/IAI.73.12.7977-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Palmer GE, Cashmore A, Sturtevant J. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryotic Cell. 2003;2:411–21. doi: 10.1128/EC.2.3.411-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Palmer GE, Kelly MN, Sturtevant JE. The Candida albicans vacuole is required for differentiation and efficient macrophage killing. Eukaryotic Cell. 2005;4:1677–86. doi: 10.1128/EC.4.10.1677-1686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. Embo Journal. 1997;16:1820–31. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nature Reviews Molecular Cell Biology. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- [7].Kullas AL, Li MC, Davis DA. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryotic Cell. 2004;3:1609–18. doi: 10.1128/EC.3.6.1609-1618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee SA, Jones J, Khalique Z, Kot J, Alba M, Bernardo S, et al. A functional analysis of the Candida albicans homolog of Saccharomyces cerevisiae VPS4. Fems Yeast Research. 2007;7:973–85. doi: 10.1111/j.1567-1364.2007.00253.x. [DOI] [PubMed] [Google Scholar]
- [9].Lee SA, Jones J, Hardison S, Kot J, Khalique Z, Bernardo SM, et al. Candida albicans VPS4 is required for secretion of aspartyl proteases and in vivo virulence. Mycopathologia. 2009;167:55–63. doi: 10.1007/s11046-008-9155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baillie GS, Douglas LJ. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. Journal of Antimicrobial Chemotherapy. 2000;46:397–403. doi: 10.1093/jac/46.3.397. [DOI] [PubMed] [Google Scholar]
- [11].Lopez-Ribot JL, Gozalbo D, Sepulveda P, Casanova M, Martinez JP. Preliminary Characterization of the Material Released to the Culture-Medium by Candida-Albicans Yeast and Mycelial Cells. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 1995;68:195–201. doi: 10.1007/BF00871815. [DOI] [PubMed] [Google Scholar]
- [12].Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- [13].Lopez-Ribot JL, McAtee RK, Kirkpatrick WR, La Valle R, Patterson TF. Low levels of antigenic variability in fluconazole-susceptible and -resistant Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Clin Diagn Lab Immunol. 1999;6:665–70. doi: 10.1128/cdli.6.5.665-670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Casanova M, Lopezribot JL, Monteagudo C, Llombartbosch A, Sentandreu R, Martinez JP. Identification of a 58-Kilodalton Cell-Surface Fibrinogen-Binding Mannoprotein from Candida-Albicans. Infect Immun. 1992;60:4221–9. doi: 10.1128/iai.60.10.4221-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ramage G, Lopez-Ribot JL. Techniques for antifungal susceptibility testing of Candida albicans biofilms. Methods in Molecular Medicine. 2005;118:71–9. doi: 10.1385/1-59259-943-5:071. [DOI] [PubMed] [Google Scholar]
- [16].Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. Journal of Antimicrobial Chemotherapy. 2002;49:973–80. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- [17].Thomas DP, Bachmann SP, Lopez-Ribot JL. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics. 2006;6:5795–804. doi: 10.1002/pmic.200600332. [DOI] [PubMed] [Google Scholar]
- [18].Bernardo SM, Khalique Z, Kot J, Jones JK, Lee SA. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genetics and Biology. 2008;45:861–77. doi: 10.1016/j.fgb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chamilos G, Lewis RE, Albert N, Kontoyiannis DP. Paradoxical effect of Echinocandins across Candida species in vitro: evidence for echinocandin-specific and candida species-related differences. Antimicrob Agents Chemother. 2007;51:2257–9. doi: 10.1128/AAC.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Melo AS, Colombo AL, Arthington-Skaggs BA. Paradoxical growth effect of caspofungin observed on biofilms and planktonic cells of five different Candida species. Antimicrob Agents Chemother. 2007;51:3081–8. doi: 10.1128/AAC.00676-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Miceli MH, Bernardo SM, Lee SA. In vitro analysis of the occurrence of a paradoxical effect with different echinocandins and Candida albicans biofilms. Int J Antimicrob Agents. 2009;34:500–2. doi: 10.1016/j.ijantimicag.2009.07.001. [DOI] [PubMed] [Google Scholar]
- [22].Walther A, Wendland J. Septation and cytokinesis in fungi. Fungal Genetics and Biology. 2003;40:187–96. doi: 10.1016/j.fgb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- [23].Velours G, Boucheron C, Manon S, Camougrand N. Dual cell wall/mitochondria localization of the ‘SUN’ family proteins. Fems Microbiology Letters. 2002;207:165–72. doi: 10.1111/j.1574-6968.2002.tb11046.x. [DOI] [PubMed] [Google Scholar]
- [24].Firon A, Aubert S, Iraqui I, Guadagnini S, Goyard S, Prevost MC, et al. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Molecular Microbiology. 2007;66:1256–75. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- [25].Hiller E, Heine S, Brunner H, Rupp S. Candida albicans sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryotic Cell. 2007;6:2056–65. doi: 10.1128/EC.00285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Norice CT, Smith FJ, Solis N, Filler SG, Mitchell AP. Requirement for Candida albicans sun41 in biofilm formation and virulence. Eukaryotic Cell. 2007;6:2046–55. doi: 10.1128/EC.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dunkler A, Walther A, Specht CA, Wendland J. Candida albicans CHT3 encodes the functional homolog of the Cts1 chitinase of Saccharomyces cerevisiae. Fungal Genetics and Biology. 2005;42:935–47. doi: 10.1016/j.fgb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- [28].Newport G, Agabian N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. Journal of Biological Chemistry. 1997;272:28954–61. doi: 10.1074/jbc.272.46.28954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.