Abstract
Collagen is an abundant, triple-helical protein comprising three strands of the repeating sequence: Xaa–Yaa–Gly. (2S)-Proline and (2S,4R)-4-hydroxyproline (Hyp) are common in the primary structure of collagen. Here, we use nonnatural proline derivatives to reveal determinants of collagen stability. Specifically, we report high-yielding syntheses of (2S,4S)-4-chloroproline (clp) and (2S,4R)-4-chloroproline (Clp). We find that the crystal structure of Ac-Clp-OMe is virtually identical to that of Ac-Hyp-OMe. In contrast, the conformational properties of Ac-clp-OMe are similar to those of Ac-Pro-OMe. Ac-Clp-OMe has a stronger preference for a trans amide bond than does Ac-Pro-OMe, whereas Ac-clp-OMe has a weaker preference. (Pro–Clp–Gly)10 forms triple helices that are significantly more stable than those of (Pro–Pro–Gly)10. Triple helices of (clp–Pro–Gly)10 have stability similar to those of (Pro–Pro–Gly)10. Unlike (Pro–Clp–Gly)10 and (clp–Pro–Gly)10, (clp–Clp–Gly)10 does not form a stable triple helix, presumably due to a deleterious steric interaction between proximal chlorines on different strands. These data, which are consistent with previous work on 4-fluoroprolines and 4-methylprolines, support the importance of stereoelectronic and steric effects in the stability of the collagen triple helix and provide another means to modulate that stability.
Keywords: 4-chloroproline, collagen, polyproline II-type helix, proline, prolyl peptide-bond isomerization, triple helix
INTRODUCTION
Collagen is the major proteinaceous component of the extracellular matrix (ECM) in vertebrates.1 The three-dimensional structure of collagen was determined in the 1950s2–6 to be a right-handed triple helix formed by the parallel coiling of three left-handed polyproline II-type (PPII) strands about a common axis. At least 28 different members of the collagen superfamily of proteins have been discovered to date, as well as a number of other proteins with triple-helical, collagenous domains.7,8
In the fibrillar collagens and in most other types of collagen, every third residue is always the smallest of the twenty common amino acids, Gly,7,9,10 which is required for the tight packing of the triple helix.11 Often, the amino acid in the Xaa position of the Xaa–Yaa–Gly repeat is Pro and that in the Yaa position is (2S,4R)-4-hydroxyproline (Hyp).12,13 Hyp is formed by the stereospecific post-translational hydroxylation of proline residues in the Yaa position by the enzyme prolyl-4-hydroxylase (P4H).14
The post-translational hydroxylation of prolines in the Yaa position is essential for the formation of a stable ECM in animals. Both Caenorhabditis elegans and mice lacking P4H experience embryonic morbidity due to weakened collagen superstructures.15,16 Why is proline hydroxylation essential to the formation of a stable ECM? It is known that collagens containing a high fraction of Hyp in the Yaa position are particularly stable,17,18 but the physicochemical basis for this stability was unclear.
Determining the basis for the impact of proline hydroxylation on collagen stability is difficult because the high molecular weight and insolubility of native collagen prevent high-resolution structural analysis of the protein in its native form. Elkan Blout and his coworkers were among the first to address this obstacle by employing the reductionist approach of using peptide mimics Shoulders, Guzei, and Raines page 4 of collagen, known as collagen-related peptides (CRPs), to reveal fundamental aspects of triple-helix stability.19–21
The study of CRPs has led to a series of landmark discoveries regarding the structure and stability of natural collagen. Prockop and coworkers used CRPs to demonstrate that both the stereochemistry and location of Hyp residues are important for its stabilizing effect on collagen triple helices, as illustrated by the Tm values for triple-helix denaturation listed in Table I.22–24 CRPs with Hyp in the Yaa position are more stable than those with Pro, but Hyp in the Xaa position prevents triple-helix formation when the Yaa amino acid is Pro.24–29
Table I.
Effect of 4-Hydroxyproline, 4-Fluoroproline, and 4-Methylproline Diastereomers on the Conformational Stability of Collagen Triple Helices
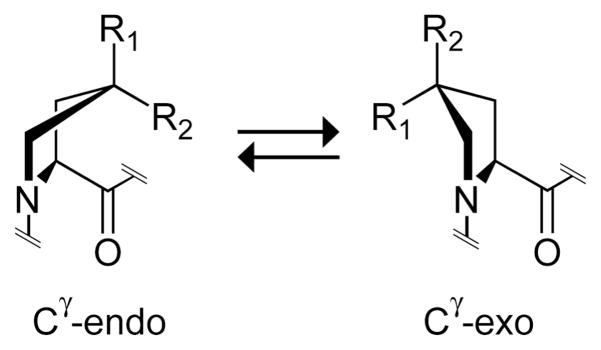
In the last decade, we have demonstrated that the stability (or instability) conferred by Hyp derives from the manifestation of previously unappreciated stereoelectronic effects. We did so by installing functional groups at Cγ that mediate such effects, as well as reciprocal steric effects (Figure 1).30,31 For example, replacing Hyp with (2S,4R)-4-fluoroproline (Flp), which has the native-like stereochemistry, but not (2S,4S)-4-fluoroproline (flp) results in triple helices with markedly enhanced stability (Table I).32–34 This and other results revealed that a gauche effect in Hyp and Flp mandates a Cγ -exo pucker in the pyrrolidine ring,34,35 which preorganizes the φ, ψ, and ω dihedral angles to those required for triple-helix assembly.34,36 (2S,4S)-4-Methylproline (Mep) achieves the same end by manifesting a steric rather than stereoelectronic effect (Figure 1; Table I).31 (The pyrrolidine ring of proline actually prefers two distinct twist, rather than envelope, conformations.37 As Cγ experiences a large out-of-plane displacement in these twisted rings, we refer to pyrrolidine ring conformations simply as “Cγ -exo” and “Cγ -endo”.)
FIGURE 1.
Ring conformations of 4-substituted prolines. The Cγ -endo conformation is favored strongly by stereoelectronic effects when R1 = H, R2 = F (flp) or Cl (clp) and by steric effects when R1 = Me (mep), R2 = H. The Cγ -exo conformation is favored strongly by stereoelectronic effects when R1 = OH (Hyp), F (Flp) or Cl (Clp), R2 = H and by steric effects when R1 = H, R2 = Me (Mep).
The Cγ -endo pucker of proline derivatives in the Xaa position is favorable for triple-helix stability.31,38,39 This finding was presaged by Zagari and coworkers, who observed that prolines in the Xaa position of high-resolution crystal structures of CRPs nearly always exhibit the Shoulders, Guzei, and Raines page 5 Cγ -endo pucker.40 Consequently, substitution of flp or (2S,4R)-4-methylproline (mep) for Pro in the Xaa position of CRPs stabilizes triple helices (Table 1).39,41,42,31 Although the Cγ -endo pucker preorganizes the φ and ψ angles to those required for triple-helix assembly, it does not preorganize the ω angle because proline derivatives with a Cγ -endo pucker have an enhanced preference for the cis peptide bond due to another stereoelectronic effect—an n→π* interaction—described elsewhere,35,43–46 whereas all peptide bonds in collagen are trans. Despite its preference for the Cγ -endo pucker, (2S,4S)-4-hydroxyproline (hyp) in the Xaa position does not allow for the formation of stable triple helices (Table I),23 perhaps due to deleterious hydration absent from flp and mep or to idiosyncratic conformational preferences.
Thus, steric and stereoelectronic effects can endow great stability on collagen triple helices.30,31 Beneficial preorganization prescribes that triple helices should be stabilized by proline derivatives in the Xaa position that prefer the Cγ -endo pucker and by proline derivatives in the Yaa position that prefer the Cγ -exo pucker. We reasoned that 4-chloro substitution, as in (2S,4S)-4-chloroproline (clp) and (2S,4R)-4-chloroproline (Clp), could control proline ring pucker and modulate collagen stability in much the same manner as does a 4-fluoro substitution (Figure 1).
Chlorine and fluorine have similar physicochemical properties. Organic fluorine has an especially weak propensity to form hydrogen bonds.47 Hydrogen bonds to organic chlorine (e.g., O–H···Cl–C) can be slightly stronger, but are still relatively weak.48 Chlorine is the third-most electronegative element after oxygen and fluorine (χO = 3.5; χF = 4.0; χCl = 3.0).49 The inductive effect manifested by a chloro group is actually greater than that of a hydroxyl group and similar to that of a fluoro group (FOH = 0.33; FF = 0.45; FCl = 0.42).50 The stereoelectronic effect that controls proline ring pucker can be regarded as a hyperconjugative effect,35 and Shoulders, Guzei, and Raines page 6 natural bond order analysis suggests that σ*C–Cl orbitals have a greater acceptor ability than do the corresponding σ*C–F orbitals.51 Nevertheless, proline ring pucker is controlled by competing stereoelectronic and steric effects,31,35 and chlorine has a significantly larger covalent radius than does fluorine (rF = 0.64 Å; rCl = 0.77 Å). Hence, it was not apparent a priori whether the electronegativity or the size of a 4-chloro substitution would dominate its effect on proline ring pucker.
4-Chloroproline residues have not been studied in detail in any context. A few chlorinated proline moieties have been found in natural products, all of which are cyclic peptides.52 Cyclochloritine53 and astins A–C,54–57 which are cyclic pentapeptide toxins from Penicillium islandicum and Aster tataricus, respectively, contain a (2R,3S,4R)-3,4-dichloroproline moiety (which is a derivative of L-proline with all three substituents on the same side of the pyrrolidine ring). Likewise, (2S,3S,4R)-3-hydroxy-4-chloroproline is a component of astin I.58 Lee and coworkers demonstrated that replacing the (2R,3S,4R)-3,4-dichloroproline moiety in cyclochlorotine with clp decreased its toxicity.59 Okumura and coworkers found that clp and especially Clp were incorporated readily into viridogrisein by Streptomyces griseoviridus G-89.60 Mauger and Thomas reported that clp or Clp have different effects on the conformation of actinomycin.61 Finally, Chiba and coworkers synthesized very late antigen-4 (VLA-4) antagonists containing a clp or Clp residue.62
To the best of our knowledge, the effects of 4-chloro substitution on proline ring pucker and peptide-bond isomerization have not been described previously. Likewise, neither clp nor Clp have been incorporated in CRPs or other acyclic peptides. Here, we report on the effect of 4-chloro substitution on the conformation of proline rings, prolyl peptide-bond isomerization, and collagen triple-helix stability.
RESULTS AND DISCUSSION
Synthesis of 4-Chloroproline Residues
Clp and clp derivatives appropriately protected for peptide synthesis were prepared from the corresponding hydroxyproline derivatives in 97% and 92% overall yield, respectively, via the route shown in Scheme 1. Boc-Hyp-OBn (1) and Boc-hyp-OBn (2) prepared as we described previously63 were converted to Boc-clp-OBn (3) and Boc-Clp-OBn (4) via an Appel reaction,64 which had been used previously for the synthesis of 4-chloroproline derivatives.62,65 Subsequent hydrogenolysis of the benzyl group and then exchange of the N-Boc protecting group for an N-Fmoc protecting group afforded Fmoc-clp-OH (7) and Fmoc-Clp-OH (8).
SCHEME 1.
Ring Pucker and Ktrans/cis of clp and Clp
Compounds of the form Ac-Xaa-OMe are useful model systems for studying proline ring conformation and peptide-bond isomerization, and have been employed both by our group and by others.31,35,45,66–68 To determine the effect of 4-chloro substitution on proline ring pucker and peptide-bond isomerization, we prepared Ac-clp-OMe (9) and Ac-Clp-OMe (10), as shown in Scheme 1. Acidic methanol was used to cleave the N-Boc group and introduce the methyl ester.69 Subsequent treatment with acetyl chloride (13C-labeled acetyl chloride was used to aid structural analysis as described in the Materials and Methods section) and N,N-dimethylaminopyridine afforded the desired N-acetylated target compounds.
Values of Ktrans/cis for molecules of the type Ac-Xaa-OMe correlate closely with proline ring pucker. Proline itself has a slight preference for the Cγ -endo ring pucker, and Ktrans/cis = 4.6 in water.34 Our group has shown that the minimum-energy conformation of 4-substituted derivatives of proline with Ktrans/cis > 4.6 is generally a Cγ -exo ring pucker, whereas those with Ktrans/cis ≤ 4.6 typically prefer a Cγ -endo ring pucker.35 Using 13C NMR spectroscopy, we determined that Ktrans/cis = 5.4 for Ac-Clp-OMe (10) and 2.2 for Ac-clp-OMe (9).
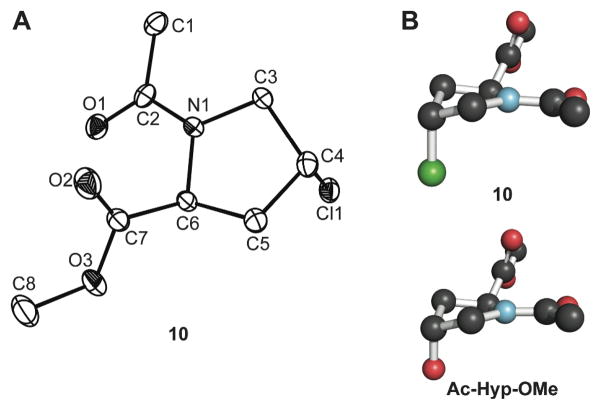
We were able to obtain a crystal structure of Ac-Clp-OMe (10). Crystalline Ac-Clp-OMe displayed the Cγ -exo ring pucker, as expected, and the same overall conformation as we observed previously in the crystal structure of Ac-Hyp-OMe (Figure 2).66 Notably, the structure of Ac-Clp-OMe indicates the presence of a strong n→π* interaction between the amide oxygen and the ester carbonyl, as the Oi−1′ ···Ci′ distance is δBD = 2.80 Å and the Oi−1′ ···Ci′=Oi angle is τBD = 94.1°. This n→π* interaction is known to stabilize the trans conformation of the amide bond.35,43–46 Structural parameters for crystalline Ac-Clp-OMe and Ac-Hyp-OMe66 are listed in Table II.
FIGURE 2.
(A) Molecular drawing of crystalline Ac–Clp–OMe (10; 50% probability ellipsoids). (B) Conformation of crystalline 10 and Ac-Hyp-OMe66 depicted with the program PyMOL (Delano Scientific, Palo Alto, CA).
Table II.
Backbone Dihedral Anglesa of Crystalline Ac-Clp–OMe (10) Derived from X-Ray Diffraction Analysis Compared to Those of Ac-Hyp-OMe69
| Compound | Ring Pucker | φ | ψ | ω | δBD (Å) | τBD (°) |
|---|---|---|---|---|---|---|
| Ac-Clp-OMe (10) | Cγ-exo | −56.0 | 147.5 | 179.2 | 2.80 | 94.1 |
| Ac-Hyp-OMe | Cγ-exo | −56.9 | 150.8 | 178.9 | 2.82 | 91.8 |
For both 10 and Ac–Hyp–OMe, there were two independent molecules in the asymmetric unit of the crystal. Values shown here are the average for these two molecules.
Synthesis of Chlorinated CRPs
With these results in hand, we suspected that Ac-Clp-OMe prefers the Cγ -exo ring pucker and Ac-clp-OMe prefers the Cγ -endo ring pucker. Thus, we expected clp to stabilize triple helices in the Xaa position and Clp to stabilize them in the Yaa position.40 To test this hypothesis, we synthesized (clp–Pro–Gly)7/10, (Pro–Clp–Gly)7/10, (Clp–Pro–Gly)10, and (clp–Clp–Gly)10. These CRPs were prepared by segment condensation of the tripeptides Fmoc-clp–Pro–Gly-OH (18), Fmoc-Pro–Clp–Gly-OH (13), Fmoc-Clp–Pro–Gly-OH (20), and Fmoc-clp–Clp–Gly-OH (15), respectively, on a solid phase. The Fmoc-protected tripeptides were synthesized as shown in Schemes 2 and 3 using PyBOP70 and PyBroP71 to effect the problematic couplings to clp and Clp derivatives.
SCHEME 2.
SCHEME 3.
Conformational Analysis of clp- and Clp-containing CRPs
Triple-helical CRPs have a signature circular dichroism (CD) spectrum with a small maximum near 225 nm and a large minimum near 205 nm. This CD spectrum is also characteristic of PPII conformations, but triple helices undergo a cooperative transition upon heating and PPII structures do not. Therefore, we used CD spectroscopy to assess the impact of clp and Clp on triple-helix structure and stability.
The peptides (Pro–Clp–Gly)7, (Pro–Clp–Gly)10, and (Clp–Pro–Gly)10 all possess the signature CD spectra of PPII and triple-helical conformations (Figure 3A). Only two of those peptides, (Pro–Clp–Gly)7 and (Pro–Clp–Gly)10, undergo cooperative transitions upon heating with Tm values of 23 and 52 °C, respectively (Figure 3B; Table III). Thus, our studies indicate that Clp does indeed stabilize triple helices in the Yaa position relative to Pro, although not quite to the same level as Hyp. When Clp is placed in the Xaa position of CRPs, even the long CRP (Clp–Pro–Gly)10 does not form a triple helix, as indicated by the linear decrease in ellipticity at 225 nm upon heating (Figure 3B and Table III). Thus, both the stereochemistry and position of Clp within CRPs are important for its stabilizing effect on triple helices. The self-association of (Pro–Clp–Gly)10 at 4 °C was confirmed by sedimentation equilibrium (see: Supporting Information).
FIGURE 3.
Conformational analysis of clp- and Clp-containing CRPs. (A, C, and E) CD spectra of peptide solutions (0.2 mM in 50 mM HOAc) at 4 °C after incubating at ≤4 °C for ≥24 h. (B, D, and F) Effect of temperature on the molar ellipticity at 225 or 226 nm. Data were recorded at 3-°C intervals after a 5-min equilibration.
Table III.
Effect of 4-Chloroproline Diastereomers on the Conformational Stability of Collagen Triple Helices
The conformation and stability of clp-containing CRPs was also studied by CD spectroscopy. The effect of clp in the Xaa position on triple-helix stability is similar to that of Pro. Neither (Pro–Pro–Gly)7 nor (clp–Pro–Gly)7 forms a stable triple helix. The failure of (clp–Pro–Gly)7 to form a triple helix is indicated by the linear decrease in ellipticity at 225 nm upon heating (Figure 3D), despite its CD spectrum demonstrating PPII structure at low temperature (Figure 3C). Nonetheless, the longer CRPs (Pro–Pro–Gly)10 and (clp–Pro–Gly)10 form triple Shoulders, Guzei, and Raines page 10 helices with Tm values of 31–4133,72 and 33 °C, respectively (Figures 3C and 3D and Table III). The self-association of (clp–Pro–Gly)10 at 4 °C was confirmed by sedimentation equilibrium (see: Supporting Information).
Two issues must be considered when analyzing the impact of a 4-chloro substitution on proline ring conformation. We have shown that proline ring pucker, and thus Ktrans/cis for the peptide bond, is modulated by reciprocal steric and stereoelectronic effects.31,35 Chlorine is both more electronegative than methyl and larger than fluorine. Therefore, its impact on proline ring pucker should lie somewhere between the extremes of the small, electron-withdrawing fluorine moiety and the large, electron-donating methyl moiety, both of which strongly enforce proline pucker and thus strongly stabilize (or destabilize) a triple helix. 4-Chloroproline derivatives cannot be expected to endow the same degree of conformational stability on triple helices as do mep/Mep and flp/Flp or even Hyp, because of the competing effects of chlorine’s large size and high electronegativity on proline ring conformation. Thus, the results for triple-helix stability of clp- and Clp-containing CRPs (Table III) are in gratifying agreement with our ideas about collagen strand preorganization.
Conformational Analysis of (clp–Clp–Gly)10
As clp and Clp can be accommodated in the Xaa or Yaa position, respectively, of stable collagen triple helices, it might be expected that the CRP (clp–Clp–Gly)10 would form a stable triple helix as well. Surprisingly, however, CRPs of the type (flp–Flp–Gly)7/10 form significantly less stable triple helices than do the analogous (Pro–Hyp–Gly)7/10 CRPs.63,73 Previously, we suggested that this antagonism was due to a deleterious steric interaction between the fluorines of flp and Flp in neighboring strands (Figure 4A).63 In contrast, Kobayashi and coworkers argued that the instability of triple helices formed from (flp–Flp–Gly) repeats disproved the preorganization theory of triple-helix formation, at least for doubly-substituted CRPs.73 We have since shown that peptides of the form (mep–Mep–Gly)7 are more stable than both of the mono-substituted variants (mep–Pro–Gly)7 and (Pro–Mep–Gly)7,31 substantiating the importance of preorganization in doubly-substituted CRPs. We sought to probe deleterious steric interactions in doubly-substituted CRPs again, now with chlorinated CRPs.
FIGURE 4.
Space-filling models of segments of triple helices constructed from the three-dimensional structure of a (Pro-Hyp-Gly)n triple helix (PDB entry 1CAG11) by replacing the H or OH on Pro and Hyp with F or Cl, respectively, using the program SYBYL (Tripos, St. Louis, MO) and depicting the images with the program PyMOL. (A) Segment of a (flp-Flp-Gly)n triple helix (rF···F = 2.4 Å).63 (B) Segment of a (clp-Clp-Gly)n triple helix (rCl···Cl = 1.9 Å).
We found that (clp–Clp–Gly)10 triple helices do not form, even at low temperatures. The (clp–Clp–Gly)10 CRP does possess a PPII-type structure at low temperature (Figure 3E), but does not fold into a stable triple helix. This instability is demonstrated by the absence of a cooperative transition at 225 nm upon heating (Figure 3F) and by sedimentation equilibrium, which showed no self-assembly at low temperature (see: Supporting Information). In contrast, (flp–Flp–Gly)10 triple helices have a Tm of 30 °C.73 These findings suggest that the large size of chlorine relative to fluorine exacerbates the deleterious steric interaction between neighboring strands, as depicted in Figure 4B. 4-Chloroproline derivatives are likely to have greater conformational flexibility than 4-fluoroproline derivatives, so if “locking” the proline rings into a particular deleterious conformation were responsible for the instability of (flp–Flp–Gly)10,73 then (clp–Clp–Gly)10 triple helices should be stable. They are not, supporting our hypothesis63 rather than that of Kobayashi and coworkers.73
CONCLUSIONS
Clp has a strong preference for the Cγ -exo ring pucker and the trans amide bond, wheras clp prefers the Cγ -endo ring pucker and has a lower proportion of the trans amide bond. These results could explain the contrasting effects of clp and Clp on the biosynthesis and activity of natural products.59–61 They further suggest that clp has an appropriate conformation to stabilize polyproline I-type strands whereas Clp should stabilize PPII strands, as was found previously for flp and Flp.74,75 Additionally, we have demonstrated that Clp in the Yaa position of CRPs confers nearly the same degree of stability to triple helices as does Hyp, but it prevents triple helix formation when placed in the Xaa position. In contrast, clp behaves much like Pro in a triple-helical context, as (clp–Pro–Gly)10 and (Pro–Pro–Gly)10 triple helices have similar thermal stabilities. Triple helices do not form from the doubly-substituted CRP (clp–Clp–Gly)10, in contrast to (Pro–Pro–Gly)10 and (flp–Flp–Gly)10 triple helices. This instability is likely due to a strongly deleterious steric interaction between the chlorines in neighboring strands. An interesting attribute of clp- and Clp-containing triple helices is that they could be modified covalently by the SN2 attack of nucleophiles at the halogenated carbon. Finally, we note the similarity of the active site and reactivity of mammalian P4H, the enzyme responsible for hydroxylation of Pro residues in natural collagen,76 and the non-haem iron halogenase SyrB2,77 an enzyme responsible for the chlorination of unactivated carbon atoms.78 As Clp confers thermal stability on collagen triple helices at a level similar to that of Hyp, it is possible that organisms evolving in an environment rich in chloride ions could evolve stable chlorinated collagens analogous to the hydroxylated collagens found in modern animals. The impact of chlorination rather than hydroxylation on the supramolecular structure of native collagen is a subject for future investigation.
MATERIALS AND METHODS
General
Commercial chemicals were of reagent grade or better, and were used without further purification. Anhydrous DMF and CH2Cl2 were obtained from CYCLE-TAINER® solvent delivery systems (J. T. Baker, Phillipsburg, NJ). Other anhydrous solvents were obtained in septum-sealed bottles. In all reactions using anhydrous solvents, glassware was either oven- or flame-dried. “NaHCO3(aq)” and “brine” (i.e., NaCl) refer to saturated aqueous solutions unless specified otherwise. Flash chromatography was performed with columns of silica gel 60, 230–400 mesh (Silicycle, Québec City, Canada). HPLC was performed with gradients of solvent A (0.1% v/v TFA in water) and solvent B (0.1% v/v TFA in acetonitrile), as indicated.
The term “concentrated under reduced pressure” refers to the removal of solvents and other volatile materials using a rotary evaporator at water aspirator pressure (<20 torr) while maintaining the water-bath temperature below 50 °C. Residual solvent was removed from samples at high vacuum (<0.1 torr). The term “high vacuum” refers to vacuum achieved by a mechanical belt-drive oil pump.
NMR spectra were acquired with a Bruker DMX-400 Avance spectrometer unless specified otherwise (1H, 400 MHz; 13C, 100.6 MHz) at the National Magnetic Resonance Facility at Madison (NMRFAM). NMR spectra were obtained at ambient temperatures on samples dissolved in CDCl3 or MeOH-d4. Coupling constants J are provided in Hertz. Compounds with a carbamate protecting group (e.g., Boc or Fmoc) exist as mixtures of Z and E isomers that do not interconvert on the NMR time scale at ambient temperatures. Accordingly, these compounds exhibit two sets of NMR signals.
Mass spectrometry was performed with either a Micromass LCT (electrospray ionization, ESI) in the Mass Spectrometry Facility in the Department of Chemistry or an Applied Biosystems Voyager DE-Pro (matrix-assisted laser desorption/ionization, MALDI) mass spectrometer in the University of Wisconsin Biophysics Instrumentation Facility.
Synthesis of Boc-clp-OBn (3) and Boc-Clp-OBn (4)
General Protocol
The appropriate protected 4-hydroxyproline derivative 1 or 2 (33.8 g, 105 mmol) was dissolved in anhydrous CH2Cl2 (115 mL) under Ar(g) and cooled to 0 °C. Triphenylphosphine (49.6 g, 189 mmol) and then CCl4 (153.8 g, 96 mL) were added. The reaction mixture was stirred at 0 °C for 3 h, then heated to 35 °C for 2 h, and then cooled to rt and stirred for 45 min. The reaction mixture was concentrated under reduced pressure, and the residue was purified by flash chromatography over silica gel (20% v/v EtOAc in hexane).
N-tert-Butyloxycarbonyl-(2S,4S)-4-chloroproline Benzyl Ester (3)
Boc-clp-OBn (3) was obtained in 92% yield as a colorless liquid. 1H NMR δ: 1.34 and 1.46 (s, 9H), 2.38 (dt, J = 5.2, 13.9, 1H), 2.64–2.78 (m, 1H), 3.65 (td, J = 4.7, 12.7, 1H), 3.88–4.01 (m, 1H), 4.37 and 4.51 (m, 2H), 5.06–5.32 (m, 2H), 7.29–7.40 (m, 5H); 13C NMR δ: 28.2, 28.4, 39.5, 40.4, 53.7, 54.7, 55.0, 55.4, 57.7, 58.0, 67.1, 80.6, 128.2, 128.5, 128.6, 128.6, 135.4, 135.6, 153.3, 153.8, 171.3, 171.6. ESI–MS (m/z): [2M + Na]+ calcd for C34H44Cl2N2O8Na 701.2; found 701.9 (2 Cl).
N-tert-Butyloxycarbonyl-(2S,4R)-4-chloroproline Benzyl Ester (4)
Boc-Clp-OBn (4) was obtained in 97% yield as a colorless liquid. 1H NMR δ: 1.36 and 1.47 (s, 9H), 2.28–2.57 (m, 2H), 3.64–3.90 (m, 2H), 4.47 (m, 1H), 4.50 and 4.59 (t, J = 7.6, 1H), 5.08–5.31 (m, 2H), 7.35 (m, 5H); 13C NMR (125 MHz) δ: 28.3, 28.5, 39.8, 40.8, 55.3, 55.5, 55.6, 55.7, 57.8, 58.0, 67.2, 80.8, 80.9, 128.3, 128.5, 128.6, 128.7, 128.8, 135.4, 135.7, 153.7, 154.2, 172.1, 172.4; HRMS–ESI (m/z): [2M + Na]+ calcd for C34H44Cl2N2O8Na 701.2372; found 701.2343 (2 Cl).
Synthesis of Boc-clp-OH (5) and Boc-Clp-OH (6)
General Protocol
MeOH (400 mL) was added carefully to a mixture of the appropriate 4-chloroproline derivative 3 or 4 (31.8 g, 93.6 mmol) and Pd/C (10% w/w, 10.4 g) under Ar(g), and the resulting black suspension was stirred under H2(g) for 23 h. The suspension was filtered through a pad of Celite and concentrated under reduced pressure.
N-tert-Butyloxycarbonyl-(2S,4S)-4-chloroproline (5)
Boc-clp-OH (5) was obtained in quantitative yield as a white solid. 1H NMR δ: 1.44 and 1.49 (s, 9H), 2.37–2.85 (m, 2H), 3.56–3.72 (m, 1H), 3.84–4.04 (m, 1H), 4.32–4.51 (m, 2H), 8.22 (bs, 1H); 13C NMR (125 MHz) δ: 28.3, 28.4, 38.8, 40.3, 53.6, 54.5, 54.9, 55.7, 57.9, 81.3, 81.8, 153.6, 155.3, 175.2, 177.2; HRMS–ESI (m/z): [M - H]− calcd for C10H15ClNO2 248.0690; found 248.0694 (1 Cl).
N-tert-Butyloxycarbonyl-(2S,4R)-4-chloroproline (6)
Boc-Clp-OH (6) was obtained in quantitative yield as a white solid. 1H NMR δ: 1.44 and 1.50 (s, 9H), 2.38–2.72 (m, 2H), 3.70–3.83 (m, 2H), 4.49 (m, 1H), 4.57 (t, J = 7.2, 1H); 13C NMR (125 MHz) δ: 28.3, 28.4, 38.9, 40.7, 55.3, 55.4, 55.9, 57.8, 58.0, 81.3, 82.5, 153.6, 156.4, 174.4, 177.9; HRMS–ESI (m/z): [M - H]− calcd for C10H15ClNO2 248.0690; found 248.0680 (1 Cl).
Synthesis of Fmoc-clp-OH (7) and Fmoc-Clp-OH (8)
General Protocol
The appropriate 4-chloroproline derivative 5 or 6 (22.6 g, 90.3 mmol) was dissolved in 4 N HCl in dioxane (550 mL) under Ar(g) and stirred for 3 h. The resulting solution was concentrated under reduced pressure. The free amine was dissolved in 10% w/v NaHCO3(aq) (375 mL). A solution of Fmoc-OSu (33.5 g, 99.3 mmol) in dioxane (600 mL) was added, and the resulting white suspension was stirred for 19 h. The reaction mixture was concentrated under reduced pressure, and the aqueous solution was diluted with water (300 mL) and washed with ether (3 × 500 mL). The aqueous layer was acidified to pH 1.5 with 12 N HCl, extracted with ether (3 × 500 mL), dried over anhydrous MgSO4(s), and concentrated under reduced pressure.
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-chloroproline (7)
Fmoc-clp-OH (7) was obtained in quantitative yield as a white solid. 1H NMR (500 MHz) δ: 2.42–2.77 (m, 2H), 3.63–3.76 (m, 1H), 3.85–4.04 (m, 1H), 4.13–4.62 (m, 5H), 7.27–7.45 (m, 4H), 7.50–7.62 (m, 2H), 7.69–7.80 (m, 2H); 13C NMR (125 MHz) δ: 38.7, 40.4, 47.2, 53.8, 54.6, 55.7, 57.4, 58.2, 67.8, 68.3, 120.2, 125.0, 125.1, 127.3, 127.9, 128.0, 141.5, 143.6, 143.8, 154.3, 155.5, 174.6, 176.0; HRMS–ESI (m/z): [M + Na]+ calcd for C20H18ClNO4Na 394.0822; found 394.0828 (1 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S,4R)-4-chloroproline (8)
Fmoc-Clp-OH (8) was obtained in quantitative yield as a white solid. 1H NMR (500 MHz) δ: 2.35–2.68 (m, 2H), 3.83 (m, 2H), 4.11–4.67 (m, 5H), 7.13 (bs, 1H), 7.23–7.45 (m, 4H), 7.49–7.62 (m, 2H), 7.74 (m, 2H); 13C NMR (125 MHz) δ: 39.3, 40.8, 47.2, 47.3, 55.0, 55.5, 55.7, 56.0, 57.3, 58.0, 68.0, 68.3, 120.1, 120.2, 124.9, 125.1, 127.2, 127.3, 127.8, 128.0, 141.4, 143.6, 146.7, 144.0, 154.5, 155.8, 175.2, 176.8; HRMS–ESI (m/z): [M + Na]+ calcd for C20H18ClNO4Na 394.0822; found 394.0812 (1 Cl).
Synthesis of Ac-clp-OMe (9) and Ac-Clp-OMe (10)
General Protocol
The appropriate 4-chloroproline derivative 5 or 6 (107 mg, 0.4 mmol) was dissolved in anhydrous MeOH (8 mL) and cooled to 0 °C. Acetyl chloride (8 mL) was added dropwise and the resulting solution was stirred for 7 h at rt. The solution was concentrated under reduced pressure and dried overnight under high vacuum. The residue was dissolved in anhydrous CH2Cl2 (20 mL) under Ar(g). N,N-dimethylaminopyridine (397 mg, 3.3 mmol) and acetyl chloride (250 mg, 3.1 mmol) were added, and the resulting solution was stirred for 22 h. The reaction mixture was concentrated under reduced pressure, and the residue was dissolved in 10% w/v aqueous citric acid (40 mL). The aqueous solution was extracted with CH2Cl2 (2 × 75 mL), which was then dried over anhydrous MgSO4(s), and concentrated under reduced pressure. The residue was purified by flash chromatography over silica gel (10% v/v hexane in EtOAc to elute byproducts and then 15% v/v MeOH in EtOAc to elute product).
N-(Acetyl)-(2S,4S)-4-chloroproline Methyl Ester (9)
Ac-clp-OMe (9) was obtained in 82% yield as a fragrant, colorless oil. 1H NMR δ: 2.02 and 2.08 (2 s, 3H), 2.30–2.38 (m, 0.7H), 2.62– 2.79 (m, 1.3H), 3.73 and 3.78 (2 s, 3H), 3.72–3.82 (m, 1H), 3.97–4.06 (m, 1H), 4.38–4.49 (m, 1.3H), 4.58 (dd, J = 5.2, 8.8, 0.7H); 13C NMR δ: 22.1, 22.4, 39.0, 41.1, 52.6, 52.9, 54.2, 54.5, 55.8, 56.2, 57.4, 58.8, 169.2, 170.0, 171.4, 171.4; HRMS–ESI (m/z): [M + H]+ calcd for C8H13NO3 205.0584; found 205.0582 (1 Cl).
N-(Acetyl)-(2S,4R)-4-chloroproline Methyl Ester (10)
Ac-Clp-OMe (10) was obtained in 76% yield as a fragrant, colorless oil. 1H NMR δ: 1.99 and 2.08 (2 s, 3H), 2.31–2.69 (m, 2H), 3.74 and 3.78 (2 s, 3H), 3.69–3.85 (m, 1H), 4.03 (dd, J = 5.0, 11.4, 1H), 4.45–4.67 (m, 2H); 13C NMR δ: 21.7, 22.3, 39.3, 41.4, 52.6, 53.0, 54.4, 55.2, 55.7, 56.7, 57.4, 58.6, 169.4, 170.0, 172.1, 172.4; HRMS–ESI (m/z): [M + Na]+ calcd for C8H12NO3Na 205.0403; found 205.0403 (1 Cl).
N-tert-Butyloxycarbonyl-(2S,4R)-4-chloroprolyl–glycine Benzyl Ester (11)
Boc-Clp-OH (6) (23.3 g, 93.2 mmol), glycine benzyl ester tosylate (40.9 g, 121.2 mmol), and PyBOP (48.5 g, 93.2 mmol) were dissolved in anhydrous CH2Cl2 (400 mL) under Ar(g). DIEA (30.1 g, 233 mmol) was added slowly, and the resulting solution was stirred for 16 h. The reaction mixture was washed with 10% w/v aqueous citric acid (2 × 1.0 L), dried over anhydrous MgSO4(s), and concentrated under reduced pressure. The crude oil was purified by flash chromatography over silica gel (gradient: 33% v/v EtOAc in hexane to 50% v/v EtOAc in hexane) to afford Boc-Clp–Gly-OBn (11) (29.8 g, 75.1 mmol, 81%) as a colorless, sticky paste. 1H NMR δ: 1.47 (s, 9H), 2.25–2.80 (m, 2H), 3.70 and 3.92 (m, 2H), 4.01–4.16 (m, 2H), 4.41–4.59 (m, 2H), 5.18 (s, 2H), 6.61 (m, 0.3H), 7.36 (m, 5H); 13C NMR δ: 28.4, 38.2, 41.0, 41.3, 41.6, 55.7, 55.9, 58.6, 59.8, 67.3, 67.5, 81.6, 128.5, 128.7, 128.8, 135.3, 157.1, 166.8, 169.5, 171.3; HRMS–ESI (m/z): [M + Na]+ calcd for C19H25ClN2O5Na, 419.1350; found, 419.1335 (1 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S)-prolyl–(2S,4R)-4-chloroprolyl–glycine Benzyl Ester (12)
Boc-Clp–Gly-OBn (11) (1.00 g, 2.5 mmol) was dissolved in 4 N HCl in dioxane (30 mL) under Ar(g) and stirred for 2 h. The resulting solution was concentrated under reduced pressure and the residue dissolved in anhydrous DMF (50 mL) under Ar(g). DIEA (1.20 g, 9.3 mmol) was added, followed by Fmoc-Pro-OPfp (2.67 g, 5.3 mmol). The solution was stirred for 18 h and then concentrated by rotary evaporation under high vacuum. Flash chromatography over silica gel (gradient: 20% v/v EtOAc in hexane to 30% v/v EtOAc in hexane) afforded 12 (1.10 g, 1.8 mmol, ~71%) as a white solid containing an impurity that was removed after the subsequent step.
PyBroP Couplings
General Protocol
The appropriate Boc-Xaa–Gly-OBn derivative (11) or (16)79 (52.5 mmol) was dissolved in 4 N HCl in dioxane (700 mL) under Ar(g) and stirred for 2 h. The resulting solution was concentrated under reduced pressure, dried under high vacuum and the residue dissolved in anhydrous CH2Cl2 (800 mL) under Ar(g). The appropriate 4-chloroproline derivative (7) or (8) (52.5 mmol) was added, and the resulting solution was cooled to 0 °C. PyBroP (24.5 g, 52.5 mmol) and DIEA (23.8 g, 184 mmol) were added. The resulting solution was allowed towarm slowly to room temperature and then stirred for 18 h. The reaction mixture was washed with 10% w/v aqueous citric acid (1.0 L), NaHCO3(aq) (1.0 L), and brine (1.0 L). The organic layer was dried over anhydrous MgSO4(s), and concentrated under reduced pressure. The residue was purified by flash chromatography over silica gel (gradient: 30% v/v EtOAc in hexane to 90% v/v EtOAc in hexane).
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-chloroprolyl–(2S,4R)-4-chloroprolyl–glycine Benzyl Ester (14)
Fmoc-clp–Clp–Gly-OBn (14) was obtained in 67% yield as a white solid. HRMS–ESI (m/z): [M + Na]+ calcd for C34H33Cl2N3O6Na, 672.1644; found, 672.1646 (2 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-chloroprolyl–(2S)-prolyl–glycine Benzyl Ester (17)
Fmoc-clp–Pro–Gly-OBn (17) was obtained in 82% yield as a white solid. HRMS–ESI (m/z): [M + Na]+ calcd for C34H34ClN3O6Na, 638.2034; found, 638.2039 (1 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S,4R)-4-chloroprolyl–(2S)-prolyl–glycine Benzyl Ester (19)
Fmoc-Clp–Pro–Gly-OBn (19) was obtained in 72% yield as a white solid. HRMS–ESI (m/z): [M + Na]+ calcd for C34H34ClN3O6Na, 638.2034; found, 638.2009 (1 Cl).
Hydrogenolysis of Benzyl Groups on Protected Tripeptides
General Protocol
MeOH (400 mL) was added carefully to a mixture of the appropriate Fmoc-protected tripeptide benzyl ester (12), (14), (17), or (19) (34.6 mmol) and Pd/C (10% w/w, 9.6 g) under Ar(g). The resulting black suspension was stirred under H2(g) for ~4 h. Careful monitoring by TLC was necessary to prevent hydrogenolysis of the Fmoc group. The suspension was filtered through a pad of Celite and concentrated under reduced pressure. The crude product was purified by flash chromatography over silica gel (EtOAc to elute byproducts, then 12% v/v MeOH in CH2Cl2 containing 0.1% v/v formic acid). The fractions containing the reaction product were concentrated under reduced pressure and the formic acid was removed by dissolving the residue in 10% v/v MeOH in toluene and concentrating under reduced pressure.
N-9-Fluorenylmethoxycarbonyl-(2S)-prolyl–(2S,4R)-4-chloroprolyl–glycine (13)
Fmoc-Pro–Clp–Gly-OH (13) was obtained in 67% yield as a white solid. 13C NMR (125 MHz, MeOH-d4) δ: 24.1, 25.2, 30.1, 31.0, 40.1, 42.1, 47.9, 56.7, 57.0, 57.3, 57.4, 59.3, 59.7, 59.9, 60.2, 60.3, 68.4, 68.7, 120.9, 126.1, 126.2, 126.3, 128.2, 128.4, 128.8, 142.5, 142.6, 145.0, 145.1, 145.4, 145.5, 156.2, 156.6, 173.3, 173.3, 173.5, 173.7; HRMS–ESI (m/z): [M − H]− calcd for C27H27ClN3O6, 524.1589; found, 524.1586 (1 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-chloroprolyl–(2S,4R)-4-chloroprolyl–glycine (15)
Fmoc-clp–Clp–Gly-OH (15) was obtained in 66% yield as a white solid. 13C NMR (125 MHz, MeOH-d4) δ: 39.6, 40.0, 40.1, 40.5, 41.9, 53.2, 53.6, 55.6, 56.1, 56.8, 57.1, 57.4, 58.5, 58.8, 60.3, 60.4, 68.8, 68.9, 120.9, 126.1, 126.3, 128.2, 128.4, 128.7, 128.7, 142.5, 142.5, 142.6, 142.7, 144.9, 145.0, 145.3, 145.4, 156.7, 156.0, 172.1, 172.2, 172.6, 173.4, 173.6; HRMS–ESI (m/z): [M + Na]+ calcd for C27H27Cl2N3O6Na, 582.1175; found, 582.1157 (2 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S,4S)-4-chloroprolyl–(2S)-prolyl–glycine (18)
Fmoc-clp–Pro–Gly-OH (18) was obtained in 57% yield as a white solid. 13C NMR (125 MHz, MeOH-d4) δ: 24.1, 25.7, 25.9, 28.6, 30.1, 30.3, 37.8, 40.3, 41.8, 46.5, 53.0, 53.5, 55.2, 55.5, 56.0, 56.3, 58.3, 58.7, 59.8, 61.3, 61.6, 67.7, 68.9, 120.9, 125.7, 125.9, 126.1, 128.2, 128.9, 142.5, 142.6, 144.9, 145.0, 145.3, 145.5, 155.6, 155.9, 167.7, 169.2, 171.9, 172.1, 172.6, 174.4, 174.6; HRMS–ESI (m/z): [M + Na]+ calcd C27H28ClN3O6Na, 548.1564; found, 548.1561 (1 Cl).
N-9-Fluorenylmethoxycarbonyl-(2S,4R)-4-chloroprolyl–(2S)-prolyl–glycine (20)
Fmoc-Clp–Pro–Gly-OH (20) was obtained in 58% yield as a white solid. 13C NMR (125 MHz, MeOH-d4) δ: 24.2, 25.7, 25.9, 28.6, 30.1, 30.4, 39.1, 40.4, 41.0, 42.1, 46.3, 47.9, 56.1, 56.8, 57.1, 57.3, 57.4, 57.7, 58.0, 58.2, 59.9, 61.4, 61.7, 67.7, 68.9, 121.0, 125.7, 125.9, 126.1, 128.2, 128.3, 128.9, 142.6, 142.7, 144.8, 145.0, 145.2, 145.5, 156.1, 156.4, 168.0, 168.6, 172.2, 172.4, 174.2, 174.5; HRMS–ESI (m/z): [M + Na]+ calcd C27H28ClN3O6Na, 548.1564; found, 548.1578 (1 Cl).
Measurement of Ktrans/cis Values of Ac-clp-OMe (9) and Ac-Clp-OMe (10)
13C-labeled versions of Ac-clp-OMe (9) and Ac-Clp-OMe (10) (5–10 mg) were dissolved in D2O with sufficient MeOH-d4 added to solubilize the compound (<20% of total volume). The 13C NMR spectra were recorded using an inverse-gated decoupling pulse program with a relaxation delay of 100 s and a pulse width of 10 μs. The spectral baselines were corrected and peaks corresponding to the labeled carbon were integrated. Values of Ktrans/cis were determined by the relative areas of the trans and cis peaks for the labeled carbons.
General Protocol for Attachment of Fmoc-Pro–Clp–Gly-OH (13), Fmoc-clp–Clp–Gly-OH (15), Fmoc-clp–Pro–Gly-OH (18), and Fmoc-Clp–Pro–Gly-OH (20) to 2-Chlorotrityl Resin
Under Ar(g), 23 mg of 2-chlorotrityl resin (loading: 1.6 mmol/g) was swelled in anhydrous CH2Cl2 (0.7 mL) for 5 min. A solution of the appropriate Fmoc-protected tripeptide (13), (15), (18), or (20) (0.034 mmol) and DIEA (17 mg, 0.13 mmol) in anhydrous CH2Cl2 (0.7 mL) was added by syringe. Additional anhydrous CH2Cl2 (0.5 mL) was used to ensure complete transfer. After 2 h, anhydrous MeOH (0.2 mL) was added to cap any remaining active sites on the resin. The resin-bound peptide was isolated by gravity filtration, washed with several portions of anhydrous CH2Cl2 (~25 mL), and dried under high vacuum. Loadings were measured by an Fmoc-deprotection ultraviolet spectroscopy assay to be 0.44 mmol/g for (13), 0.55 mmol/g for (15), 0.35 mmol/g for (18), and 0.48 mmol/g for (20).
Synthesis of (Pro–Clp–Gly)7, (clp–Pro–Gly)7, (Pro–Clp–Gly)10, (clp–Pro–Gly)10, (clp–Clp–Gly)10, and (Clp–Pro–Gly)10
The two 21-mer peptides and the four 30-mer peptides were synthesized by segment condensation of their corresponding Fmoc-tripeptides (13, 15, 18, and 20) on a solid phase using an Applied Biosystems Synergy 432A Peptide Synthesizer at the University of Wisconsin–Madison Biotechnology Center. The first trimer was loaded onto the resin as described above. Fmoc-deprotection was achieved by treatment with 20% (v/v) piperidine in DMF. The trimers (3 equiv) were converted to active esters by treatment with HBTU, DIEA, and HOBt. Extended couplings (30–60 min) were employed at room temperature.
Peptides were cleaved from the resin in 95:3:2 TFA:triisopropylsilane:H2O (1.5 mL), precipitated from t-butylmethylether at 0 °C, isolated by centrifugation, and lyophilized. Semi-preparative HPLC was used to purify the peptides (Pro–Clp–Gly)7 (Dynamax C-18 column, gradient: 10% B to 65% B over 50 min), (clp–Pro–Gly)7 (Dynamax C-18 column, gradient: 10% B to 65% B over 50 min), (Pro–Clp–Gly)10 (Zorbax C-8 column, gradient: 10% B to 65% B over 60 min), (clp–Pro–Gly)10 (Zorbax C-8 column, gradient: 10% B to 90% B over 60 min), (clp–Clp–Gly)10 (Zorbax C-8 column, gradient: 10% B to 90% B over 60 min), and (Clp–Pro–Gly)10 (Zorbax C-8 column, gradient 10% B to 65% B over 60 min). All six peptides were >90% pure by analytical HPLC and MALDI–TOF mass spectrometry (m/z) [M + H]+ calcd for C84H122Cl7N21O22 2020; found 2019 for (Pro–Clp–Gly)7; [M + Na]+ calcd for C84H121Cl7N21NaO22 2044; found 2041 for (clp–Pro–Gly)7; [M + Na]+ calcd for C120H162Cl10N30O31Na 2900; found 2900 for (Pro–Clp–Gly)10, 2897 for (clp–Pro–Gly)10, and 2899 (Clp–Pro–Gly)10; [M + Na]+ calcd for C120H153Cl20N30O31Na 3244; found 3242 for (clp–Clp–Gly)10.
Circular Dichroism Spectroscopy of (Pro–Clp–Gly)7, (clp–Pro–Gly)7, (Pro–Clp–Gly)10, (clp–Pro–Gly)10, (clp–Clp–Gly)10, and (Clp–Pro–Gly)10
Peptides were dried under vacuum for at least 24 h before being weighed and dissolved to 0.2 mM in 50 mM acetic acid (pH 3.0). These solutions were incubated at ≤4 °C for ≥24 h before CD spectra were acquired with an Aviv 202SF spectrometer at the University of Wisconsin Biophysics Instrumentation Facility. Spectra were measured with a 1-nm band-pass in cuvettes with a 0.1-cm pathlength. The signal was averaged for 3 s during wavelength scans and 5 s during denaturation experiments using a 0.6-°C temperature deadband. During denaturation experiments, CD spectra were acquired at intervals of 3 °C. At each temperature, solutions were equilibrated for 5 min before data acquisition. Values of Tm were determined by fitting the molar ellipticity at 225 nm for (Pro-Clp-Gly)7 and (clp-Pro-Gly)7 or 226 nm for the other four peptides to a two-state model.80 Tm values were determined in triplicate.
X-Ray Crystallography
The crystals of Ac-Clp-OMe (10) used for X-ray structure determination were obtained by dissolving the colorless oil in a minimum of ethyl acetate and equilibrating with a reservoir of hexanes. Crystals suitable for X-ray crystallography grew slowly over the course of two months. The experimental procedure for the structure determination and tables of atomic coordinates, bond lengths, bond angles, and torsion angles are provided in the Supporting Information.
Sedimentation Equilibrium Experiments on (clp–Pro–Gly)10, (Pro–Clp–Gly)10, and (clp–Clp–Gly)10
Sedimentation equilibrium experiments were performed with a Beckman XL-A analytical ultracentrifuge at the University of Wisconsin Biophysics Instrumentation Facility to evaluate the self-assembly of the peptides (clp–Pro–Gly)10, (Pro–Clp–Gly)10, and (clp–Clp–Gly)10. Detailed experimental procedures are provided in the Supporting Information.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Elkan R. Blout (1919–2006). We are grateful to D. Gottlieb, J. Kalia, and D.R. McCaslin for helpful discussion. MDS was supported by a Department of Homeland Security Graduate Fellowship. This work was supported by Grant AR44276 (NIH). CD, MALDI-TOF, and sedimentation equilibrium experiments were performed at the University of Wisconsin–Madison Biophysics Instrumentation Facility, which was established by Grants BIR-9512577 (NSF) and RR13790 (NIH). NMRFAM was supported by Grant P41RR02301 (NIH).
References
- 1.Myllyharju J, Kivirikko KI. Ann Med. 2001;33:7–21. doi: 10.3109/07853890109002055. [DOI] [PubMed] [Google Scholar]
- 2.Ramachandran GN, Kartha G. Nature. 1954;174:269–270. doi: 10.1038/174269c0. [DOI] [PubMed] [Google Scholar]
- 3.Ramachandran GN, Kartha G. Nature. 1955;176:593–595. doi: 10.1038/176593a0. [DOI] [PubMed] [Google Scholar]
- 4.Rich A, Crick FHC. Nature. 1955;176:915–916. doi: 10.1038/176915a0. [DOI] [PubMed] [Google Scholar]
- 5.Cowan PM, McGavin S, North ACT. Nature. 1955;176:1062–1064. doi: 10.1038/1761062a0. [DOI] [PubMed] [Google Scholar]
- 6.Rich A, Crick FHC. J Mol Biol. 1961;3:483–506. doi: 10.1016/s0022-2836(61)80016-8. [DOI] [PubMed] [Google Scholar]
- 7.Ricard-Blum S, Ruggiero F, van der Rest M. Top Curr Chem. 2005;247:35–84. [Google Scholar]
- 8.Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R. J Biol Chem. 2006;281:3494–3504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins CL, Raines RT. Nat Prod Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 10.Brodsky B, Persikov AV. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 11.Bella J, Eaton M, Brodsky B, Berman HM. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 12.Fietzek PP, Kühn K. Mol Cell Biochem. 1975;8:141–157. doi: 10.1007/BF01792765. [DOI] [PubMed] [Google Scholar]
- 13.Engel J, Bächinger HP. Top Curr Chem. 2005;247:7–33. [Google Scholar]
- 14.Myllyharju J. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 15.Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, Kimble J. Proc Natl Acad Sci USA. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivrikko KI, Myllyharju J. J Biol Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- 17.Berg RA, Prockop DJ. Biochem Biophys Res Commun. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 18.Burjanadze TV. Biopolymers. 2000;53:523–528. doi: 10.1002/(SICI)1097-0282(200005)53:6<523::AID-BIP8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Brown FR, III, Carver JP, Blout ER. J Mol Biol. 1969;39:307–313. doi: 10.1016/0022-2836(69)90319-2. [DOI] [PubMed] [Google Scholar]
- 20.Brown FR, III, di Corato A, Lorenzi GP, Blout ER. J Mol Biol. 1972;63:85–99. doi: 10.1016/0022-2836(72)90523-2. [DOI] [PubMed] [Google Scholar]
- 21.Doyle BB, Bendit EG, Blout ER. Biopolymers. 1975;14:937–957. doi: 10.1002/bip.1975.360140505. [DOI] [PubMed] [Google Scholar]
- 22.Sakakibara S, Inouye K, Shudo K, Kishida Y, Kobayashi Y, Prockop DJ. Biochim Biophys Acta. 1973;303:198–202. doi: 10.1016/0005-2795(73)90164-5. [DOI] [PubMed] [Google Scholar]
- 23.Inouye K, Sakakibara S, Prockop DJ. Biochim Biophys Acta. 1976;420:133–141. doi: 10.1016/0005-2795(76)90352-4. [DOI] [PubMed] [Google Scholar]
- 24.Inouye K, Kobayashi Y, Kyogoku Y, Kishida Y, Sakakibara S, Prockop DJ. Arch Biochem Biophys. 1982;219:198–203. doi: 10.1016/0003-9861(82)90149-7. [DOI] [PubMed] [Google Scholar]
- 25.Bann JG, Bächinger HP. J Biol Chem. 2000;275:24466–24469. doi: 10.1074/jbc.M003336200. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno K, Hayashi T, Bächinger HP. J Biol Chem. 2003;278:32373–32379. doi: 10.1074/jbc.M304741200. [DOI] [PubMed] [Google Scholar]
- 27.Mizuno K, Hayashi T, Peyton DH, Bächinger HP. J Biol Chem. 2004;279:38072–38078. doi: 10.1074/jbc.M402953200. [DOI] [PubMed] [Google Scholar]
- 28.Kawahara K, Nishi Y, Nakamura S, Uchiyama S, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. Biochemistry. 2005;44:15812–15822. doi: 10.1021/bi051619m. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher M, Mizuno K, Bächinger HP. J Biol Chem. 2005;280:20397–20403. doi: 10.1074/jbc.M501453200. [DOI] [PubMed] [Google Scholar]
- 30.Raines RT. Protein Sci. 2006;15:1219–1225. doi: 10.1110/ps.062139406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoulders MD, Hodges JA, Raines RT. J Am Chem Soc. 2006;128:8112–8113. doi: 10.1021/ja061793d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Nature. 1998;392:666–667. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- 33.Holmgren SK, Bretscher LE, Taylor KM, Raines RT. Chem Biol. 1999;6:63–70. doi: 10.1016/S1074-5521(99)80003-9. [DOI] [PubMed] [Google Scholar]
- 34.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 35.DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. J Am Chem Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]
- 36.Improta R, Benzi C, Barone V. J Am Chem Soc. 2001;123:12568–12577. doi: 10.1021/ja010599i. [DOI] [PubMed] [Google Scholar]
- 37.Giacovazzo C, Monaco HL, Artioli G, Viterbo D, Ferraris G, Gilli G, Zanotti G, Catti M. Fundamentals of Crystallography. 2. Oxford University Press; Oxford, UK: 2002. [Google Scholar]
- 38.Umashankara M, Babu IR, Ganesh KN. Chemical Commun. 2003:2606–2607. doi: 10.1039/b308581c. [DOI] [PubMed] [Google Scholar]
- 39.Hodges JA, Raines RT. J Am Chem Soc. 2003;125:9262–9263. doi: 10.1021/ja035881z. [DOI] [PubMed] [Google Scholar]
- 40.Vitagliano L, Berisio R, Mazzarella L, Zagari A. Biopolymers. 2001;58:459–464. doi: 10.1002/1097-0282(20010415)58:5<459::AID-BIP1021>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 41.Doi M, Nishi Y, Uchiyama S, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. J Am Chem Soc. 2003;125:9922–9923. doi: 10.1021/ja035997v. [DOI] [PubMed] [Google Scholar]
- 42.Barth D, Milbradt AG, Renner C, Moroder L. ChemBioChem. 2004;5:79–86. doi: 10.1002/cbic.200300702. [DOI] [PubMed] [Google Scholar]
- 43.Hinderaker MP, Raines RT. Protein Sci. 2003;12:1188–1194. doi: 10.1110/ps.0241903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodges JA, Raines RT. Org Lett. 2006;8:4695–4697. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonntag LS, Schweizer S, Ochsenfeld C, Wennemers H. J Am Chem Soc. 2006;128:14697–14703. doi: 10.1021/ja0654938. [DOI] [PubMed] [Google Scholar]
- 46.Gorske BC, Bastian BL, Geske GD, Blackwell HE. J Am Chem Soc. 2007;129:8928–8929. doi: 10.1021/ja071310l. [DOI] [PubMed] [Google Scholar]
- 47.Dunitz JD, Taylor R. Chem Eur J. 1997;3:89–98. [Google Scholar]
- 48.Banerjee R, Desiraju GR, Mondal R, Howard JAK. Chem Eur J. 2004;10:3373–3383. doi: 10.1002/chem.200400003. [DOI] [PubMed] [Google Scholar]
- 49.Pauling L. The Nature of the Chemical Bond. Cornell University Press; Ithaca, NY: 1960. [Google Scholar]
- 50.Hansch C, Leo A, Taft RW. Chem Rev. 1991;91:165–195. [Google Scholar]
- 51.Alabugin IV, Zeidan TA. J Am Chem Soc. 2002;124:3175–3185. doi: 10.1021/ja012633z. [DOI] [PubMed] [Google Scholar]
- 52.Mauger AB. J Nat Prod. 1996;59:1205–1211. doi: 10.1021/np9603479. [DOI] [PubMed] [Google Scholar]
- 53.Yoshioka H, Nakatsu K, Sato M, Tatsuno T. Chem Lett. 1973:1319–1322. [Google Scholar]
- 54.Morita H, Nagashima S, Takeya K, Itokawa H. Chem Pharm Bull. 1993;41:992–993. doi: 10.1248/cpb.41.992. [DOI] [PubMed] [Google Scholar]
- 55.Morita H, Nagashima S, Takeya K, Itokawa H. Tetrahedron. 1994;50:11613–11622. [Google Scholar]
- 56.Morita H, Nagashima S, Takeya K, Itokawa H, Iitaka Y. Tetrahedron. 1995;51:1121–1132. [Google Scholar]
- 57.Tan NH, Zhou J. Chem Rev. 2006;106:840–895. doi: 10.1021/cr040699h. [DOI] [PubMed] [Google Scholar]
- 58.Morita H, Nagashima S, Takeya K, Itokawa H. Chem Lett. 1994:2009–2010. [Google Scholar]
- 59.Lee S, Noda K, Aoyagi H, Kato T, Izumiya N. Int J Pept Protein Res. 1986;27:44–50. [Google Scholar]
- 60.Okumara Y, Okamoto R, Ishikura T. Agric Biol Chem. 1984;48:543–544. [Google Scholar]
- 61.Mauger AB, Thomas WA. Org Magn Reson. 1981;17:186–191. [Google Scholar]
- 62.Chiba J, Takayama G, Takashi T, Yokoyama M, Nakayama A, Baldwin JJ, McDonald E, Moriarty KJ, Sarko CR, Saionz KW, Swanson R, Hussain Z, Wong A, Machinaga N. Bioorg Med Chem. 2006;14:2725–2746. doi: 10.1016/j.bmc.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 63.Hodges JA, Raines RT. J Am Chem Soc. 2005;127:15923–15932. doi: 10.1021/ja054674r. [DOI] [PubMed] [Google Scholar]
- 64.Appel R. Angew Chem Int Ed Engl. 1975;14:801–811. [Google Scholar]
- 65.Berger Y, Dehmlow H, Blum-Kaelin D, Kitas EA, Löffler BM, Aebi JD, Juillerat-Jeanneret L. J Med Chem. 2005;48:483–498. doi: 10.1021/jm040857x. [DOI] [PubMed] [Google Scholar]
- 66.Panasik N, Jr, Eberhardt ES, Edison AS, Powell DR, Raines RT. Int J Pept Protein Res. 1994;44:262–269. doi: 10.1111/j.1399-3011.1994.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 67.Eberhardt ES, Panasik N, Jr, Raines RT. J Am Chem Soc. 1996;118:12261–12266. doi: 10.1021/ja9623119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jenkins CL, McCloskey AI, Guzei IA, Eberhardt ES, Raines RT. Biopolymers: Pept Sci. 2005;80:1–8. doi: 10.1002/bip.20164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nudelman A, Bechor Y, Falb E, Fischer B, Wexler BA, Nudelman A. Synth Commun. 1998;28:471–474. [Google Scholar]
- 70.Coste J, Le-Nguyen D, Castro B. Tetrahedron Lett. 1990;31:205–208. [Google Scholar]
- 71.Coste J, Frérot E, Jouin P, Castro B. Tetrahedron Lett. 1991;32:1967–1970. [Google Scholar]
- 72.Nishi Y, Uchiyama S, Doi M, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. Biochemistry. 2005;44:6034–6042. doi: 10.1021/bi047887m. [DOI] [PubMed] [Google Scholar]
- 73.Doi M, Nishi Y, Uchiyama S, Nishiuchi Y, Nishio H, Nakazawa T, Ohkubo T, Kobayashi Y. J Pept Sci. 2005;11:609–616. doi: 10.1002/psc.671. [DOI] [PubMed] [Google Scholar]
- 74.Horng JC, Raines RT. Protein Sci. 2006;15:74–83. doi: 10.1110/ps.051779806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruzza P, Siligardi G, Donella-Deana A, Calderan A, Hussain R, Rubini C, Cesaro L, Osler A, Guiotto A, Pinna LA, Borin G. J Pept Sci. 2006;12:462–471. doi: 10.1002/psc.750. [DOI] [PubMed] [Google Scholar]
- 76.Myllyharju J. Top Curr Chem. 2005;247:115–147. [Google Scholar]
- 77.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 78.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 79.Jenkins CL, Vasbinder MM, Miller SJ, Raines RT. Org Lett. 2005;7:2619–2622. doi: 10.1021/ol050780m. [DOI] [PubMed] [Google Scholar]
- 80.Becktel WJ, Schellman JA. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.