Abstract
Recently, a novel class of global gene regulators called microRNAs (miRNAs), were identified in both plants and animals. MiRNAs can reduce protein levels of their target genes with a minor impact on the target genes' mRNAs. Accumulating evidence demonstrates the importance of miRNAs in cancer. MiRNAs that are overexpressed in cancer may function as oncogenes, and miRNAs with tumour suppressor activity in normal tissue may be downregulated in cancer. Although major advances have been achieved in our understanding of cancer biology, as well as in the development of new targeted therapies, the progress in developing improved early diagnosis and screening tests has been inadequate. This results in most cancers being diagnosed in advanced stages, delaying timely treatment and leading to poor outcomes. There is intense research seeking specific molecular changes that are able to identify patients with early cancer or precursor lesions. MiRNA expression data in various cancers demonstrate that cancer cells have different miRNA profiles compared with normal cells, thus underscoring the tremendous diagnostic and therapeutic potential of miRNAs in cancer. These unique properties of miRNAs make them extremely useful potential agents for clinical diagnostics as well as in personalised care for individual patients in the future.
INTRODUCTION
MicroRNAs (miRNAs), as the name suggests, are small single-stranded RNA molecules, about 22 nucleotides (nt) long, that regulate gene expression. The first microRNA was identified in 1993 in the roundworm Caenorhabditis elegans by Lee et al.1 Ever since then it has become evident that miRNAs are naturally abundant and evolutionarily conserved non-coding RNA molecules found in both plants and animals.2 They function as a novel class of global gene regulators3 by binding to partially complementary sequences in 3′ untranslated regions (UTRs) of downstream target mRNAs. Various cloning and bioinformatics studies predict that the human genome may contain up to 1000 miRNAs, and to date 706 human miRNAs are listed in miRBase (release 13).4 5 MiRNA genes are spread throughout the genome and it is estimated that they account for 2–5% of human genes.6 MiRNAs exhibit unique temporal and spatial expression patterns that are specific for developmental stage and tissue. It is believed that similarly to mRNA expression, miRNA expression is determined by both intrinsic cellular factors and diverse environmental variables.7
MiRNA biogenesis
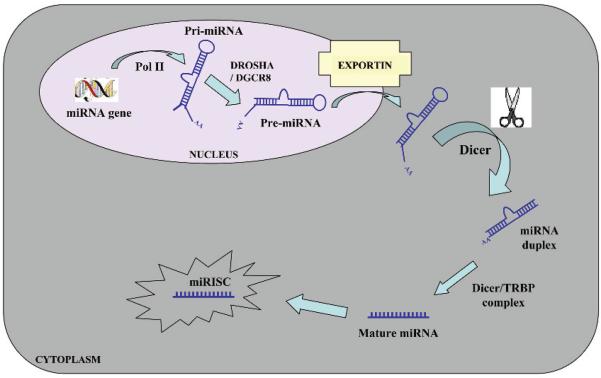
MiRNA production begins in the cell's nucleus and involves a series of RNA processing steps. Intergenic miRNA genes are commonly clustered and, along with those located in the introns of protein-coding genes, are transcribed by RNA polymerase II.8 These transcripts, known as primiRNAs, are capped, polyadenylated and are usually several thousand bases in length.9 PrimiRNAs are then cleaved by an RNase III enzyme Drosha10 in association with its cofactor Pasha (in flies) or DGCR8 (in humans) to generate ~70–90 nt long precursor miRNA (pre-miRNA) which folds into an imperfect stem–loop hairpin structure. These pre-miRNAs are transported to the cytoplasm by exportin 5, where they are further processed by Dicer to form a transient 22 nt mature double stranded (ds) miRNA (miRNA duplex). One strand of this duplex is preferentially incorporated into a miRNA-associated RNA-induced silencing complex (miRISC).11 The mature miRNA guides RISC to target mRNAs containing a sequence partially complementary (miRNA target site) to the miRNA (fig 1).
Figure 1.
Biogenesis of microRNAs (miRNAs). MiRNA genes are generally transcribed by RNA polymerase II (Pol II) within the nucleus to form large capped and polyadenylated pri-miRNA transcripts. These pri-miRNA transcripts are processed by the RNase III enzyme Drosha and its cofactor, DGCR8, to a pre-miRNA precursor product. The pre-miRNA is then transported to the cytoplasm by exportin 5. Subsequently, another RNase III enzyme, Dicer, processes the pre-miRNA to generate a transient ~22 nucleotide miRNA:miRNA* duplex. This duplex is then loaded into the miRNA-associated RNA-induced silencing complex (miRISC), which includes the Argonaute proteins, and the mature single-stranded miRNA is preferentially retained in this complex.
▶ MicroRNAs are 22 nt untranslated RNA molecules
▶ Are evolutionarily conserved
▶ Are found in diverse species
▶ Are estimated to regulate 10–30% of all protein-coding genes
▶ Are involved in biological processes such as cell proliferation, differentiation, apoptosis, metabolism, development, ageing and cancer
MiRNA function
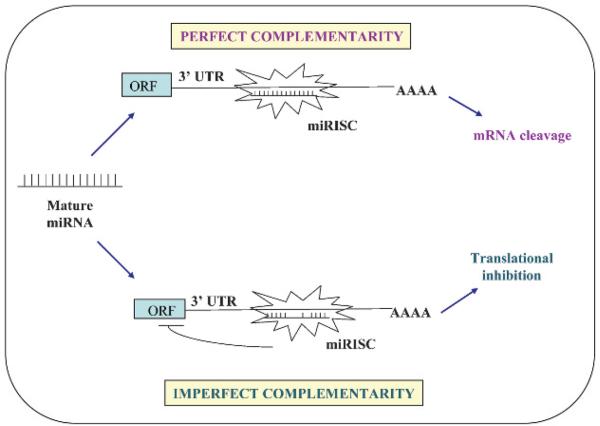
It is estimated that miRNAs regulate 10–30% of all protein-coding genes. They do this in two ways; first, miRNAs that bind to protein-coding mRNA sequences that are exactly complementary to the miRNA induce the RNA-mediated interference (RNAi) pathway, leading to cleavage of mRNA by Argonaute in the RISC.12-14 This mechanism is commonly observed in plants, although some studies do report it in animals. In the second and more common mechanism, miRNAs exert their effect by binding to imperfect complementary sites within the 3′UTRs of their target protein-coding mRNAs, leading to repression of expression of these genes at the level of translation1 15-20 (fig 2). Consistent with translational control, miRNAs can reduce protein levels of their target genes with low impact on the genes' mRNA levels. In humans, miRNAs mainly inhibit protein translation of their target genes and infrequently cause degradation or cleavage of the mRNA.2
Figure 2.
Mechanism of action of microRNAs (miRNAs). The mature miRNA negatively regulates gene expression by binding to complementary sites in the target mRNA in one of two ways depending on the degree of complementarity between the miRNA and its target. MiRNAs that bind to mRNA targets with imperfect complementarity at the sites located within the 3′ untranslated region (UTR) of the mRNA gene block target gene expression at the level of protein translation. MiRNAs that bind to their mRNA targets with perfect (or nearly perfect) complementarity at the sites generally found in the coding sequence or open reading frame (ORF) of the mRNA target induce target mRNA cleavage. miRISC, miRNA-associated RNA-induced silencing complex.
Biological roles of miRNAs
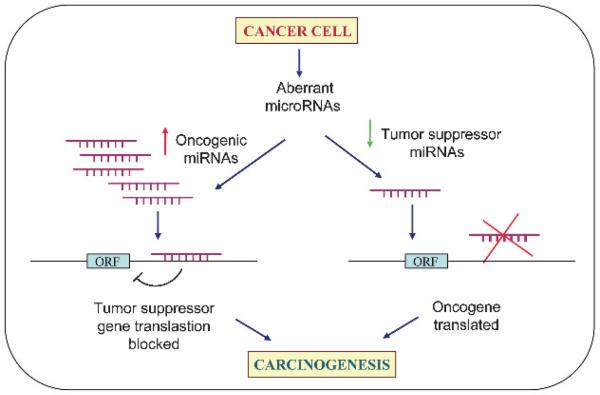
A large number of studies have demonstrated that miRNAs are key regulators of a variety of fundamental biological processes such as development, cell proliferation, apoptosis, fat metabolism, haematopoiesis, stress resistance, neural development, death and, importantly, tumourigenesis.21 Accumulating evidence demonstrates the importance of miRNAs in cancer. In contrast to the tight regulation during development and in normal tissues it is now well established that miRNAs are misregulated in cancer. MiRNAs that are overexpressed in cancer may function as oncogenes, and miRNAs with tumour suppressor activity in normal tissue may be down-regulated in cancer (fig 3).22
Figure 3.
MicroRNAs (miRNAs) as tumour suppressors and oncogenes. Downregulation or loss of miRNAs with tumour suppressor function may increase translation of oncogenes and hence formation of excess oncogenic proteins, leading to tumour formation. On the other hand, upregulation of oncogenic miRNAs may block tumour suppressor genes and also lead to tumour formation.
Diagnostics in cancer
Cancer is the second leading cause of death, exceeded only by heart diseases, and accounts for nearly one-quarter of deaths in the USA. Worldwide, cancer is the third leading cause of mortality after cardiovascular and infectious diseases. Although we have made major advances in the understanding of cancer biology and pathogenesis as well as in the development of new targeted therapies, the progress in developing improved early diagnosis and screening tests has been inadequate. As a result, most cancers are diagnosed in advanced stages, leading to poor outcomes. Intense research today is focused on seeking specific molecular changes that are able to identify patients with early cancer or precursor lesions. Biological samples such as blood, serum, stool, pancreatic juice or urine, as well as both DNA and RNA, have been analysed for tumour-specific changes. Additionally genomic DNA alterations, circulating viral DNA or RNA, various mutations such as KRAS, p16 and/or APC either in serum, blood or circulating cancer cells in blood samples have been evaluated to allow the early diagnosis of cancer patients. Recently, epigenetic changes such as gene methylation in biological samples from cancer patients have also been analysed. In addition, serum levels of certain proteins involved in tumour biology such as cathepsin B, E-cadherin, hepatocyte growth factor, interleukins, and other cytokines and hormones have been measured in the serum of cancer patients. However, none of these analysis methods has yet shown adequate sensitivities and specificities to facilitate the detection of cancer in its early stages.23
▶MicroRNAs are misregulated in almost all human cancers
▶They can function as tumour suppressor genes or oncogenes
▶MicroRNAs display tissue and cell lineage specificity
▶Their small size and resistance to RNase degradation render them superior to mRNAs as molecular markers
▶MicroRNA expression patterns provide more pathognomonic information than mRNA expression
The unique patterns of aberrant miRNA expression in each type of cancer, their stability in serum and their role as biomarkers of disease risk due to inherited polymorphisms suggest that miRNAs may potentially serve as novel molecular biomarkers for clinical cancer diagnosis. This review will focus on miRNAs as potential diagnostics for cancers of the lower bowel.
MIRNA PROFILING FOR DIFFERENTIATING BOWEL CANCERS VERSUS NORMAL TISSUE
One possible application of miRNAs is in establishing a diagnosis of cancer, and a large amount of miRNA profiling data from many independent studies in various cancers show that miRNA expression patterns can help to differentiate tumour from benign tissue accurately. A pioneering study by Lu et al very successfully demonstrated this.24 They report that 129 of 217 miRNAs were expressed to a lower extent in all tumour samples compared with the normal tissue. Further, they showed that expression profiles of only a few hundred miRNAs correctly classified the tumours of different origin. Their miRNA classifier clustered the tumours from an endothelial lineage such as colon, liver, pancreas and stomach together, and tumours of haematopoietic origin were clustered as a separate group. In contrast, profiling of > 15 000 mRNA genes failed to group the tumors accurately.24
A study by Volinia et al was the first to show that 26 miRNAs were overexpressed and 17 downregulated in six kinds of solid cancers including stomach.25 Subsequent studies by different groups demonstrated that several miRNAs are associated with gastric cancer. miR-21, miR-20b, miR-20a, miR-17-5p, miR-106a, miR-18a, miR-106b, miR-18b, miR-135b, miR-183, miR-421, miR-340*, miR-19a and miR-658 are all overexpressed in gastric cancer compared with adjacent non-tumourous tissue. Findings by the Chan group demonstrated that miR-21 is over-expressed in 92% of gastric cancer, and those by Xiao et al showed 1.625-fold increased expression of miR-106a in 55 gastric carcinoma samples compared with 17 normal samples, and this was also confirmed in gastric cancer cell lines.26-28 Recently Schetter et al reported that specific miRNA signatures could distinguish colon cancer from normal colon and particularly miR-21 was found to be overexpressed in 87% of patients with colon cancer.28 On the other hand, miR-143, miR-145,29 let-7a-1,30 miR-16, miR-125b, miR-31, miR-133b, miR-96 and miR-14531 were found to be significantly downregulated in colorectal cancers (CRCs). The level of another tumour suppressor miRNA, miR-34a, is also shown to be dramatically lower in human CRC tissue. These results are suggestive of potential diagnostic roles for specific miRNAs in gastric cancers/CRCs.
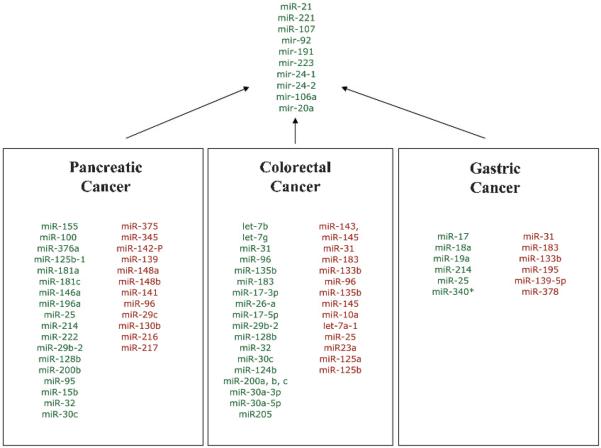
One example of the power of miRNAs in differentiating normal from tumour tissue is evidence that miRNA expression patterns can successfully separate pancreatic cancer from benign pancreatic tissue. Pancreatic cancer is a lethal disease, and early diagnosis is limited due to the paucity of specific markers. Twenty-one miRNAs are differentially overexpressed and four are underexpressed relative to chronic pancreatitis and adjacent benign pancreas tissue, indicating the diagnostic role of miRNAs in pancreatic cancer.32 Szafranska et al found that miR-196a and miR-196b levels were significantly increased in pancreatic ductal adenocarcinoma (PDAC) compared with normal tissue, as well as normal pancreatic lines and acute pancreatitis specimens. MiR-217 exhibited opposite expression patterns, suggesting its potential diagnostic power.33 34 Another study by Bloomston et al has demonstrated that aberrant expression of 22 miRNAs could accurately distinguish benign and pancreatic cancer from normal tissue in 90% of cases and, separately from chronic pancreatitis, with 93% accuracy.32 In 2007 Schmittgen's group reported that expression of miR-301 and miR-376a is also upregulated in PDAC; in contrast miR-345, miR-139 and miR-142-P were found to be significantly downregulated.33 Another miRNA profiling study by Chen et al in 2008 found that eight miRNAs, miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b and miR-95, were upregulated >3.3-fold in the pancreatic cancer tissue and cell lines compared with that in normal tissue and human pancreatic ductal epithelium cells. Additionally several studies independently observed that miR-155 and miR-221 were upregulated in PDAC, suggesting a potential diagnostic utility. Figure 4 depicts the miRNAs found to be deregulated in gastric, colorectal and pancreatic cancers in separate studies conducted by different groups.
Figure 4.
MicroRNAs (miRNAs) misregulated in pancreatic, colorectal and gastric cancers. Upregulated miRNAs in each cancer type are shown in green, and the downregulated miRNAs are shown in red. MicroRNAs that are upregulated in all three cancers are shown in green at the top.
MIRNAS IN EARLY DETECTION OF CANCERS IN BLOOD
The development of biomarkers that help detect cancer at an early stage is important since early detection has a direct impact on prognosis and clinical outcome, as evidenced by a higher (49%) 5-year survival in lung cancer patients diagnosed at an early stage compared with those diagnosed later (15%). Currently employed tests for tumour biomarkers are cumbersome, time consuming, labour intensive and offer a relatively limited number of targets. Considering the simplicity and minimal invasive nature, the development of blood, serum or plasma biomarkers is of considerable value.
The small size, relative stability and resistance to RNase degradation make the miRNAs more superior molecular markers than mRNAs.35 The recent advances in quantitative real-time PCR (qRT-PCR) methods have improved the sensitivity of miRNA detection to a few nanograms of total RNA, thus making it possible to quantify miRNA by qRTPCR on fine-needle aspiration biopsy samples.36 Furthermore, a few recent studies have demonstrated that miRNAs are extremely stable in human plasma and serum where they are protected from RNases. Their stability and predictive properties make them ideal candidates to be tested in patient serum and plasma samples. This provides a means of direct measurement of the miRNAs from patients' blood, surpassing the need for invasive procedures. A report by Chen et al showed that miRNA expression profiles from serum of healthy individuals were significantly different from that of patients with non-small cell lung cancer (NSCLC), CRC and type 2 diabetes.37
A very recent study by Ng et al found profound overexpression of five miRNAs in both plasma and tissue samples of patients with CRC compared with that in healthy controls. Of these, miR-17-3p and miR-92 were significantly elevated in patients with CRC. Furthermore, these researchers demonstrated that miR-92 plasma levels correctly discriminated CRC from gastric cancer, inflammatory bowel disease and normal subjects in an independent set of plasma samples.38 CRC is the third most common cancer worldwide. The current colonoscopic screening is both invasive and costly, while the fecal occult blood test is limited by low sensitivity and meticulous dietary restriction. Therefore, the finding that miR-92 can serve as a potential non-invasive molecular marker for CRC screening is very encouraging.
Several other studies have shown the diagnostic potential of miRNAs that can be measured in serum. Lawrie et al demonstrated that miR-21 levels are high in the serum from patients with diffuse B cell lymphoma, and this was associated with relapse-free survival.39 Also studies in mice implanted with human prostate cancer cells demonstrated high circulating levels of tumour-derived miRNAs. High serum miR-141 levels were detected in patients with metastatic prostate cancer and were also found to identify patients with prostate cancer with high accuracy.40 A similar study investigating the expression of miRNAs in plasma demonstrated that miR-141 is a biomarker for prostate cancer.40 In a study conducted by Chen and colleagues, unique serum miRNA profiles were identified in patients with lung cancer, CRC and diabetes. Furthermore, 63 miRNAs not present in healthy individuals were observed in the serum from patients with lung cancer. Specifically miR-25 and miR-223 with high expression in the sera from patients with lung cancer were found to be biomarkers for NSCLC. Expression of miR-485-5p, miR-361-3p, miR-326 and miR-487b was found to be specific to patients with CRC.37
An interesting study carried out by Tayler et al profiled miRNAs in circulating tumour-derived exosomes in ovarian cancer. Exosomes are small (50–100 nm) membrane vesicles of endocytic origin in the peripheral circulation of women with ovarian cancer. This group found that levels of the eight specific miRNAs were similar between cellular and exosomal miRNAs. These exosomal miRNAs displayed similar profiles in patients with ovarian cancer but were significantly distinct from that in benign disease, and exosomal miRNAs were not found in normal controls.41 Recently the same group demonstrated that in patients with lung adenocarcinoma, the circulating exosomal miRNA and tumour-derived miRNA patterns were similar but different from those of healthy controls. These results suggest that circulating exosomal miRNAs can serve as a potential diagnostic tool in ovarian cancer and as a screening test for lung adenocarcinoma.
The findings from all these studies build a foundation and provide a rationale to delve further into the potential of miRNAs as circulating cancer biomarkers for different cancers. However, most of these studies have been conducted in small and limited patient populations. Therefore, in order to prove the practical utility of miRNAs in clinical diagnosis, further validation in larger and independent cohorts is necessary. Since individual miRNA levels in one type of cancer can vary within different patients, designing a fingerprint comprising a signature of multiple miRNAs as opposed to a single miRNA or or couple of miRNAs would be a more reliable, accurate and sensitive tool for determining cancer status. Moreover, all these studies measured circulating miRNAs in populations after they developed cancer, and so conducting prospective studies to profile miRNA signatures in normal populations with a family history of cancer beforehand would also prove useful. Furthermore, to enable the use of miRNAs as early cancer detectors in a true sense it will be important to investigate when exactly these miRNAs become significantly expressed during the development of cancer.
MIRNAS CAN DISTINGUISH BETWEEN TUMOUR SUBTYPES
Around 15% of CRCs develop through microsatellite instability pathway and are as such molecularly classified as microsatellite-stable and microsatellite-unstable (MSS or MSI) cancers. The two are similar histologicaly; however, MSI cancers have more favourable clinical outcomes than MSS tumours. Also, MSI cancers usually follow a benign disease course and do not respond well to chemotherapy. Lanza et al first demonstrated that 14 miRNAs were differentially expressed between the two CRC subtypes.42 Interestingly, significant overexpression of the oncogenic miR-17-92 family members, miR-17-5p, miR-20, miR-25, miR-92-1, miR-92-2, miR-93-1 and miR-106a, was observed in MSS versus MSI colon cancer. Consistent with this report, another group also found that MSS and MSI cancers have distinct miRNA profiles, and a signature of specific miRNAs (miR-142-3p, miR-212, miR-151 and miR-144) correctly classified the majority of colon cancers as either MSI or MSS with 84% accuracy, 81% specificity and 92% sensitivity.42 43
The strong association of deregulated miRNA expression with breast cancer has been well documented by a vast amount of data from numerous studies. In a genome-wide miRNA expression profiling study of a large set of breast cancer and normal tissue, Iorio et al demonstrated that 29 miRNAs were differentially expressed in breast cancer versus normal tissues; miR-21 and miR-155 were upregulated, whereas miR-10b, miR-125b and miR-145 were downregulated, suggesting that these miRNAs may potentially act as diagnostic markers. Subsequent studies showed that expression patterns of miR-21 and miR-145 could discriminate between cancer and normal tissues.44 45 Another group has demonstrated the potential of miR-145 as a novel biomarker for breast cancer diagnosis.46 Blenkiron et al have developed a bead-based flow cytometric method and showed that miRNA profiling of 93 primary human breast tumours accurately classified those as luminal A, luminal B, basal-like, HER2+ and normal-like. Particularly they found that miR-155 could discriminate ER− and ER+ tumours.47 In addition, a study led by Scott et al profiling miRNAs in a cohort of 20 different breast tumours followed by supervised analysis identified a unique subset of miRNAs that distinguished HER2+ from HER2− and ER+ from ER− breast cancers.48
Findings from these studies suggest that miRNAs possibly influence the pathogenesis of different cancer subtypes and hence function as effective diagnostic markers differentiating them.
MIRNAS IN DIAGNOSING TUMOURS OF UNKNOWN ORIGIN
An ever-increasing number of tumour profiling studies in different cancers have shown that each cancer and normal tissue has a unique miRNA signature that distinguishes normal from neoplastic tissue, premalignant lesions from malignant ones, and primary tumours from one organ system from others. Moreover, miRNAs are differentially expressed across different tumour types.49 Furthermore, the downregulation or overexpression of certain miRNAs has been correlated with aggressive or metastatic phenotypes. Metastatic cancers of unknown primary origin represent a unique class with a frustrating diagnosis and huge treatment challenge for the patient as well as the oncologist. Over the past few years, cancer chemotherapy has become more targeted according to tumour type and primary site of origin. Cancers of unknown primary origin are defined as histologicaly confirmed metastatic tumours for which no known primary site has been identified. Autopsy studies have revealed the lungs, pancreas, liver, kidney, gut, bone, etc. to be the possible primary sites.50 These cancers of unknown primary site represent approximately 3–6% of aggressive malignancies, ranking among the 10 most common malignancies with poor prognosis and ill-defined therapeutic strategies. In addition, previous attempts to classify them better and determine their tissue of origin by conventional mRNA-based expression profiling have failed. As a result all cancer types in this category tend to be treated equivalently with only consideration for the cell type, leading to reduced antineoplastic efficacy and poor outcome.
Using miRNA-based classification, Rosenfeld has recently attempted to define tumour identity in tumours of unknown origin. MiRNA microarrays run on 22 different tumour tissues and metastases were used to construct a classifier based on 48 miRNAs. This classifier, when used on a blinded test set of 83 samples, predicted tissue type with approximately 90% accuracy. The classification system developed in these studies could therefore be utilised to identify the tissue of origin in cancers of unknown primaries. Moreover, despite the smaller number of miRNAs (a few hundred) than the protein-coding genes (several thousands), the unique yet differential miRNA expression patterns correlated more accurately with cancer type, stage and clinicopathological variables than gene profiling.51
An analogous study by Lu and colleagues profiled miRNA expression in 17 poorly differentiated tumours with non-diagnostic histological appearance and demonstrated that their smaller miRNA classifier made a superior diagnosis of the samples compared with that by the large mRNA classifier.24 This miRNA classifier was also found to be more informative with better predictive power for diagnosis of cancer of unknown primary than the traditional profiling of several thousands of mRNA genes.24 This could be a significant step towards the use of miRNA-based classification and identification of cancers of unknown origin, a major clinical problem, and can pave the way to the use of more personalised and targeted therapeutic strategies.
MIRNAS IN DIAGNOSING CANCER PREDISPOSITION
The notion that single nucleotide polymorphisms (SNPs) in protein-coding genes can affect the functions of proteins and in turn influence the individual susceptibility to cancers has been well documented. However, the role of miRNA-associated SNPs in disease is just emerging. Because small variations in the quantity of miRNAs may have an effect on thousands of target mRNAs and result in diverse functional consequences, the most common genetic variation, SNPs, in miRNA sequences may also be functional and therefore may represent ideal candidate biomarkers for cancer diagnosis, prognosis and outcome. SNPs that disrupt miRNA gene sequences have been associated with cancer risk. Inherited mutations or rare SNPs in the primary transcripts of hsa-mir-15a and hsa-mir-16-1 have been linked to familial chronic lymphocytic leukaemia and familial breast cancer.52 Several miRNA-associated SNPs have been shown to increase breast cancer susceptibility—for example, an SNP, rs11614913 located in the pre-miRNA of miR-196a2, has been identified in the miRNA hsa-mir-196a2. The variant geno-types CC/CT were associated with significantly increased breast cancer risks in a case–control study of 1009 breast cancer cases and 1093 cancer-free controls in a population of Chinese women. Similarly the subjects carrying variant homozygous genotypes hsa-mir-499rs3746444: A> G displayed significantly increased risks of breast cancer (odds ratio (OR) 1.75; 95% CI 1.07 to 2.85 for rs3746444 GG, respectively) compared with their wild-type homozygotes.53 Another study conducted by Shen et al has identified a G to C polymorphism (rs2910164) within the sequence of the mir-146a precursor and demonstrated that a variant C allele led to increased levels of mature miR-146 in patients with breast and ovarian cancer and predisposed them to an earlier age of onset of familial breast and ovarian cancer.54 All of these findings suggest, for the first time, that common SNPs in miRNAs may contribute to breast cancer susceptibility and may serve as novel biomarkers for breast cancer diagnosis.
On the other hand, in a cancer association study of 479 hepatocellular carcinoma (HCC) and 504 control subjects, Xu et al demonstrated that male individuals with the GG genotype in rs2910164 were twofold more susceptible to HCC (OR 2.016, 95% CI 1.056 to 3.848, p = 0.034) compared with those with the CC genotype.55 Furthermore, in a case–control association study for papillary thyroid carcinoma (PTC) Jazdzewski et al found that individuals heterozygous (GC genotype) for the same SNP had an increased risk of acquiring PTC (OR 1.62, 95% CI 1.3 to 2.0, p = 0.000007).56
A case–control study of 346 Caucasian patients with oesophageal cancer conducted by Wu et al57 provides the first evidence that miRNAs may affect oesophageal cancer risk in general and that the specific genetic variants in miRNA-related genes may affect oesophageal cancer risk individually and jointly. Seven SNPs were found to be significantly associated with oesophageal cancer risk, the most prominent being the homozygous wild-type geno-type of the SNP rs6505162 located in the pre-miR-423 region.57 Two other SNPs in the miRNA processing pathway genes XPO5 and RAN were also identified and associated with an increased oesophageal cancer risk. The XPO5 SNP has also been reported to be associated with an increased risk of renal cell carcinoma.57 58
A recent genotyping study by Horikawa et al genotyping 40 SNPs from 11 miRNA processing genes and 15 miRNA genes in 279 Caucasian patients with renal cell carcinoma and 278 matched controls reported that two SNPs in the GEMIN4 gene were significantly associated with altered renal cell carcinoma risks, indicating a possible putative role for the genetic polymorphisms of the miRNA machinery genes in the diagnosis of renal cell carcinoma.58 There is also evidence that miRNA-binding site SNPs can influence cancer risk. Two recent papers report SNPs in miRNA target sites in human cancer genes59 and show that allele frequencies vary between normal people and patients with cancer.60 A recent case–control study in a cohort from the Czech Republic, a population with the highest worldwide incidence of CRC, found that two miRNA-binding site polymorphisms, rs17281995 and rs1051690 in the 3′UTRs of CD86 and INSR genes, respectively, were significantly associated with increased CRC risk.61 An SNP identified in a miRNA-binding site in the kit oncogene was associated with increased gene expression in papillary thyroid carcinoma.62 Another SNP has been identified in a let-7-binding site in the KRAS oncogene, which disrupts let-7 regulation of KRAS and is also associated with altered cellular miRNA levels. This SNP let-7 complementary site (LCS6SNP) has been shown to be a biomarker of an increased risk of developing NSCLC in two independent case–control studies.63 Taken together, the findings from all these studies, summarised in Table 1, demonstrate the valuable utility of miRNA-associated SNP evaluation in cancer predisposition.
Table 1.
MiRNA-associated SNPs in various cancers
| Disease | miRNA | SNP | Risk allele | Target gene | SNP in |
|---|---|---|---|---|---|
| Colorectal cancer | miR-337 | rs17281995 | C | CD86 | Target site |
| miR-582 | |||||
| miR-200a* | |||||
| miR-184 | |||||
| miR-212 | |||||
| Colorectal cancer | miR-618 | rs1051690 | A | INSR | Target site |
| miR-612 | |||||
| Lung cancer | let-7 | rs61764370 | G | KRAS | KRAS 3′ UTR |
| miR-196a2 | rs11614913 | T (Chinese population) | pre-miR | ||
| Papillary thyroid carcinoma | miR-221/222 | rs17084733 | A | KIT | KIT 3′ UTR |
| miR-146a | rs2910164 | GC (heterozygous genotype) | Pre-miR-146a | ||
| Hepatocellular carcinoma | miR-146a | rs2910164 | G | RAF1 | MiRNA gene |
| Breast cancer | miR-196a2 | rs11614913 | C (Chinese population) | Pre-miR | |
| hsa-mir-499 | rs3746444 | G | Pre-miR | ||
| Oesophgeal cancer | miR-423 | rs6505162 | C | Pre-miR |
miRNA, microRNA; SNP, single nucleotide polymorphism; UTR, untranslated region.
Although these results demonstrate the possible application of miRNA-associated SNPs in cancer diagnosis, one must be prudent in interpreting the data on account of the small sample size. Therefore, large, independent, well-characterised, family and population-based case–control and additional validation studies are warranted. In order to take a step closer to the implementation of miRNA-associated SNP-based analysis in the clinic, one would have to characterise the miRNA-related SNPs further, develop novel in vivo molecular functional assays and thus gain a better insight into the biological significance of these SNPs and their contribution to cancer progression.
CONCLUSIONS
The current diagnostic methods in cancer lack sufficient sensitivity and specificity to facilitate the detection of cancer in its early stages. In the case of colon cancer, the available diagnostic tests are invasive, uncomfortable for the patient, as well as expensive. The studies reviewed here clearly demonstrate that several aspects of miRNAs including their intricate nature of interaction with multiple targets and multiple pathways make them extremely useful potential agents for clinical diagnostics (fig 5). Profiling of blood miRNAs as a diagnostic test for colon cancer would be a huge advance in the field, and a non-invasive as well as an easy alternative to colonoscopy. Diagnosis of metastatic cancers with an unknown primary site of origin is very frustrating both for the patient and for the physician, and also poses huge treatment challenges. The power of miRNAs in accurately identifying the tissue of origin in such cases is a significant advance. The study of miRNA SNPs and miRNA-binding site SNPs as biomarkers of cancer risk is another way in which miRNAs may open up new avenues, allowing early cancer detection. By identifying those at greatest risk, the application of improved diagnostic tests becomes more cost-effective. To harness the true potential miRNAs as diagnostic markers in clinical settings, and to ensure and advance their practical utility, it will be necessary to perform both prospective and retrospective miRNA profiling studies in multiple cohorts representing all patient populations using highly efficient and optimised methods. Furthermore, to improve the accuracy and efficiency of miRNA in cancer diagnostics, a combined evaluation of miRNAs and selective mRNA as well as protein markers will help in developing a more complete classifier. Although the field of miRNAs and their roles in diseases are in their infancy, more information becomes available about the function of miRNAs and their roles in various gene regulatory pathways with every passing week, leading to better insights into their role as diagnostic biomarkers. Additionally, with continuing technological advances facilitating easy and cost-effective methods for the detection of miRNAs, the idea of harnessing the tremendous potential of miRNAs as novel diagnostic biomarkers looks very promising.
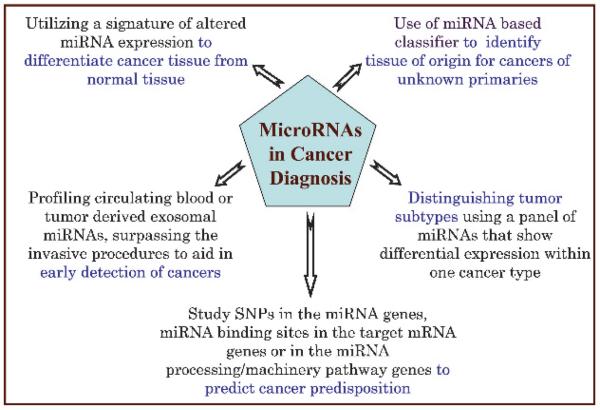
Figure 5.
MicroRNAs (miRNAs) as potential diagnostic biomarkers. Various aspects of miRNAs provide novel ways of utilising these in disease diagnosis. The unique miRNA signatures of different tumours distinguish the cancer from normal tissue. MiRNA classifiers can accurately identify the tissue of origin in the case of cancers of unknown primaries. Blood-based miRNA profiling as a diagnostic test provides a non-invasive and fast alternative to traditional methods. MiRNA patterns can also differentiate various tumour subtypes. Testing for miRNA-associated SNPs is another novel approach towards predicting cancer predisposition.
Acknowledgments
Funding: TP was supported by a grant from the Yale Center for Clinical Investigation. FS and JW were supported by NIH CA131301-01A1.
Footnotes
Competing interests: None.
Provenance and peer review: Commissioned; externally peer reviewed.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 4.Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 5.Berezikov E, Guryev V, van de Belt J, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Mirnezami AH, Pickard K, Zhang L, et al. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur J Surg Oncol. 2009;35:339–47. doi: 10.1016/j.ejso.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM, Bernstein E, Beach D, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–2. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart BJ, Weinstein EG, Rhoades MW, et al. MicroRNAs in plants. Genes Dev. 2002;16:1616–26. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 15.Abrahante JE, Daul AL, Li M, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–37. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 16.Lin SY, Johnson SM, Abraham M, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–50. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 17.Slack FJ, Basson M, Liu Z, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 18.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 19.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–46. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 20.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–80. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 21.Osaki M, Takeshita F, Ochiya T. MicroRNAs as biomarkers and therapeutic drugs in human cancer. Biomarkers. 2008;13:658–70. doi: 10.1080/13547500802646572. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Pan X, Cobb GP, et al. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 23.Ebert MP, Korc M, Malfertheiner P, et al. Advances, challenges, and limitations in serum-proteome-based cancer diagnosis. J Proteome Res. 2006;5:19–25. doi: 10.1021/pr050271e. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo J, Miao Y, Xiao B, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 27.Xiao B, Guo J, Miao Y, et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta. 2009;400:97–102. doi: 10.1016/j.cca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michael MZ, O'Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 30.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biolog Pharm Bull. 2006;29:903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 31.Bandres E, Cubedo E, Agirre X, et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 33.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 35.Waldman SA, Terzic A. Translating MicroRNA discovery into clinical biomarkers in cancer. JAMA. 2007;297:1923–5. doi: 10.1001/jama.297.17.1923. [DOI] [PubMed] [Google Scholar]
- 36.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 38.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of colorectal cancer patients: a potential marker for colorectal cancer screening. Gut. 2009 doi: 10.1136/gut.2008.167817. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 39.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 42.Lanza G, Ferracin M, Gafa R, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–24. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 44.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 45.Iorio MV, Casalini P, Tagliabue E, et al. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer. 2008;44:2753–9. doi: 10.1016/j.ejca.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 46.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 47.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–60. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 50.Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer. 2007;43:2026–36. doi: 10.1016/j.ejca.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 51.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 53.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 54.Shen J, Ambrosone CB, DiCioccio RA, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–6. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 55.Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–31. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 56.Jazdzewski K, Murray EL, Franssila K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila, Pa) 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Z, Li Z, Jolicoeur N, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–41. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landi D, Gemignani F, Barale R, et al. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2008;27:35–43. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- 61.Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 62.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Rs. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]