Abstract
Epigenetic mechanisms are essential for normal development and maintenance of tissue-specific gene expression patterns in mammals. Disruption of epigenetic processes can lead to altered gene function and malignant cellular transformation. Global changes in the epigenetic landscape are a hallmark of cancer. The initiation and progression of cancer, traditionally seen as a genetic disease, is now realized to involve epigenetic abnormalities along with genetic alterations. Recent advancements in the rapidly evolving field of cancer epigenetics have shown extensive reprogramming of every component of the epigenetic machinery in cancer including DNA methylation, histone modifications, nucleosome positioning and non-coding RNAs, specifically microRNA expression. The reversible nature of epigenetic aberrations has led to the emergence of the promising field of epigenetic therapy, which is already making progress with the recent FDA approval of three epigenetic drugs for cancer treatment. In this review, we discuss the current understanding of alterations in the epigenetic landscape that occur in cancer compared with normal cells, the roles of these changes in cancer initiation and progression, including the cancer stem cell model, and the potential use of this knowledge in designing more effective treatment strategies.
Introduction
Chromatin structure defines the state in which genetic information in the form of DNA is organized within a cell. This organization of the genome into a precise compact structure greatly influences the abilities of genes to be activated or silenced. Epigenetics, originally defined by C.H.Waddington (1) as ‘the causal interactions between genes and their products, which bring the phenotype into being’, involves understanding chromatin structure and its impact on gene function. Waddington's definition initially referred to the role of epigenetics in embryonic development; however, the definition of epigenetics has evolved over time as it is implicated in a wide variety of biological processes. The current definition of epigenetics is ‘the study of heritable changes in gene expression that occur independent of changes in the primary DNA sequence’. Most of these heritable changes are established during differentiation and are stably maintained through multiple cycles of cell division, enabling cells to have distinct identities while containing the same genetic information. This heritability of gene expression patterns is mediated by epigenetic modifications, which include methylation of cytosine bases in DNA, posttranslational modifications of histone proteins as well as the positioning of nucleosomes along the DNA. The complement of these modifications, collectively referred to as the epigenome, provides a mechanism for cellular diversity by regulating what genetic information can be accessed by cellular machinery. Failure of the proper maintenance of heritable epigenetic marks can result in inappropriate activation or inhibition of various signaling pathways and lead to disease states such as cancer (2,3).
Recent advances in the field of epigenetics have shown that human cancer cells harbor global epigenetic abnormalities, in addition to numerous genetic alterations (3,4). These genetic and epigenetic alterations interact at all stages of cancer development, working together to promote cancer progression (5). The genetic origin of cancer is widely accepted; however, recent studies suggest that epigenetic alterations may be the key initiating events in some forms of cancer (6). These findings have led to a global initiative to understand the role of epigenetics in the initiation and propagation of cancer (7). The fact that epigenetic aberrations, unlike genetic mutations, are potentially reversible and can be restored to their normal state by epigenetic therapy makes such initiatives promising and therapeutically relevant (8).
In this review, we take a comprehensive look at the current understanding of the epigenetic mechanisms at work in normal mammalian cells and their comparative aberrations that occur during carcinogenesis. We also discuss the idea of cancer stem cells as the originators of cancer and the prospect of epigenetic therapy in designing efficient strategies for cancer treatment.
Epigenetic mechanisms in normal cells
Chromatin is made of repeating units of nucleosomes, which consist of ∼146 base pairs of DNA wrapped around an octamer of four core histone proteins (H3, H4, H2A and H2B) (9). Epigenetic mechanisms that modify chromatin structure can be divided into four main categories: DNA methylation, covalent histone modifications, non-covalent mechanisms such as incorporation of histone variants and nucleosome remodeling and non-coding RNAs including microRNAs (miRNAs). These modifications work together to regulate the functioning of the genome by altering the local structural dynamics of chromatin, primarily regulating its accessibility and compactness. The interplay of these modifications creates an ‘epigenetic landscape’ that regulates the way the mammalian genome manifests itself in different cell types, developmental stages and disease states, including cancer (4,10–14). The distinct patterns of these modifications present in different cellular states serve as a guardian of cellular identity (Table I). Here, we will discuss the important aspects of the key epigenetic mechanisms present in normal cells.
Table I.
Epigenetic mechanisms involved in regulating gene expression and chromatin structure in normal mammalian cells
| DNA methylation |
| All cell types |
| Stable heritable modification |
| Gene silencing |
| Chromatin organization |
| Imprinting, X-chromosome inactivation, silencing of repetitive elements |
| Mediated by DNMTs |
| ES cells |
| Bimodal distribution pattern |
| Global CpG methylation |
| CpG islands unmethylated |
| Pluripotency gene promoters unmethylated |
| Somatic cells |
| Tissue-specific methylation of some CpG islands and most non-CpG island promoters |
| Pluripotency gene promoters methylated |
| Covalent histone modifications |
| All cell types |
| Labile heritable modification |
| Both gene silencing (H3K9me, H3K27me etc.) and gene activation (H3K4me, acetylation etc.) |
| Specific distribution patterns of histone marks contribute to chromatin organization |
| Mediated by HMTs, HDMs, HATs and HDACs etc. |
| ES cells |
| Bivalent domains—coexistence of active and repressive marks (H3K4me and H3K27me) at promoters of developmentally important genes |
| Plastic epigenome |
| Somatic cells |
| Loss of bivalency and restricted epigenome |
| Establishment of tissue-specific monovalent H3K27me and H3K4me domains |
| Presence of large organized chromatin K9 modifications |
| Nucleosome positioning and histone variants |
| All cell types |
| Labile epigenetic regulatory mechanism |
| Both gene silencing and gene activation by modulating chromatin accessibility |
| Mediated by ATP-dependent chromatin-remodeling complexes |
| Both sliding of existing and incorporation of new nucleosomes |
| H2A.Z and H3.3 preferentially localized to gene promoters that are active or poised for activation |
| Acetylated H2A.Z associates with euchromatin and ubiquitylated H2A.Z with facultative heterochromatin |
| miRNAs |
| All cell types |
| Labile epigenetic regulatory mechanism |
| Gene silencing |
| Tissue-specific expression |
| Can be epigenetically regulated |
Epigenetic mechanisms including DNA methylation, covalent histone modifications, nucleososme positioning and miRNAs are essential for normal mammalian development and regulation of gene expression. These epigenetic modifications display unique properties and distribution patterns in different mammalian cells. The distinct combinatorial patterns of these modifications, collectively termed the epigenome, are key determinants of cell fate and gene activity. ES cells maintain a more plastic epigenome required for developmental processes. In contrast, the epigenome of differentiated tissue displays a relatively restricted structure that is stably maintained through multiple cell divisions.
DNA methylation
DNA methylation is perhaps the most extensively studied epigenetic modification in mammals. It provides a stable gene silencing mechanism that plays an important role in regulating gene expression and chromatin architecture, in association with histone modifications and other chromatin associated proteins. In mammals, DNA methylation primarily occurs by the covalent modification of cytosine residues in CpG dinucleotides. CpG dinucleotides are not evenly distributed across the human genome but are instead concentrated in short CpG-rich DNA stretches called ‘CpG islands’ and regions of large repetitive sequences (e.g. centromeric repeats, retrotransposon elements, rDNA etc.) (15,16). CpG islands are preferentially located at the 5′ end of genes and occupy ∼60% of human gene promoters (17). While most of the CpG sites in the genome are methylated, the majority of CpG islands usually remain unmethylated during development and in differentiated tissues (11). However, some CpG island promoters become methylated during development, which results in long-term transcriptional silencing. X-chromosome inactivation and imprinted genes are classic examples of such naturally occurring CpG island methylation during development (15). Some tissue-specific CpG island methylation has also been reported to occur in a variety of somatic tissues, primarily at developmentally important genes (18,19). In contrast, the repetitive genomic sequences that are scattered all over the human genome are heavily methylated, which prevents chromosomal instability by silencing non-coding DNA and transposable DNA elements (11). DNA methylation can lead to gene silencing by either preventing or promoting the recruitment of regulatory proteins to DNA. For example, it can inhibit transcriptional activation by blocking transcription factors from accessing target-binding sites e.g. c-myc and MLTF (20,21). Alternatively, it can provide binding sites for methyl-binding domain proteins, which can mediate gene repression through interactions with histone deacetylases (HDACs) (22,23). Thus, DNA methylation uses a variety of mechanisms to heritably silence genes and non-coding genomic regions.
The precise DNA methylation patterns found in the mammalian genome are generated and heritably maintained by the cooperative activity of the de novo methyltransferases—DNMT3A and DNMT3B, which act independent of replication and show equal preference for both unmethylated and hemimethylated DNA and the maintenance DNA methyltransferase—DNMT1, which acts during replication preferentially methylating hemimethylated DNA (24,25).
While the role of CpG island promoter methylation in gene silencing is well established, much less is known about the role of methylation of non-CpG island promoters. Recent studies have shown that DNA methylation is also important for the regulation of non-CpG island promoters. For example, tissue-specific expression of MASPIN, which does not contain a CpG island within its promoter, is regulated by DNA methylation (26). Similarly, methylation of the non-CpG island Oct-4 promoter, strongly influences its expression level (27). Since CpG islands occupy only ∼60% of human gene promoters, it is essential to elucidate the role of non-CpG island methylation in order to fully understand the global role of DNA methylation in normal tissue (17).
Covalent histone modifications
Histone proteins, which comprise the nucleosome core, contain a globular C-terminal domain and an unstructured N-terminal tail (9). The N-terminal tails of histones can undergo a variety of posttranslational covalent modifications including methylation, acetylation, ubiquitylation, sumoylation and phosphorylation on specific residues (12). These modifications regulate key cellular processes such as transcription, replication and repair (12). The complement of modifications is proposed to store the epigenetic memory inside a cell in the form of a ‘histone code’ that determines the structure and activity of different chromatin regions (28). Histone modifications work by either changing the accessibility of chromatin or by recruiting and/or occluding non-histone effector proteins, which decode the message encoded by the modification patterns. The mechanism of inheritance of this histone code, however, is still not fully understood.
Unlike DNA methylation, histone modifications can lead to either activation or repression depending upon which residues are modified and the type of modifications present. For example, lysine acetylation correlates with transcriptional activation (12,29), whereas lysine methylation leads to transcriptional activation or repression depending upon which residue is modified and the degree of methylation. For example, trimethylation of lysine 4 on histone H3 (H3K4me3) is enriched at transcriptionally active gene promoters (30), whereas trimethylation of H3K9 (H3K9me3) and H3K27 (H3K27me3) is present at gene promoters that are transcriptionally repressed (12). The latter two modifications together constitute the two main silencing mechanisms in mammalian cells, H3K9me3 working in concert with DNA methylation and H3K27me3 largely working exclusive of DNA methylation (Figure 1). A vast array of active and repressive histone modifications have been identified, which constitute a complex gene regulatory network essential for the physiological activities of cells (10,12). Genome-wide studies showing distinct localization and combinatorial patterns of these histone marks in the genome have significantly increased our understanding of how these diverse modifications act in a cooperative manner to regulate global gene expression patterns (31,32).
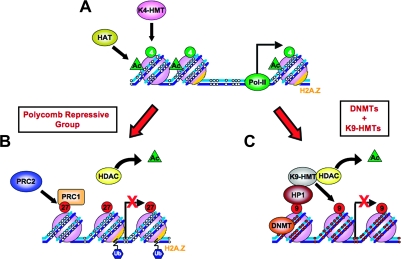
Fig. 1.
Epigenetic gene silencing mechanisms in mammals. (A) An active gene shows an open chromatin structure consisting of an unmethylated promoter region (small white circles on DNA strands), with no nucleosome upstream of the transcription start site (thick black arrow), an enrichment of active histone marks such as acetylation (green triangle, Ac) and H3K4 methylation (green circles, 4) and high levels of H2A.Z on nucleosomes (orange) surrounding the transcription start site. The open chromatin structure is permissible for binding of transcription factors and RNA Pol-II, which mediates active transcription on such promoters. Repression of such active genes (indicated by red arrows) can be achieved in normal cells by two main mechanisms: (B) Gene repression by the action of PRC1 and PRC2 that mediate the repressive H3K27 methylation (red circles, 27) is accompanied by the removal of acetylation by HDACs, loss of H3K4 methylation, chromatin compaction, nucleosome occupancy in the NFR and ubiquitylation of H2A.Z; (C) Long-term silencing through DNA methylation is performed by DNA methyltransferases. DNA methylation (small red circles on DNA strands) is often accompanied by the repressive H3K9 methylation (red circles, 9), on promoters, which leads to chromatin compaction by recruitment of HP1. DNA Methylated-silenced promoters show a depletion of H2A.Z, loss of H3K4 methylation and histone de-acetylation. Ac, acetylation; EZH2, enhancer of zeste homolog 2; HP1, heterochromatin protein 1; K4-HMT, Histone H3 lysine 4 histone methyltransferase; K9-HMT, Histone H3 lysine 9 histone methyltransferase; Pol-II, RNA polymerase II; PRC1 and PRC2, polycomb repressive complex 1 and 2; Ub, ubiquitination.
Specific patterns of histone modifications are present within distinct cell types and are proposed to play a key role in determining cellular identity (33,34). For example, embryonic stem (ES) cells possess ‘bivalent domains’ that contain coexisting active (H3K4me3) and repressive (H3K27me3) marks at promoters of developmentally important genes (35,36). Such bivalent domains are established by the activity of two critical regulators of development in mammals: the polycomb group that catalyzes the repressive H3K27 trimethylation mark and is essential for maintaining ES cell pluripotency through silencing cell fate-specific genes and potentially the trithorax group that catalyzes the activating H3K4 trimethylation mark and is required for maintaining active chromatin states during development (34). This bivalency is hypothesized to add to phenotypic plasticity, enabling ES cells to tightly regulate gene expression during different developmental processes. Differentiated cells lose this bivalency and acquire a more rigid chromatin structure, which may be important for maintaining cell fate during cellular expansion (33). This hypothesis is supported by the recent discovery of large condensed chromatin regions containing the repressive H3K9me2 mark, termed ‘LOCKs’ (large organized chromatin K9 modifications), in differentiated ES cells that can maintain silencing of large genomic regions in differentiated tissues (37).
Histone modification patterns are dynamically regulated by enzymes that add and remove covalent modifications to histone proteins. Histone acetyltransferases (HATs) and histone methyltransferases (HMTs) add acetyl and methyl groups, respectively, whereas HDACs and histone demethylases (HDMs) remove acetyl and methyl groups, respectively (38,39). A number of histone-modifying enzymes including various HATs, HMTs, HDACs and HDMs have been identified in the past decade (12). These histone-modifying enzymes interact with each other as well as other DNA regulatory mechanisms to tightly link chromatin state and transcription.
Interplay of DNA methylation and histone modifications
In addition to performing their individual roles, histone modifications and DNA methylation interact with each other at multiple levels to determine gene expression status, chromatin organization and cellular identity (40). Several HMTs, including G9a, SUV39H1 and PRMT5, can direct DNA methylation to specific genomic targets by directly recruiting DNA methyltransferases (DNMTs) to stably silence genes (41–43). In addition to the direct recruitment of DNMTs, HMTs and demethylases also influence DNA methylation levels by regulating the stability of DNMT proteins (44,45). DNMTs can in turn recruit HDACs and methyl-binding proteins to achieve gene silencing and chromatin condensation (22,23). DNA methylation can also direct H3K9 methylation through effector proteins, such as MeCP2, thereby establishing a repressive chromatin state (46). The interactions between DNA methylation machinery and histone modifying enzymes further enhance the complexity of epigenetic regulation of gene expression, which determines and maintains cellular identity and function.
Nucleosome positioning and histone variants
Non-covalent mechanisms, such as nucleosome remodeling and replacement of canonical histone proteins with specialized histone variants, also play an important role in how chromatin structure regulates gene activity. In addition to serving as the basic modules for DNA packaging within a cell, nucleosomes regulate gene expression by altering the accessibility of regulatory DNA sequences to transcription factors (14). Genome-wide nucleosome mapping data for various eukaryotic organisms reveal a common organizational theme with precise positioning of nucleosomes around gene promoters, compared with the relatively random pattern found in gene bodies (47). Nucleosome-free regions (NFRs) present at the 5′ and 3′ ends of genes are thought to provide the sites for assembly and disassembly of the transcription machinery (48). The loss of a nucleosome directly upstream of the transcription start site is tightly correlated with gene activation (49,50). Furthermore, the presence of an NFR at gene promoters with basal level of transcription correlates with their ability for rapid activation upon stimulation (51). In contrast, occlusion of the transcription start site within the NFR by a nucleosome is associated with gene repression (52). Modulation of the NFRs is orchestrated by ATP-dependent chromatin-remodeling complexes, which modify the accessibility of DNA regulatory sites through both sliding and ejection of nucleosomes (53). The interaction of nucleosome remodeling machinery with DNA methylation and histone modifications plays a pivotal role in establishing global gene expression patterns and chromatin architecture (Figures 1 and 2) (54,55).
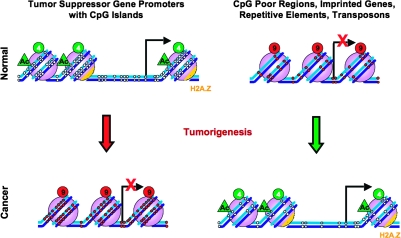
Fig. 2.
DNA methylation changes in cancer. In normal cells, CpG island promoters are generally unmethylated and when active, as in the case of tumor suppressor genes, are accompanied by active histone marks such as acetylation and H3K4 methylation (green circles, 4) allowing for a transcriptionally active open chromatin structure. However, repetitive regions, transposons, CpG poor intergenic regions and imprinted gene promoters are heavily methylated and accompanied by repressive histone marks such as H3K9 methylation (red circles, 9) that together form a silent chromatin state. During tumorigenesis, tumor suppressor gene promoters with CpG islands become methylated, resulting in the formation of silent chromatin structure and aberrant silencing (indicated by the red arrow). In contrast, the repetitive sequences, transposons and imprinted gene promoters become hypomethylated resulting in their aberrant activation (indicated by the green arrow).
In addition to physical alterations in nucleosomal positioning via nucleosome remodelers, the incorporation of histone variants, e.g. H3.3 and H2A.Z, into nucleosomes also influences nucleosome occupancy and thus gene activity (56,57). Unlike the major histone subtypes whose synthesis and incorporation is coupled to DNA replication in S phase, these variants are synthesized and incorporated into chromatin throughout the cell cycle (56). H3.3 and H2A.Z are preferentially enriched at promoters of active genes or genes poised for activation and can mediate gene activation by altering the stability of nucleosomes (58). H2A.Z incorporation may also contribute to gene activation by protecting genes against DNA methylation (59). In ES cells, H2A.Z colocalizes with bivalent domains where it may assist in maintaining key developmental genes in a poised state (60). Like canonical histones, histone variants undergo various posttranslational modifications, which determine their nuclear localization and function. For example, acetylated H2A.Z primarily associates with active genes in euchromatin, whereas ubiquitylated H2A.Z associates with facultative heterochromatin (61,62). Taken together, the inclusion of histone variants within nucleosomes provides an additional epigenetic mechanism utilized by cells to modify chromatin structure according to the needs of diverse cellular processes.
miRNAs
miRNAs are small, ∼22 nt, non-coding RNAs that regulate gene expression through posttranscriptional silencing of target genes. Sequence-specific base pairing of miRNAs with 3′ untranslated regions of target messenger RNA within the RNA-induced silencing complex results in target messenger RNA degradation or inhibition of translation (63). miRNAs are expressed in a tissue-specific manner and control a wide array of biological processes including cell proliferation, apoptosis and differentiation. The list of miRNAs identified in the human genome and their potential target genes is growing rapidly, demonstrating their extensive role in maintaining global gene expression patterns (13). Like normal genes, the expression of miRNAs can be regulated by epigenetic mechanisms (64). In addition, miRNAs can also modulate epigenetic regulatory mechanisms inside a cell by targeting enzymes responsible for DNA methylation (DNMT3A and DNMT3B) and histone modifications (EZH2) (65,66). Such interaction among the various components of the epigenetic machinery re-emphasizes the integrated nature of epigenetic mechanisms involved in the maintenance of global gene expression patterns.
Aberrant reprogramming of the epigenome in cancer
The precise epigenomic landscape present in normal cells undergoes extensive distortion in cancer (4). These epimutations, along with widespread genetic alterations, play an important role in cancer initiation and progression (3). The cancer epigenome is characterized by global changes in DNA methylation and histone modification patterns as well as altered expression profiles of chromatin-modifying enzymes. These epigenetic changes result in global dysregulation of gene expression profiles leading to the development and progression of disease states (2). Epimutations can lead to silencing of tumor suppressor genes independently and also in conjunction with deleterious genetic mutations or deletions; thus, serving as the second hit required for cancer initiation according to the ‘two-hit’ model proposed by Alfred Knudson (5). In addition to inactivating tumor suppressors, epimutations can also promote tumorigenesis by activating oncogenes. The events that lead to initiation of these epigenetic abnormalities are still not fully understood. Nevertheless, since epigenetic alterations, like genetic mutations, are mitotically heritable, they are selected for in a rapidly growing cancer cell population and confer a growth advantage to tumor cells resulting in their uncontrolled growth.
DNA methylation aberrations in cancer
Cancer initiation and progression are accompanied by profound changes in DNA methylation that were the first epigenetic alterations identified in cancer (67,68). A cancer epigenome is marked by genome-wide hypomethylation and site-specific CpG island promoter hypermethylation (Figure 2) (3). While the underlying mechanisms that initiate these global changes are still under investigation, recent studies indicate that some changes occur very early in cancer development and may contribute to cancer initiation (6).
Global DNA hypomethylation plays a significant role in tumorigenesis and occurs at various genomic sequences including repetitive elements, retrotransposons, CpG poor promoters, introns and gene deserts (69). DNA hypomethylation at repeat sequences leads to increased genomic instability by promoting chromosomal rearrangements (3,70). Hypomethylation of retrotransposons can result in their activation and translocation to other genomic regions, thus increasing genomic instability (71). Induction of genomic instability by hypomethylation is best exemplified in patients with the immunodeficiency, centromeric region instability and facial anomalies syndrome, which have a germ line mutation in the DNMT3B enzyme resulting in hypomethylation and subsequent chromosomal instability (72). Similar loss of DNA methylation and genomic instability is implicated in a variety of human cancers (71). In addition, DNA hypomethylation can lead to the activation of growth-promoting genes, such as R-Ras and MAPSIN in gastric cancer, S-100 in colon cancer and MAGE (melanoma-associated antigen) in melanoma (73), and a loss of imprinting (LOI) in tumors (74). In Wilms’ tumor, hypomethylation-induced LOI of IGF2, an important autocrine growth factor, results in its pathological biallelic expression (75). LOI of IGF2 has also been linked with an increased risk of colorectal cancer (76). Thus, DNA hypomethylation leads to aberrant activation of genes and non-coding regions through a variety of mechanisms that contributes to cancer development and progression.
In contrast to hypomethylation, which increases genomic instability and activates proto-oncogenes, site-specific hypermethylation contributes to tumorigenesis by silencing tumor suppressor genes. Since the initial discovery of CpG island hypermethylation of the Rb promoter (a tumor suppressor gene associated with retinoblastoma) (77), various other tumor suppressor genes, including p16, MLH1 and BRCA1, have also been shown to undergo tumor-specific silencing by hypermethylation (3,4,78). These genes are involved in cellular processes, which are integral to cancer development and progression, including DNA repair, cell cycle, cell adhesion, apoptosis and angiogenesis. Epigenetic silencing of such tumor suppressor genes can also lead to tumor initiation by serving as the second hit in the Knudson's two-hit model (5). In addition to direct inactivation of tumor suppressor genes, DNA hypermethylation can also indirectly silence additional classes of genes by silencing transcription factors and DNA repair genes. Promoter hypermethylation-induced silencing of transcription factors, such as RUNX3 in esophageal cancer (79) and GATA-4 and GATA-5 in colorectal and gastric cancers (80), leads to inactivation of their downstream targets. Silencing of DNA repair genes (e.g. MLH1, BRCA1 etc.) enables cells to accumulate further genetic lesions leading to the rapid progression of cancer.
While the ability of DNA hypermethylation to silence tumor suppressor genes in cancer is well established, how genes are targeted for this aberrant DNA methylation is still unclear. One possibility is that silencing specific genes by hypermethylation provides a growth advantage to cells resulting in their clonal selection and proliferation. Tumor-specific CpG island methylation can occur through a sequence-specific instructive mechanism by which DNMTs are targeted to specific genes by their association with oncogenic transcription factors. Aberrant hypermethylation and silencing of specific target gene promoters by the PML–RAR fusion protein in acute promyelocytic leukemia is an example of such a mechanism (81). Large stretches of DNA can become abnormally methylated in cancer (82) causing some CpG islands to be hypermethylated as a result of their location inside such genomic regions that have undergone large-scale epigenetic reprogramming. Another interesting mechanism proposes a role of histone marks in the tumor-specific targeting of de novo methylation and will be discussed in detail in the next section. Interestingly, regions that are hypermethylated in cancer are often pre-marked with H3K27me3 polycomb mark in ES cells (Figure 3) (83–85) suggesting a link between the regulation of development and tumorigenesis. This observation also partially explains the theory of ‘CpG island methylator phenotype’ or CIMP that hypothesizes that there is coordinated methylation of a subset of CpG islands in tumors since many of these CIMP loci are known polycomb targets (84,86). Further understanding of how specific genomic regions are targeted for DNA hypermethylation in cancer will potentially lead to additional therapeutic targets.
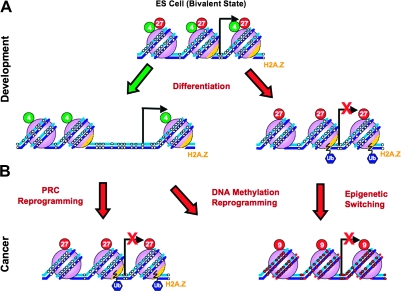
Fig. 3.
Reprogramming of the epigenome during development and tumorigenesis. (A) In ES cells, developmentally important genes are marked by a unique ‘bivalent domain’ structure, consisting of the active H3K4 methylation (green circles, 4) and repressive H3K27 methylation (red circles, 27) marks together with H2A.Z. Such bivalent domains are important for maintaining epigenomic plasticity that is required during development. During differentiation, the bivalent domains are lost, giving way to the establishment of a more rigid ‘monovalent domain’ structure that is either active (indicated by the green arrow) or repressive (indicated by the red arrow) depending upon which mark is maintained. (B) In cancer, cells undergo aberrant somatic reprogramming that results in gene silencing through formation of a compact chromatin structure. Silencing can occur through PRC (Polycomb Repressive Complex) reprogramming—silencing of active genes by the polycomb group; DNA methylation reprogramming—silencing through de novo hypermethylation (small red circles on DNA strands) accompanied by H3K9 methylation (red circles, 9) or epigenetic switching—replacement of gene repression by the polycomb mark with long-term silencing through DNA methylation; Ub, ubiquitylation.
Changes in histone modifications in cancer
Recent advances in high-throughput sequencing have enabled genome-wide mapping of chromatin changes occurring during tumorigenesis. These studies have revealed a global loss of acetylated H4-lysine 16 (H4K16ac) and H4-lysine 20 trimethylation (H4K20me3) (87). Such loss of histone acetylation, which is mediated by HDACs, results in gene repression. HDACs are often found overexpressed in various types of cancer (88,89) and thus, have become a major target for epigenetic therapy. HATs, which work in concert with HDACs to maintain histone acetylation levels, can also be altered in cancer. Aberrant formation of fusion proteins through chromosomal translocations of HAT and HAT-related genes (e.g. MOZ, MORF, CBP and p300) occurs in leukemia (90). Mistargeting of such deleterious fusion proteins contributes to global alterations in histone acetylation patterns in cancer.
In addition to changes in histone acetylation, cancer cells also display widespread changes in histone methylation patterns. Alterations in H3K9 and H3K27 methylation patterns are associated with aberrant gene silencing in various forms of cancer (91,92). Dysregulation of HMTs responsible for repressive marks results in altered distribution of these marks in cancer and leads to aberrant silencing of tumor suppressor genes. For example, EZH2, which is the H3K27 HMT, is overexpressed in breast and prostate cancer (92). Increased levels of G9a, the H3K9 HMT, has been found in liver cancer and is implicated in perpetuating malignant phenotype possibly through modulation of chromatin structure (93,94). Chromosomal translocations of MLL, the H3K4 HMT, lead to ectopic expression of various homeotic (Hox) genes and play a key role in leukemic progression (95).
In addition to HMTs, lysine specific-demethylases that work in coordination with HMTs to maintain global histone methylation patterns are also implicated in cancer progression (96). LSD1, the first identified lysine demethylase, can effectively remove both activating and repressing marks (H3K4 and H3K9 methylation, respectively) depending on its specific binding partners (97,98), thus, acting as either a corepressor or a co-activator. After LSD1, several other histone lysine demethylases have been discovered including Jumonji C domain proteins. Several of these HDMs are upregulated in prostate cancer, thus, making them potential therapeutic targets (39). However, since HDMs, like LSD1, can perform both activating and repressive functions, it is essential to first understand their precise context-dependent roles before their therapeutic inhibition can be used as an effective cancer treatment strategy. Despite these challenges, targeting HDMs is a promising treatment option for the future as revealed by a recent study which showed that inhibition of LSD1 in neuroblastoma causes decreased proliferation in vitro and inhibition of xenograft growth (99).
Epigenetic switching in cancer
As mentioned previously, DNA methylation and histone modifications work independently and in concert to alter gene expression during tumorigenesis. A key facet of such silencing mechanisms is the formation of a rigid repressive chromatin state that results in reduced cellular plasticity. The recent discovery of tumor-specific de novo methylation of polycomb target genes, which are silenced by H3K27me3 in normal cells, is another example of this phenomenon (83–85). In ES cells, developmentally important genes are reversibly silenced by polycomb proteins through the establishment of the repressive H3K27me3 mark. After differentiation, these genes continue to be repressed through the maintenance of the polycomb mark on their unmethylated promoters by EZH2. In cancer, the polycomb mark is replaced by de novo DNA methylation possibly through the recruitment of DNMTs via the polycomb complex (100). This tumor-specific ‘epigenetic switching’ of the plastic polycomb mark with more stable DNA methylation results in the permanent silencing of key regulatory genes that may contribute to cell proliferation and tumorigenesis (Figure 3) (101). However, which transformation-associated factors trigger this switch is still unclear.
Role of nucleosome positioning in cancer
The roles of DNA methylation and histone modifications in cancer initiation and progression are well established; however, the changes in chromatin structure that accompany DNA methylation and histone modification changes are less well understood. Emerging data have revealed that nucleosome remodeling works in concert with DNA methylation and histone modifications and plays a central role in tumor-specific gene silencing. DNA methylation-induced silencing of tumor suppressor genes in cancer involves distinct changes in nucleosome positioning resulting in nucleosome occupancy at transcription start site (Figure 2). Reactivation of such silenced genes using DNMT inhibitors is accompanied by a loss of nucleosomes from the promoter region (50). In addition, nucleosome remodeling can lead to aberrant gene silencing via the transmission of repressive epigenetic marks to tumor suppressor gene promoters. Recent work by Morey et al. demonstrated that nucleosome remodeling and deacetylase (NuRD) corepressor complex plays a central role in aberrant gene silencing in leukemia via the oncogenic transcription factor, PML–RARa. The NuRD complex facilitates recruitment of the polycomb repressive complex 2 and DNMT3A to PML–RARa target gene promoters leading to their permanent silencing by establishing a repressive chromatin state (102). NuRD can also be recruited to methylated promoters through its interaction with the methyl-binding domain2 protein (103). Sustained binding of NuRD to such promoters may assist in preserving their repressive state through maintenance of DNA methylation.
Alterations in the SWI–SNF complex, an ATP-dependent chromatin-remodeling complex, are also associated with cancer development (104). Abrogation of SWI–SNF function through alterations in its various subunits can result in malignant transformation. The BAF47 (hSNF5) subunit of the SWI–SNF complex is a bona fide tumor suppressor and its inhibition in rhabdoid tumors causes inactivation of the p21 and p16 pathways leading to oncogenic transformation (105). Furthermore, BRG1 and BRM, the catalytic subunits of SWI–SNF, are silenced in ∼15–20% of primary non-small-cell lung cancers (106). Treatment of BRM null cell lines with HDAC inhibitors has been shown to restore its expression, thus, making it a promising target for epigenetic therapy. However, such treatment also resulted in acetylation of BRM protein that abrogated its function (104). Development of specific HDAC inhibitors, which can circumvent BRM acetylation, is essential for successful induction of functional BRM in tumors, which can be used as a prospective therapeutic target in the future.
Interestingly, a context-dependent oncogenic role of BRG1 has also been proposed. Work by Naidu et al. reveals that BRG1 contributes to cancer development by constraining p53 activity through the destabilization of the p53 protein. Opposing roles of SWI–SNF subunits highlight the requirement for a deeper understanding of the role of nucleosome remodeling in cancer development in order to develop effective tumor-specific therapies (107).
In addition to remodeling complexes, the histone variant H2A.Z has also been implicated in tumorigenesis. H2A.Z is overexpressed in several types of cancer and has been associated with the promotion of cell cycle progression (62). Interestingly, loss of H2A.Z has also been implicated in tumor progression through possible destabilization of chromosomal boundaries resulting in spreading of repressive chromatin domains and de novo hypermethylation of tumor suppressor gene promoters (108).
Deregulation of miRNAs in cancer
Accumulating evidence from studies comparing miRNA expression profiles in tumors and corresponding normal tissues indicate widespread changes in miRNA expression during tumorigenesis (109). Since miRNAs regulate genes involved in transcriptional regulation, cell proliferation and apoptosis (the most common processes deregulated in cancer), alteration in their expression can promote tumorigenesis. miRNAs can function as either tumor suppressors or oncogenes depending upon their target genes. Many tumor suppressor miRNAs that target growth-promoting genes are repressed in cancer. For example, miR-15 and 16 that target BCL2, an antiapoptotic gene, are downregulated in chronic lymphocytic leukemia, whereas let-7 that targets the oncogene, RAS, is downregulated in lung cancer (13,110). Furthermore, miR-127, which targets BCL6, is significantly downregulated in prostate and bladder tumors (111) and mir-101, which targets polycomb group protein EZH2, is downregulated in bladder transitional cell carcinoma (66). In contrast to tumor suppressor miRNAs, oncogenic miRNAs, which target growth inhibitory pathways, are often upregulated in cancer. For example, miR-21, which targets PTEN, is upregulated in human glioblastoma (112). miRNA-155 is upregulated in breast, lung and several hematopoietic malignancies (113). While the exact mechanism of action of miR-155 is still unclear, there are suggestions that it may play a role in the class switch recombination process by targeting activation-induced cytidine deaminase (114). The oncogenic miR-17–miR-92 cluster, which targets pro-apoptotic gene Bim, is found overexpressed in several kinds of cancer (115).
Changes in miRNA expression can be achieved through various mechanisms including chromosomal abnormalities, transcription factor binding and epigenetic alterations (116). The initial report by Saito et al. demonstrated that miR-127, a tumor suppressor miRNA embedded in a CpG island, was silenced in cancer by DNA methylation and has led to subsequent discovery of several other miRNAs that are also silenced by epigenetic mechanisms in cancer (117,118). Since such epigenetic repression of tumor suppressor miRNAs can be potentially reversed by treatment with chromatin modifying drugs, they can serve as promising targets for epigenetic therapy. Saito et al. (64,111) successfully demonstrated reactivation of miR-127 in T24 bladder cancer cells following treatment with chromatin modifying drugs including DNA methylation and HDAC inhibitors. Such drug-induced activation of tumor suppressor miRNAs holds great promise for the future of cancer therapeutics.
The cancer stem cell model
Recent work suggests that the global epigenetic changes in cancer may involve the dysregulation of hundreds of genes during tumorigenesis. The mechanism by which a tumor cell accumulates such widespread epigenetic abnormalities during cancer development is still not fully understood. The selective advantage of these epimutations during tumor progression is possible, but it is unlikely that the multitude of epigenetic alterations that reside in a cancer epigenome occur in a random fashion and then accumulate inside the tumor due to clonal selection. A more plausible explanation would be that the accumulation of such global epigenomic abnormalities arises from initial alterations in the central epigenetic control machinery, which occur at a very early stage of neoplastic evolution. Such initiating events can predispose tumor cells to gain further epimutations during tumor progression in a fashion similar to accumulation of the genetic alterations that occurs following defects in DNA repair machinery in cancer. The ‘cancer stem cell’ model suggests that the epigenetic changes, which occur in normal stem or progenitor cells, are the earliest events in cancer initiation (6). The idea that these initial events occur in stem cell populations is supported by the common finding that epigenetic aberrations are some of the earliest events that occur in various types of cancer and also by the discovery that normal tissues have altered progenitor cells in cancer patients (76,119,120). This stem cell-based cancer initiation model is consistent with the observation that tumors contain a heterogenous population of cells with diverse tumorigenic properties (121). Since epigenetic mechanisms are central to maintenance of stem cell identity (33,122), it is reasonable to speculate that their disruption may give rise to a high-risk aberrant progenitor cell population that can undergo transformation upon gain of subsequent genetic gatekeeper mutations. Such epigenetic disruptions can lead to an overall increase in number of progenitor cells along with an increase in their ability to maintain their stem cell state, forming a high-risk substrate population that can readily become neoplastic on gain of additional genetic mutations (4).
Several findings have recently emerged in support of the cancer stem cell model. Mice with a LOI at the IGF2 locus and an Apc mutation show an expansion in the progenitor cell population of the intestinal epithelium, with the epithelial cells showing higher expression of progenitor cell markers and shifting toward a less-differentiated state (123). These mice were also at a higher risk for intestinal tumors relative to control mice (123). Interestingly, humans with LOI of IGF2 also show a similar dedifferentiation of normal colonic mucosa cells along with a higher risk for colorectal cancer (76). Also, stem cell-like characteristics of tumor cells were displayed through successful cloning of mouse melanoma and medulloblastoma nuclei to form blastocysts and chimeric mice (124).
DNA methylation-induced silencing of genes involved in the regulation of stem/precursor cells’ self renewal capacity, such as p16, APC, SFRPs etc., is commonly observed in the early stages of colon and other cancers (4). Aberrant silencing of these so called ‘epigenetic gatekeeper’ genes in conditions of chronic stress, such as inflammation, enables stem/precursor cells to gain infinite renewal capacity thereby becoming immortal. These preinvasive immortal stem cells are selected for and then form a pool of abnormal precursor cells that can undergo further genetic mutations leading to tumorigenesis (4,125). Human ES cells with cancer cell characteristics including higher frequency of teratoma-initiating cells, growth factor and niche independence have also been found (126). These partially transformed stem cells display a higher expression of pluripotency markers suggesting their enhanced ‘stemness’ along with high proliferative capacity (126).
Polycomb proteins, which control the silencing of developmental regulators in ES cells, provide another link between stem cell biology and cancer initiation. Polycomb proteins are commonly upregulated in various forms of cancer (92). In addition, genes that are marked by polycomb repressive mark H3K27me3 in ES cells are often methylated in cancer suggesting the presence of a shared regulatory framework, which connects cancer cells with stem/progenitor cell populations. Such findings support the hypothesis of epigenetics playing a central role in early neoplasia and cancer stem cells being the key perpetuators of cancer (83,84).
Epigenetic therapy of cancer
The reversible nature of the profound epigenetic changes that occur in cancer has led to the possibility of ‘epigenetic therapy’ as a treatment option. The aim of epigenetic therapy is to reverse the causal epigenetic aberrations that occur in cancer, leading to the restoration of a ‘normal epigenome’. Many epigenetic drugs have been discovered in the recent past that can effectively reverse DNA methylation and histone modification aberrations that occur in cancer (8). DNA methylation inhibitors were among the first epigenetic drugs proposed for use as cancer therapeutics. The remarkable discovery that treatment with cytotoxic agents, 5-azacytidine (5-aza-CR) and 5-aza-2′-deoxycytidine (5-aza-CdR), lead to the inhibition of DNA methylation that induced gene expression and caused differentiation in cultured cells led to the realization of the potential use of these drugs in cancer therapy (127). These nucleoside analogs get incorporated into the DNA of rapidly growing tumor cells during replication and inhibit DNA methylation by trapping DNA methyltransferases onto the DNA, leading to their depletion inside the cell (2). This drug-induced reduction of DNA methylation causes growth inhibition in cancer cells by activating tumor suppressor genes aberrantly silenced in cancer (8). 5-Aza-CR (azacitidine) and 5-aza-CdR (decitabine) have now been FDA approved for use in the treatment of myelodysplastic syndromes and promising results have also emerged from the treatment of other hematological malignancies such as acute myeloid leukemia and chronic myeloid leukemia using these drugs (128). The possible clinical use of other improved DNA methylation inhibitors such as zebularine, which can be orally administered, is currently under investigation (129).
The ability of these drugs to be incorporated into DNA raises concerns regarding their potential toxic effect on normal cells. However, since these drugs only act on dividing cells, one can argue that treatment with these drugs should mainly target rapidly dividing tumor cells and should have minimal effects on slowly dividing normal cells. This argument has been supported by studies demonstrating minimal side effects of long-term treatment with DNA methylation inhibitors (130). Nevertheless, an alternative approach involving the development of non-nucleoside compounds, which can effectively inhibit DNA methylation without being incorporated into DNA, is also being actively pursued. Development of several small molecule inhibitors such as SGI-1027, RG108 and MG98 is a step in that direction (131,132). These molecules can achieve their inhibitory effects by either blocking catalytic/cofactor-binding sites of DNMTs or by targeting their regulatory messenger RNA sequences; however, the weak inhibitory potential of these drugs indicates a need for the development of more potent inhibitory compounds in future.
Aberrant gene silencing in cancer is also associated with a concomitant loss of histone acetylation. Re-establishing normal histone acetylation patterns through treatment with HDAC inhibitors have been shown to have antitumorigenic effects including growth arrest, apoptosis and the induction of differentiation. These antiproliferative effects of HDAC inhibitors are mediated by their ability to reactivate silenced tumor suppressor genes (133). Suberoylanilide hydroxamic acid (SAHA), which is an HDAC inhibitor, has now been approved for use in clinic for treatment of T cell cutaneous lymphoma. Several other HDAC inhibitors such as depsipeptide and phenylbutyrate are currently under clinical trials (131).
The interaction between different components of the epigenetic machinery has led to the exploration of effective combinatorial cancer treatment strategies, which involve use of both DNA methylation and HDAC inhibitors together. Such combination treatment strategies have been found to be more effective than individual treatment approaches. For example, the derepression of certain putative tumor suppressor genes was only seen when 5-Aza-CdR and trichostatin A were combined (134). Antitumorigenic effects of depsipeptide were enhanced when leukemic cells were simultaneously treated with 5-Aza-CdR (135). Synergistic activities of DNA methylation and HDAC inhibitors were also demonstrated in a study showing greater reduction of lung tumor formation in mice when treated with phenylbutyrate and 5-Aza-CdR together (136).
Apart from DNA methylation and HDAC inhibitors, HMT inhibitors have also been actively explored recently. One such inhibitor compound, DZNep, was shown to successfully induce apoptosis in cancer cells by selectively targeting polycomb repressive complex 2 proteins, which are generally overexpressed in cancer (137). While the specificity of DZNep was challenged in a subsequent study (138), these findings reinforce the potential of HMT inhibitors and the need for further development of specific histone methylation inhibitors.
miRNAs also represent promising targets for epigenetic therapy. The finding by Saito et al. (111) demonstrated that downregulation of the oncogene BCL6 via reactivation of miR-127 following treatment with 5-Aza-CdR and 4-phenylbutyric acid strongly advocates in favor of the potential of a miRNA-based treatment strategy. In addition, the introduction of synthetic miRNAs, which mimic tumor suppressor miRNAs, can be used to selectively repress oncogenes in tumors. miRNAs, such as miR-101 that targets EZH2 (66), can be used to regulate the aberrant epigenetic machinery in cancer that may assist in restoring of the normal epigenome. However, the lack of efficient delivery methods is a major hurdle in the effective use of this strategy. Development of efficient vehicle molecules for targeted delivery of synthetic miRNAs to tumor cells is of prime importance in future.
Future prospects and challenges
The epigenetic revolution that has come about in the field of biology during the last few decades has challenged the long-held traditional view of the genetic code being the key determinant of cellular gene function and its alteration being the major cause of human diseases. Advances made in the field of cancer epigenetics have led to the realization that the packaging of the genome is potentially as important as the genome itself in regulating the essential cellular processes required for preserving cellular identity and also in giving rise to disease states like cancer. Deeper understandings of the global patterns of these epigenetic modifications and their corresponding changes in cancer have enabled the design of better treatment strategies. A combinatorial approach utilizing different epigenetic therapeutic approaches along with standard chemotherapy holds significant promise for successful treatment of cancer in future. Such approaches might also help in sensitizing cancer cells, especially cancer stem cells, which are refractory to standard chemotherapy. Further understanding of cancer stem cells along with development of more specific epigenetic drugs may hold the key to our ability to successfully reset the abnormal cancer epigenome.
Funding
National Cancer Institute (CA82422 and CA83867).
Acknowledgments
We would like to thank Gangning Liang for helpful discussions and careful reading of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- DNMT
DNA methyltransferase
- ES
embryonic stem
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDM
histone demethylase
- HMT
histone methyltransferase
- LOI
loss of imprinting
- miRNA
microRNA
- NFR
nucleosome-free region
- NuRD
nucleosome remodeling and deacetylase
References
- 1.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 2.Egger G, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, et al. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, et al. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones PA, et al. Cancer epigenetics comes of age. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, et al. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, et al. A blueprint for a Human Epigenome Project: the AACR Human Epigenome Workshop. Cancer Res. 2005;65:11241–11246. doi: 10.1158/0008-5472.CAN-05-3865. [DOI] [PubMed] [Google Scholar]
- 8.Yoo CB, et al. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein BE, et al. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki MM, et al. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, et al. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Jiang C, et al. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 16.Takai D, et al. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 18.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prendergast GC, et al. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 21.Watt F, et al. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 22.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 23.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 24.Kim GD, et al. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okano M, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 26.Futscher BW, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- 27.Hattori N, et al. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, et al. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Hebbes TR, et al. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringrose L, et al. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 36.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 37.Wen B, et al. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haberland M, et al. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 40.Cedar H, et al. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 41.Tachibana M, et al. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Q, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteve PO, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl Acad. Sci. USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 46.Fuks F, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 47.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 49.Shivaswamy S, et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin JC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gal-Yam EN, et al. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. Plos Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith CL, et al. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 54.Harikrishnan KN, et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 55.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 56.Santenard A, et al. Epigenetic reprogramming in mammalian reproduction: contribution from histone variants. Epigenetics. 2009;4:80–84. doi: 10.4161/epi.4.2.7838. [DOI] [PubMed] [Google Scholar]
- 57.Sarma K, et al. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 58.Jin C, et al. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zilberman D, et al. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Creyghton MP, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zlatanova J, et al. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 62.Svotelis A, et al. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem. Cell Biol. 2009;87:179–188. doi: 10.1139/O08-138. [DOI] [PubMed] [Google Scholar]
- 63.He L, et al. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 64.Saito Y, et al. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 65.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 67.Feinberg AP, et al. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 68.Riggs AD, et al. 5-methylcytosine, gene regulation, and cancer. Adv. Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez J, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–8468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 70.Eden A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 71.Howard G, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 72.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 73.Wilson AS, et al. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 74.Rainier S, et al. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 75.Ogawa O, et al. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature. 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 76.Cui H, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 77.Greger V, et al. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 78.Baylin SB. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(suppl. 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 79.Long C, et al. Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Invest. 2007;25:685–690. doi: 10.1080/07357900701561131. [DOI] [PubMed] [Google Scholar]
- 80.Akiyama Y, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol. Cell. Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Di Croce L, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 82.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat. Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 83.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 84.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 85.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 87.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 88.Halkidou K, et al. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 89.Song J, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 90.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nguyen CT, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 92.Valk-Lingbeek ME, et al. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 93.Kondo Y, et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kondo Y, et al. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol. Res. 2007;37:974–983. doi: 10.1111/j.1872-034X.2007.00141.x. [DOI] [PubMed] [Google Scholar]
- 95.Krivtsov AV, et al. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 96.Cloos PA, et al. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 98.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 99.Schulte JH, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 100.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 101.Gal-Yam EN, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc. Natl Acad. Sci. USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morey L, et al. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol. Cell. Biol. 2008;28:5912–5923. doi: 10.1128/MCB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feng Q, et al. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reisman D, et al. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 105.Chai J, et al. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65:10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 106.Reisman DN, et al. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 107.Naidu SR, et al. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene. 2009;28:2492–2501. doi: 10.1038/onc.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Witcher M, et al. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 110.Ventura A, et al. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 112.Chan JA, et al. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 113.Kluiver J, et al. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–1936. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- 114.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deng S, et al. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 117.Lujambio A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Toyota M, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 119.Matsubayashi H, et al. Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin. Cancer Res. 2003;9:1446–1452. [PubMed] [Google Scholar]
- 120.Peters I, et al. Adiposity and age are statistically related to enhanced RASSF1A tumor suppressor gene promoter methylation in normal autopsy kidney tissue. Cancer Epidemiol. Biomarkers Prev. 2007;16:2526–2532. doi: 10.1158/1055-9965.EPI-07-0203. [DOI] [PubMed] [Google Scholar]
- 121.Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Surani MA, et al. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 123.Sakatani T, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 124.Hochedlinger K, et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baylin SB, et al. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 126.Werbowetski-Ogilvie TE, et al. Characterization of human embryonic stem cells with features of neoplastic progression. Nat. Biotechnol. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
- 127.Constantinides PG, et al. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364–366. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- 128.Plimack ER, et al. Decitabine and its role in the treatment of hematopoietic malignancies. Leuk. Lymphoma. 2007;48:1472–1481. doi: 10.1080/10428190701471981. [DOI] [PubMed] [Google Scholar]
- 129.Cheng JC, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 130.Yang AS, et al. Comment on “Chromosomal instability and tumors promoted by DNA hypomethylation” and “Induction of tumors in nice by genomic hypomethylation”. Science. 2003;302:1153. doi: 10.1126/science.302.5648.1153a. (author reply 1153) [DOI] [PubMed] [Google Scholar]
- 131.Cortez CC, et al. Chromatin, cancer and drug therapies. Mutat. Res. 2008;647:44–51. doi: 10.1016/j.mrfmmm.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Datta J, et al. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 133.Carew JS, et al. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269:7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 134.Cameron EE, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 135.Klisovic MI, et al. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia. 2003;17:350–358. doi: 10.1038/sj.leu.2402776. [DOI] [PubMed] [Google Scholar]
- 136.Belinsky SA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 137.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miranda TB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]