Abstract
Ranpirnase (Onconase®), a cytotoxic ribonuclease from the frog Rana pipiens, is the archetype of a novel class of cancer chemotherapeutic agents that are based on homologues and variants of bovine pancreatic ribonuclease (RNase A). Ranpirnase in combination with doxorubicin is in clinical trials for the treatment of unresectable malignant mesothelioma and other cancers. The putative mechanism for the ranpirnase-mediated cytotoxicity involves binding to anionic components of the extracellular membrane, cytosolic internalization, and degradation of tRNA leading to apoptosis. The maintenance of ribonucleolytic activity in the presence of the cytosolic ribonuclease inhibitor protein is a key aspect of its cytotoxic activity. The basis for its specific toxicity for cancer cells is not known. This review describes the development of ranpirnase as a cancer chemotherapeutic agent.
RNA is the intermediate in the flow of biochemical information from genes to proteins (Fig. 1). Accordingly, intervention in the metabolism of RNA presents an opportunity for the development of chemotherapeutic agents.[1] Since the 1980’s, antisense oligonucleotides and ribozymes have been pursued as the basis for treatments of viral infections, inflammatory disorders, haematological diseases, and cancer.[2-4] In 1998, the phosphorothioate antisense oligonucleotide fomivirsen (which is also known as Vitravene®) was approved by the U.S. Food and Drug Administration (FDA) for the treatment of cytomegalovirus retinitis in immunocompromised patients.[5] More recently, manipulation of the RNA interference machinery has garnered much interest as the basis for drug development,[6-8] though the safety of this approach is a concern.[9,10]
Figure 1.
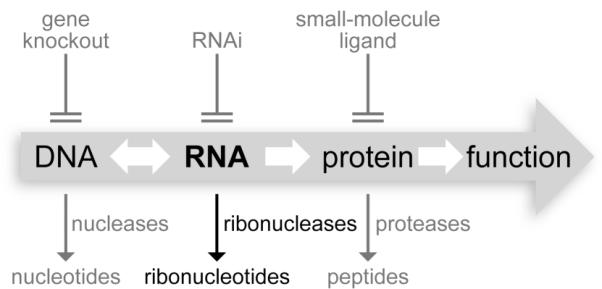
Flow of chemical information in biology. Ribonucleases can be cytotoxic because their degradation of RNA renders genetic information indecipherable.
Ribonucleases also have potential therapeutic utility.[11-15] These proteins are efficient catalysts of RNA cleavage, acting in effect as RNA depolymerases.[16] Much interest has focused on homologs and variants of bovine pancreatic ribonuclease (RNase A), which is renowned as a model system in protein biochemistry.[17] RNase A itself is not cytotoxic. In contrast, bovine seminal ribonuclease, which is a homodimer, is endowed with antitumoral, immunosuppressive, and antiviral activity.[18] Ranpirnase (which is also known as Onconase®, Pannon, or P-30 protein) is an amphibian homolog that has marked toxicity for tumor cells[19] and is the only ribonuclease in a human clinical trial.[20,21] Here, we review the structure and function of ranpirnase, which has become the archetype of a new class of cancer chemotherapeutic agent.[22]
1. History of Ranpirnase
In the early 1970’s, Shogen and Yoon discovered that extracts from embryos of the Northern leopard frog (Rana pipiens) have antitumoral activity.[23] Nearly two decades later, Ardelt, Mikulski, and Shogen attributed that activity to a basic protein, ranpirnase (meaning, Rana pipiens ribonuclease),[24] which belongs to the RNase A superfamily.[25] In oocytes, ranprinase localizes with yolk proteins.[26] It has been postulated that ranpirnase is synthesized in the liver of female frogs in a seasonal manner, and then secreted into the blood and deposited in oocytes as they mature.[27] There and in embryos, ranpirnase has been speculated to play a role in host defense.[26]
Ranpirnase is both cytotoxic and cytostatic toward cultured tumor cells and inhibits the growth of xenograft tumors in mice.[28,29] Currently, ranpirnase in combination with doxorubicin is in a confirmatory Phase IIIb clinical trial for the treatment of unresectable malignant mesothelioma, a cancer associated with exposure to asbestos.[20,21] Moreover, ranpirnase has been granted both orphan-drug and fast-track status by the FDA.
2. Biochemical Attributes of Ranpirnase
The amino-acid sequence of ranpirnase was determined in 1991,[24] and its three-dimensional structure was reported three years later.[30] Ranpirnase is a relatively small enzyme with a molecular formula of C520H810N142O155S9 and molecular mass of 11,820 Da. The active site of ranpirnase contains the catalytic triad (His10, Lys31, and His97) that is characteristic of the RNase A superfamily.[31] Ranpirnase possesses two additional active-site residues: Lys9 and an N-terminal pyroglutamate residue, which is formed by the co-translational cyclization of its encoded glutamine in the endoplasmic reticulum.[32] Like other members of the RNase A superfamily, ranpirnase catalyzes the cleavage of P–O5′ bond on the 3′ side of a pyrimidine nucleobase in an RNA strand.
The ribonucleolytic activity of ranpirnase is necessary for its cytotoxicity. A decrease in ribonucleolytic activity leads to a corresponding reduction in cytotoxicity.[24] Although ranpirnase assumes the kidney-shaped tertiary structure that is typical of the RNase A superfamily (Fig. 2)[33,30] and has the key catalytic residues, its value of kcat/KM (which is the second-order rate constant and thus a measure of catalytic efficiency) is 104-fold less than that of RNase A for cleavage of their best known substrates under similar conditions.[34] A low affinity for its substrate contributes to its low kcat/KM value. In addition, NMR spectroscopy and molecular dynamics simulations have revealed that ranpirnase has an extremely rigid β-sheet,[35,36] which could deter an “induced fit”[37] necessary for substrate binding and turnover.
Figure 2.
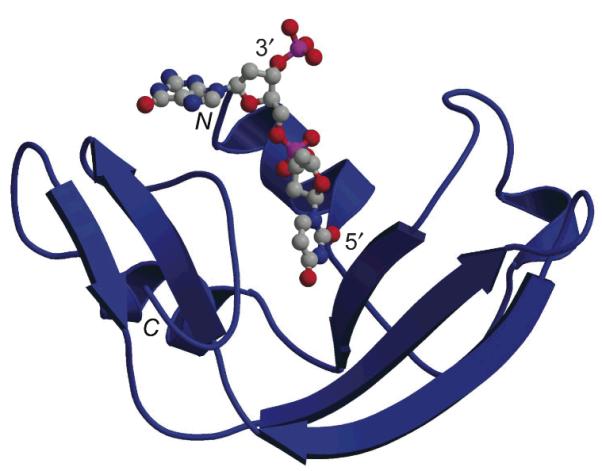
Ribbon diagram of the three-dimensional structure of a crystalline ranpirnase·nucleic acid (dAdUdGdA) complex (PDB entry 2I5S).[40] The N- and C-termini of the protein, and 3′- and 5′-termini of the nucleic acid are noted explicitly. The image was created with the programs MOLSCRIPT (Avatar Software AB, Stockholm, Sweden) and RASTER3D.[33]
The substrate specificity of ranpirnase can be considered on two levels. On the nucleobase level, ranpirnase prefers to cleave the phosphodiester bond on the 5′ side of a guanine nucleobase. This preference is in marked contrast to that of RNase A, which has little preference for guanine versus adenine at this position. In the cell, tRNA has been reported to be the main target for ranpirnase.[38] The cleavage of tRNA occurs at the guanosine–guanosine bond in the variable loop or D-arm.[39] The revelation of the atomic structure of a ranpirnase·nucleic acid complex has provided insight into the structural basis for this substrate specificity.[40]
A notable feature of ranpirnase is its extraordinary conformational stability. Ranpirnase has a Tm value of 87 °C (which is the temperature at the midpoint of the thermal transition between folded and unfolded states and thus a measure of conformational stability) and resists degradation by various proteases.[41] The exceptional conformational stability of ranpirnase is largely due to its tethered C-terminus, created by a C-terminal half-cystine residue.[42] This synapomorphic C-terminal disulfide bond is conserved in amphibian ribonucleases but absent from mammalian homologs.[31] The hydrogen-bond network at the N-terminus[41] and the absence of a cis-proline residue[43] also contribute to the conformational stability of the enzyme. This exceptional stability is critical for cytotoxicity. Variants of ranpirnase with reduced conformational stability have less cytotoxic activity.[44,41] On the other hand, glycosylation of ranpirnase at its consensus N-linked glycoyslation site (Asn69–Val70–Thr71) increases both conformational stability and cytotoxic activity.[45]
3. Mechanism of Ranpirnase-mediated Cytotoxicity
Ranpirnase is an atypical biodrug in that it is administered extracellularly but acts intracellularly. To exert its antitumoral effect, ranpirnase must reach the cytosol and there cleave RNA substrates. The generally accepted mechanism of ranpirnase-mediated cytotoxicity is divided into two major stages: cytosolic internalization and catalytic degradation of RNA, as depicted in Fig. 3.[46]
Figure 3.
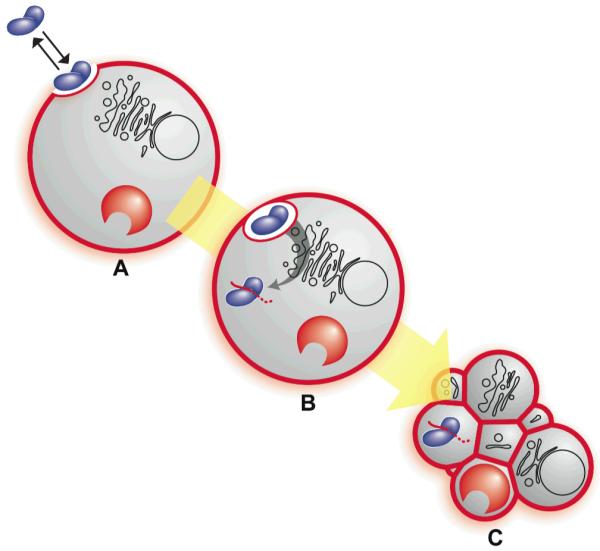
Putative mechanism of ranpirnase-mediated cytotoxicity.[46] Cationic and anionic biomolecules are depicted in blue and red, respectively. (A) Ranpirnase (blue) forms an extracellular equilibrium complex with cell-surface heparan sulfate (red). (B) Ranpirnase is internalized by endocytosis, translocates to the cytosol, evades the ribonuclease inhibitor protein (red horseshoe), and degrades tRNA (red line). (C) tRNA degradation leads to apoptosis.
3.1 Cytosolic Internalization
The first step for the cytosolic internalization of ranpirnase is its binding to the cell surface. The existence of low-affinity and high-affinity ranpirnase receptors on the cell surface has been reported,[47] but other findings contradict their existence.[48] The cell surface is highly anionic due to the abundant sulfate, phosphate, and carboxylate groups of its carbohydrates and lipids. It is probable that ranpirnase, which is a highly cationic protein with calculated pI > 9.5,[24] binds to the cell surface through favorable Coulombic interactions.
After binding to the cell surface, ranpirnase is internalized by energy-dependent endocytosis. The role of dynamin in this process is under investigation.[48,49] Internalized ranpirnase is routed to endosomes. Drugs that disrupt retrograde transport from the trans-Golgi network to the endoplasmic reticulum potentiate the cytotoxicity of ranpirnase.[50,48,49] These and other results imply that the trans-Golgi network is an inefficient site for the translocation of ranpirnase, and that endosomes are a key compartment for cytotosic delivery.
The means by which ranpirnase, which is extremely hydrophilic, ultimately crosses a lipid bilayer is not understood. To facilitate successful entry into the cell, the proteins diphtheria toxin and ricin utilize a distinct translocation domain, which dissociates from a catalytic domain upon cytosolic entry. In contrast, ranpirnase is a hyperstable, single-domain protein, which remains intact during its endocytosis. The mechanism of ranpirnase translocation could be related to that used by cationic peptides, such as residues 47–57 of the HIV-1 TAT protein and nonaarginine.[51]
3.2 Degradation of Cellular RNA and Induction of Apoptosis
Once in the cytosol, ranpirnase degrades cellular RNA. Ranpirnase is an unusual homologue of RNase in that it seems to evade completely the cytosolic ribonuclease inhibitor protein (RI).[52,53] RI is a 50-kD protein present in every surveyed mammalian cell. RI is composed of 15 leucine-rich repeats (LRRs), a motif that often participates in protein–protein interactions.[54] RI binds to certain members of the RNase A superfamily with femtomolar affinity, and renders them inactive. The complex formed by human RI and human pancreatic ribonuclease is among the tightest known in biology (Kd = 2.9 × 10−16 M).[55] The ability of ranpirnase to evade RI is likely to be necessary for its cytotoxic activity, as non-cytotoxic mammalian ribonucleases become cytotoxic by incorporating residues that enable RI evasion.[56,55] Moreover, the cytotoxicity of variants correlates with their RI-evading ability.[57]
In the cell, the ribonucleolytic activity of ranpirnase is directed predominantly on tRNA, leaving rRNA and mRNA largely intact.[38] The basis for this specificity is not understood, though bound proteins could protect rRNA and mRNA from ranpirnase cleavage. The susceptibility of non-coding RNA, such as microRNA or small-interfering RNA (siRNA), to ranpirnase cleavage is unknown.
Degradation of tRNA by ranpirnase inhibits protein synthesis in the cell, and leads to apoptosis.[58,59] The cytotoxic effect of ranpirnase becomes noticeable after a longer incubation (~48 h, in vitro) than drugs that block translation, such as cycloheximide (~2 h). In addition, ranpirnase-induced apoptosis does not require the high level of translation inhibition observed with cycloheximide, suggesting that the inhibition of protein synthesis is not the sole cause of ranpirnase-induced apoptosis.[60] In HeLa cells, ranpirnase-induced cytotoxicity is initiated with the activation of the stress-activated c-Jun N-terminal kinase (JNK), followed by the activation of caspase-9, which activates the executioner caspase-3 and -7. Caspase-8 or the tumor-suppressor protein p53 are not required in this pathway.[61] Other studies with the HL-60 leukemic cell line indict the activation of serine proteases along with these caspases.[62] The induced apoptosis is enhanced by mild hyperthermia.[63]
3.3 Basis for Therapeutic Index
Ranpirnase is more toxic to tumor cells than to normal cells in vitro and in vivo. The mechanism for this selectivity is unknown. A promising hypothesis is that ranpirnase is selectively internalized by tumor cells. In general, tumor cells are more negatively charged than are homologous normal cells.[64,65] Moreover, the level of sialic acid-rich gangliosides is greater and the phosphoplipid content is altered in certain tumor cells.[66,67] The elevated anionic character of tumor cells could promote their interaction with the highly cationic ranpirnase. Other viable hypotheses include a different and more efficient intracellular routing of ranpirnase to the cytosol in tumor cells, and a greater susceptibility of rapidly growing tumor cells to RNA degradation.
4. Therapeutic Applications
In vitro, ranpirnase has been shown to be cytotoxic/cytostatic to 9L rat glioma,[47] K-562 human leukemia,[44,34] Colo 320 CM human colon adenocarcinoma,[68] HL-60 human leukemia,[69] LNCaP and JCA-1 human prostate cancer,[69] HT-29 human colorectal cancer,[70] and U937 human lymphoma cell lines.[71] Typical IC50 values for the proliferation of 9L rat glioma[68,47] and K-562 human leukemia cells[34] are near 10−7 M. Concomitant administration of ranpirnase with tamoxifen,[72,73] cisplatin,[73] or vincristine[70] results in increased toxicity. In combination with vincristine, ranpirnase showed toxicity against multi-drug resistant tumors.[70] In vivo, ranpirnase treatment prolongs the survival of mice transplanted with human[70,74] and murine tumors.[75,76]
Ranpirnase has been adminstered as a single agent in two Phase I clinical studies to determine the optimum dose and schedule in patients with various solid tumors.[77,20] These Phase I studies indicated that ranpirnase is well tolerated. The maximum tolerated dose was 960 μg/m2, and the recommended Phase II dose was 480 μg/m2/week. Ranpirnase has been evaluated in Phase II clinical trials as a single agent in patients with non-small cell lung cancer,[78] breast cancer,[79] renal cell cancer,[80] and malignant mesothelioma.[81] The largest Phase II trial for ranpirnase was in patients with malignant mesothelioma. Among 81 patients who were evaluated for tumor response, 41 patients showed a decrease in tumor progression, which justified subsequent Phase III studies on this tumor. In the initial Phase III studies, 154 patients were treated with either ranpirnase (84 patients) or doxorubicin (70 patients). In these studies, ranpirnase treatment provided markedly increased survival compared to doxorubicin treatment.[20,82] Reversible renal toxicity was the major side effect. The current confirmatory Phase IIIb study is open-label, multi-center, and international, with the goal of comparing the efficacy of ranpirnase and doxorubicin together with doxorubicin alone.[20,82]
5. Engineering Ranpirnase and Future Directions
There have been attempts to endow ranpirnase with increased toxicity towards tumor cells. Nearly all non-Hodgkin’s lymphoma cells display a specific cell-surface receptor, CD-22. A human monoclonal antibody against CD-22 has been covalently linked to ranpirnase.[83] This fusion protein was 104-fold more toxic to non-Hodgkin’s lymphoma cells than was wild-type ranpirnase due to increased binding to the tumor cells. In addition, this protein showed enhanced potency and specificity along with decreased systemic toxicity in mice.
The ribonucleolytic activity of ranpirnase is 104-fold lower than that of other mammalian homologues.[34] Accordingly, it could be both possible and advantageous to engender ranpirnase with greater ribonucleolytic activity without compromising other attributes required for its cytotoxicity, such as cationicity, RI evasion, and conformational stability. Either enhancing substrate binding or alleviating β-sheet rigidity could yield variants with increased ribonucleolytic activity and, hence, chemotherapeutic efficacy.
6. Conclusions
Ranpirnase is a cytotoxic ribonuclease that affords a novel strategy for cancer chemotherapy. Ranpirnase is internalized by tumor cells and degrades tRNA, which leads to the inhibition of protein synthesis and apoptosis. The cationicity and maintenance of ribonucleolytic activity in the presence of RI are critical for its cytotoxicity. The efficacy of ranpirnase can be augmented with other cytotoxic agents such as doxorubicin, and a confirmatory Phase IIIb clinical trial for the treatment of malignant mesothelioma is ongoing. Emerging knowledge on the mechanism of action of ranpirnase could aid in the development of other ribonucleases, including those from mammals, as cancer chemotherapeutic agents.
Acknowledgments
Work in the Raines laboratory on ribonucleases is supported by grant CA73808 (NIH). We are grateful to R. F. Turcotte and T.-Y. Chao for comments on the manuscript.
References
- 1.Tafech A, Bassett T, Sparanese D, Lee CH. Destroying RNA as a therapeutic approach. Curr Med Chem. 2006;13:863–81. doi: 10.2174/092986706776361021. [DOI] [PubMed] [Google Scholar]
- 2.Sloud M. Ribozymes and siRNAs: From structure to preclinical applications. Handb Exp Pharmacol. 2006;173:223–42. doi: 10.1007/3-540-27262-3_11. [DOI] [PubMed] [Google Scholar]
- 3.Schubert S, Kurreck J. Oligonucleotide-based antiviral strategies. Handb Exp Pharmacol. 2006;173:261–87. doi: 10.1007/3-540-27262-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayburn ER, Wang H, Zhang R. Antisense-based cancer therapeutics: Are we there yet? Expert Opin Emerg Drugs. 2006;11:337–52. doi: 10.1517/14728214.11.2.337. [DOI] [PubMed] [Google Scholar]
- 5.Roehr B. Forivirsen approved for CMV retinitis. J Int Assoc Physicians AIDS Care. 1998;4:14–6. [PubMed] [Google Scholar]
- 6.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: A potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aagaard L, Rossi JJ. RNAi therapeutics: Principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–84. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 9.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 10.Marsden PA. RNA interference as potential therapy—not so fast. New Engl J Med. 2006;355:953–4. doi: 10.1056/NEJMcibr063864. [DOI] [PubMed] [Google Scholar]
- 11.Matoušek J. Ribonucleases and their antitumor activity. Comp Biochem Physiol. 2001;129C:175–91. doi: 10.1016/s1532-0456(01)90202-9. [DOI] [PubMed] [Google Scholar]
- 12.Leland PA, Raines RT. Cancer chemotherapy—ribonucleases to the rescue. Chem Biol. 2001;8:405–13. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarov AA, Ilinskaya ON. Cytotoxic ribonucleases: Molecular weapons and their targets. FEBS Lett. 2003;540:15–20. doi: 10.1016/s0014-5793(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 14.Benito A, Ribó M, Vilanova M. On the track of antitumor ribonucleases. Mol Biosyst. 2005;1:294–302. doi: 10.1039/b502847g. [DOI] [PubMed] [Google Scholar]
- 15.Arnold U, Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol Lett. 2006;28:1615–22. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 16.D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1997. [Google Scholar]
- 17.Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–65. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 18.D’Alessio G, Di Donato A, Mazzarella L, Piccoli R. Seminal ribonuclease: The importance of diversity. In: D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1997. pp. 383–423. [Google Scholar]
- 19.Matoušek J, Souček J, Slavík T, Tománek M, Lee JE, Raines RT. Comprehensive comparison of the cytotoxic activities of onconase and bovine seminal ribonuclease. Comp Biochem Physiol. 2003;136C:343–56. doi: 10.1016/j.cca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro-apoptotic anticancer strategy: Review of the preclinical and clinical data for ranpirnase. Cancer Invest. 2005;23:643–50. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 21.Pavlakis N, Vogelzang NJ. Ranpirnase—an antitumour ribonuclease: Its potential role in malignant mesothelioma. Expert Opin Biol Ther. 2006;6:391–9. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 22.Saxena SK, Shogen K, Ardelt W. ONCONASE® and its therapeutic potential. Lab Med. 2003;34:380–7. [Google Scholar]
- 23.Shogen K, Yoan WK. Antitumor activity in extracts of Leopard frog (Rana pipiens) embryos; 27th Annual Eastern Colleges Science Conference; The Pennsylvania State University, State College, PA. April 28, 1973. [Google Scholar]
- 24.Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. J Biol Chem. 1991;266:245–51. [PubMed] [Google Scholar]
- 25.Dyer KD, Rosenberg HF. The RNase A superfamily: Generation of diversity and innate host defense. Mol Divers. 2006;10:585–97. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 26.Liao YD, Wang JJ. Yolk granules are the major compartment for bullfrog (Rana catesbeiana) oocyte-specific ribonuclease. Eur J Biochem. 1994;222:215–20. doi: 10.1111/j.1432-1033.1994.tb18859.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Le SY, Newton DL, Maizel JV, Jr., Rybak SM. A gender-specific mRNA encoding a cytotoxic ribonuclease contains a 3′ UTR of unusual length and structure. Nucleic Acids Res. 2000;28:2375–82. doi: 10.1093/nar/28.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt WJ, Shogen K. Cytostatic and cytotoxic effect of Pannon (P-30 Protein), a novel anticancer agent. Cell Tissue Kinet. 1988;21:169–82. doi: 10.1111/j.1365-2184.1988.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee I, Kalota A, Gewirtz AM, Shogen K. Antitumor efficacy of the cytotoxic RNase, ranpirnase, on A549 human lung cancer xenografts of nude mice. Anticancer Res. 2007;27:299–307. [PubMed] [Google Scholar]
- 30.Mosimann SC, Ardelt W, James MNG. Refined 1.7 Å X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with anti-tumor activity. J Mol Biol. 1994;236:1141–53. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 31.Raines RT. Active site of ribonuclease A. In: Zenkova MA, editor. Artificial Nucleases. Springer–Verlag; Heidelberg, Germany: 2004. pp. 19–32. [Google Scholar]
- 32.Welker E, Hathaway L, Xu G, Narayan M, Pradeep L, Shin HC, Scheraga HA. Oxidative folding and N-terminal cyclization of onconase. Biochemistry. 2007;46:5485–93. doi: 10.1021/bi602495a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merritt EA, Murphy MEP. Raster3D Version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr D Biol Crysallogr. 1994;50:869–73. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 34.Lee JE, Raines RT. Contribution of active-site residues to the function of onconase, a ribonuclease with antitumoral activity. Biochemistry. 2003;42:11443–50. doi: 10.1021/bi035147s. [DOI] [PubMed] [Google Scholar]
- 35.Gorbatyuk VY, Tsai CK, Chang CF, Huang TH. Effect of N-terminal and Met23 mutations on the structure and dynamics of onconase. J Biol Chem. 2004;279:5772–80. doi: 10.1074/jbc.M311233200. [DOI] [PubMed] [Google Scholar]
- 36.Merlino A, Mazzarella L, Carannante A, Di Fiore A, Di Donato A, Notomista E, Sica F. The importance of dynamic effects on the enzyme activity: X-ray structure and molecular dynamics of onconase mutants. J Biol Chem. 2005;280:17953–60. doi: 10.1074/jbc.M501339200. [DOI] [PubMed] [Google Scholar]
- 37.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxena SK, Sirdeshmukh R, Ardelt W, Mikulski SM, Shogen K, Youle RJ. Entry into cells and selective degradation of tRNAs by a cytotoxic member of the RNase A family. J Biol Chem. 2002;277:15142–6. doi: 10.1074/jbc.M108115200. [DOI] [PubMed] [Google Scholar]
- 39.Suhasini AN, Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J Biol Chem. 2006;281:12201–9. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- 40.Lee JE, Bae E, Bingman CA, Phillips GN, Jr., Raines RT. Structural basis for catalysis by onconase. J Mol Biol. 2007;372:xxx–xxx. doi: 10.1016/j.jmb.2007.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notomista E, Catanzano F, Graziano G, Dal Piaz F, Barone G, D’Alessio G, Di Donato A. Onconase: An unusually stable protein. Biochemistry. 2000;39:8711–8. doi: 10.1021/bi000415x. [DOI] [PubMed] [Google Scholar]
- 42.Leland PA, Staniszewski KE, Kim B-M, Raines RT. A synapomorphic disulfide bond is critical for the conformational stability and cytotoxicity of an amphibian ribonuclease. FEBS Lett. 2000;477:203–7. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- 43.Arnold U, Schulenburg C, Schmidt D, Ulbrich-Hofmann R. Contribution of structural peculiarities of onconase to its high stability and folding kinetics. Biochemistry. 2006;45:3580–7. doi: 10.1021/bi0525223. [DOI] [PubMed] [Google Scholar]
- 44.Leland PA, Staniszewski KE, Kim B, Raines RT. A synapomorphic disulfide bond is critical for the conformational stability and cytotoxicity of an amphibian ribonuclease. FEBS Lett. 2000;477:203–7. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- 45.Kim B-M, Kim H, Raines RT, Lee Y. Glycosylation of onconase increases it conformational stability and toxicity for cancer cells. Biochem Biophys Res Commun. 2004;315:976–83. doi: 10.1016/j.bbrc.2004.01.153. [DOI] [PubMed] [Google Scholar]
- 46.Johnson RJ, Chao T-Y, Lavis LD, Raines RT. Cytotoxic ribonucleases: The dichotomy of Coulombic forces. Biochemistry. 2007;46:xxx–xxx. doi: 10.1021/bi700857u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Mikulski SM, Ardelt W, Rybak SM, Youle RJ. A cytotoxic ribonuclease. Study of the mechanism of onconase cytotoxicity. J Biol Chem. 1993;268:10686–93. [PubMed] [Google Scholar]
- 48.Haigis MC, Raines RT. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J Cell Sci. 2003;116:313–24. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez M, Torrent G, Bosch M, Rayne F, Dubremetz J-F, Ribó M, Benito A, Vilanova M, Beaumelle B. Intracellular pathway of Onconase that enables its delivery to the cytosol. J Cell Sci. 2007;120:1405–11. doi: 10.1242/jcs.03427. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Saxena SK, Ardelt W, Gadina M, Mikulski SM, De Lorenzo C, D’Alessio G, Youle RJ. A study of the intracellular routing of cytotoxic ribonucleases. J Biol Chem. 1995;270:17476–81741. doi: 10.1074/jbc.270.29.17476. [DOI] [PubMed] [Google Scholar]
- 51.Fuchs SM, Raines RT. Internalization of cationic peptides: The road less (or more?) traveled. Cell Mol Life Sci. 2006;63:1819–22. doi: 10.1007/s00018-006-6170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–32. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: Structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–74. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–27. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 55.Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Jr., Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007;367:434–49. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leland PA, Staniszewski KE, Kim B-M, Raines RT. Endowing human pancreatic ribonuclease with toxicity for cancer cells. J Biol Chem. 2001;276:43095–102. doi: 10.1074/jbc.M106636200. [DOI] [PubMed] [Google Scholar]
- 57.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J Mol Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 58.Deptala A, Halicka HD, Ardelt B, Ardelt W, Mikulski SM, Shogen K, Darzynkiewicz Z. Potentiation of tumor necrosis factor induced apoptosis by onconase. Int J Oncol. 1998;13:11–6. doi: 10.3892/ijo.13.1.11. [DOI] [PubMed] [Google Scholar]
- 59.Iordanov MS, Ryabinina OP, Wong J, Dinh TH, Newton DL, Rybak SM, Magun BE. Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis. Cancer Res. 2000;60:1983–94. [PubMed] [Google Scholar]
- 60.Ardelt B, Ardelt W, Darzynkiewicz Z. Cytotoxic ribonucleases and RNA interference (RNAi) Cell Cycle. 2003;2:22–4. doi: 10.4161/cc.2.1.232. [DOI] [PubMed] [Google Scholar]
- 61.Iordanov MS, Wong J, Newton DL, Rybak SM, Bright RK, Flavell RA, Davis RJ, Magun BE. Differential requirement for the stress-activated protein kinase/c-Jun NH2-terminal kinase in RNA damage-induced apoptosis in primary and in immortalized fibroblasts. Mol Cell Biol Res Commun. 2000;4:122–8. doi: 10.1006/mcbr.2000.0266. [DOI] [PubMed] [Google Scholar]
- 62.Grabarek J, Ardelt B, Du L, Darzynkiewicz Z. Activation of caspases and serine proteases during apoptosis induced by onconase (Ranpirnase) Exp Cell Res. 2002;278:61–71. doi: 10.1006/excr.2002.5568. [DOI] [PubMed] [Google Scholar]
- 63.Halicka HD, Ardelt B, Shogen K, Darzynkiewicz Z. Mild hyperthermia predisposes tumor cells to undergo apoptosis upon treatment with onconase. Int J Oncol. 2007;30:841–7. [PubMed] [Google Scholar]
- 64.James AM, Ambrose EJ, Lowick JH. Differences between the electrical charge carried by normal and homologous tumour cells. Nature. 1956;177:576–7. doi: 10.1038/177576a0. [DOI] [PubMed] [Google Scholar]
- 65.Slivinsky GG, Hymer WC, Bauer J, Morrison DR. Cellular electrophoretic mobility data: A first approach to a database. Electrophoresis. 1997;18:1109–19. doi: 10.1002/elps.1150180715. [DOI] [PubMed] [Google Scholar]
- 66.Kojima K. Molecular aspects of the plasma membrane in tumor cells. Nagoya J Med Sci. 1993;56:1–18. [PubMed] [Google Scholar]
- 67.Fredman P. Glycosphingolipid tumor antigens. Adv Lipid Res. 1993;25:213–34. [PubMed] [Google Scholar]
- 68.Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt W, Shogen K. Cytostatic and cytotoxic effect of Pannon (P-30 Protein), a novel anticancer agent. Cell Tissue Kinet. 1988;21:169–82. doi: 10.1111/j.1365-2184.1988.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 69.Halicka HD, Murakami T, Papageorgio CN, Mittelman A, Mikulski SM, Shogen K, Darzynkiewicz Z. Induction of differentiation of leukaemic (HL-60) or prostate cancer (LNCaP, JCA-1) cells potentiates apoptosis triggered by onconase. Cell Prolif. 2000;33:407–17. doi: 10.1046/j.1365-2184.2000.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rybak SM, Pearson JW, Fogler WE, Volker K, Spence SE, Newton DL, Mikulski SM, Ardelt W, Riggs CW, Kung HF, Longo DL. Enhancement of vincristine cytotoxicity in drug-resistant cells by simultaneous treatment with onconase, an antitumor ribonuclease. J Natl Cancer Inst. 1996;88:747–53. doi: 10.1093/jnci/88.11.747. [DOI] [PubMed] [Google Scholar]
- 71.Juan G, Ardelt B, Li X, Mikulski SM, Shogen K, Ardelt W, Mittelman A, Darzynkiewicz Z. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation. Leukemia. 1998;12:1241–8. doi: 10.1038/sj.leu.2401100. [DOI] [PubMed] [Google Scholar]
- 72.Mikulski SM, Viera A, Ardelt W, Menduke H, Shogen K. Tamoxifen and trifluoroperazine (Stelazine) potentiate cytostatic/cytotoxic effects of P-30 protein, a novel protein possessing anti-tumor activity. Cell Tissue Kinet. 1990;23:237–46. doi: 10.1111/j.1365-2184.1990.tb01119.x. [DOI] [PubMed] [Google Scholar]
- 73.Mikulski SM, Viera A, Darzynkiewicz Z, Shogen K. Synergism between a novel amphibian oocyte ribonuclease and lovastatin in inducing cytostatic and cytotoxic effects in human lung and pancreatic carcinoma cell lines. Br J Cancer. 1992;66:304–10. doi: 10.1038/bjc.1992.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee I, Lee YH, Mikulski SM, Shogen K. Effect of ONCONASE +/− tamoxifen on ASPC-1 human pancreatic tumors in nude mice. Adv Exp Med Biol. 2003;530:187–96. doi: 10.1007/978-1-4615-0075-9_18. [DOI] [PubMed] [Google Scholar]
- 75.Mikulski SM, Ardelt W, Shogen K, Bernstein EH, Menduke H. Striking increase of survival of mice bearing M109 Madison carcinoma treated with a novel protein from amphibian embryos. J Natl Cancer Inst. 1990;82:151–3. doi: 10.1093/jnci/82.2.151-a. [DOI] [PubMed] [Google Scholar]
- 76.Lee I, Lee YH, Mikulski SM, Lee J, Shogen K. Enhanced cellular radiation sensitivity of androgen-independent human prostate tumor cells by onconase. Anticancer Res. 2000;20:1037–40. [PubMed] [Google Scholar]
- 77.Mikulski SM, Grossman A, Carter P, Shogen K, Costanzi J. Phase I human clinical trial of Onconase (P-30 protein) administered intravenously on a weekly schedule in cancer patients with solid tumors. Int J Oncol. 1993;3:57–64. doi: 10.3892/ijo.3.1.57. [DOI] [PubMed] [Google Scholar]
- 78.Mikulski SM, Chun H, Mittelman A, Panella T, Puccio C, Shogen K, Costanzi J. Relationship between response rate and median survival in patients with advanced non-small cell lung cancer: Comparison of ONCONASE with other cancer agents. Int J Oncol. 1995;6:889–97. doi: 10.3892/ijo.6.4.889. [DOI] [PubMed] [Google Scholar]
- 79.Puccio C, Mittelman A, Chun H, Costanzi J, Panella T, Coombe N, Shogen K, Mikulski S. A new anticancer Rnase (Onconase): Clinical trial in patients (pts) with breast cancer (BC); American Society of Clinical Oncology Annual Meeting; Philadelphia, PA. May 1996; Abstract 242. [Google Scholar]
- 80.Vogelzang NJ, Aklilu M, Stadler WM, Dumas MC, Mikulski SM. A phase II trial of weekly intravenous ranpirnase (Onconase), a novel ribonuclease in patients with metastatic kidney cancer. Invest New Drugs. 2001;19:255–60. doi: 10.1023/a:1010633004157. [DOI] [PubMed] [Google Scholar]
- 81.Mikulski SM, Costanzi JJ, Vogelzang NJ, McCachren S, Taub RN, Chun H, Mittelman A, Panella T, Puccio C, Fine R, Shogen K. Phase II trial of a single weekly intravenous dose of ranpirnase in patients with unresectable malignant mesothelioma. J Clin Oncol. 2002;20:274–81. doi: 10.1200/JCO.2002.20.1.274. [DOI] [PubMed] [Google Scholar]
- 82.Pavlakis N, Vogelzang NJ. Ranpirnase—an antitumour ribonuclease: Its potential role in malignant mesothelioma. Expert Opin Biol Ther. 2006;6:391–9. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 83.Newton DL, Hansen HJ, Mikulski SM, Goldenberg DM, Rybak SM. Potent and specific antitumor effects of an anti-CD22-targeted cytotoxic ribonuclease: Potential for the treatment of non-Hodgkin lymphoma. Blood. 2001;97:528–35. doi: 10.1182/blood.v97.2.528. [DOI] [PubMed] [Google Scholar]


