Abstract
The reaction of DNA-damaging agents with the genome results in a plethora of lesions, commonly referred to as adducts. Adducts may cause DNA to mutate, they may represent the chemical precursors of lethal events and they can disrupt expression of genes. Determination of which adduct is responsible for each of these biological endpoints is difficult, but this task has been accomplished for some carcinogenic DNA-damaging agents. Here, we describe the respective contributions of specific DNA lesions to the biological effects of low molecular weight alkylating agents.
Introduction
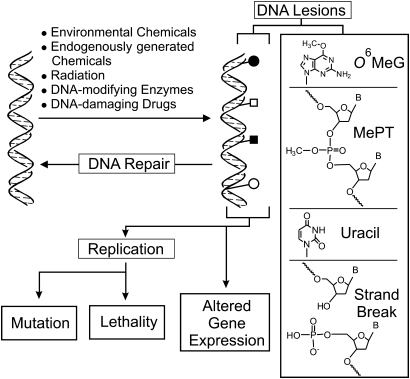
DNA damage can be caused by radiation, by organic and inorganic chemical agents and by enzymes that have the roles of promoting natural methylation and deamination, such as members of the S-adenosylmethionine (SAM)-dependent methyltransferases, the activation-induced deaminase and the apolipoprotein B editing complex (1,2). Because DNA is abundantly equipped with nucleophilic sites, reaction with extracellularly generated and endogenously produced electrophiles results in an amazingly diverse array of covalent chemical-DNA adducts. These lesions compromise cellular welfare in three major ways (Figure 1). First, misreplication or misrepair of the lesions triggers mutations, which can be the initiating lesions of genetic diseases, including cancer. Second, the lesions can jeopardize the epigenetic program imprinted by natural enzymatic DNA modifications. Finally, the lesions can block RNA and DNA polymerases and can lead directly or indirectly to DNA strand breaks, which tend to be lethal in most cells. The biological importance of DNA damage is evidenced by the large commitment of the genome to protection of informational integrity; such genoprotective networks include electrophile scavengers, recombination complexes that permit DNA lesion tolerance, specialized polymerases that afford lesion bypass and a large battery of DNA repair proteins. Loss of one or more of these networks results in loss of informational integrity and, ultimately, the onset of disease (1).
Fig. 1.
Pathways by which DNA-damaging agents induce biologically relevant events. Agents from the environment, chemically reactive natural species generated within cells and the misdirected action of natural intracellular enzymatic systems can result in the formation of a collection of DNA lesions (symbols attached to the helix). These lesions can be formal chemical-DNA adducts, such as O6MeG, which is a miscoding lesion during replication. They can be modifications of the sugar-phosphate backbone, such as MePT, which triggers a change in gene expression. They can be bases such as uracil, which can appear as the enzymatic deamination product of cytosine. Or, they can be lethal strand breaks, as would form after treatment of a cell with ionizing radiation or certain anticancer agents. Finding the relationships between the structures of each lesion and the biological endpoints of mutation, lethality and gene expression is the subject of this review.
Once it was appreciated that DNA lesions cause mutagenic and toxic events, researchers sought to understand the relationships between the structure of each lesion in DNA and the biological endpoints indicated above (3). For example, discovery of the mutagenic lesion of a carcinogenic DNA-damaging agent might lead to strategies to reduce the level of that lesion in DNA and hence reduce the likelihood of carcinogenesis. Studies on the DNA adducts of aflatoxin B1 led to intervention strategies at the population level that offer promise of reducing liver cancer burden (4). As a second example, knowledge of the relationship between the structures of DNA adducts of anticancer drugs and cytotoxicity endpoints can aid drug development efforts in clinical pharmacology. While it is obvious that establishing the relationships between DNA adducts and their biological endpoints is important, it proved very difficult to develop an experimental strategy to address the problem. Even a single simple DNA-damaging agent such as the aforementioned aflatoxin results in nearly a dozen DNA adducts, which frustrated early attempts to determine which adducts are the biologically important ones (5).
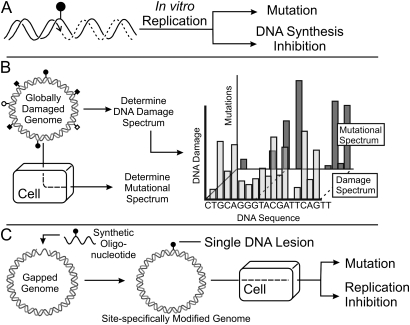
Dissection of the relative biological importance of individual DNA lesions proved to be a tractable problem with the advent of methodology whereby investigators could place one lesion at a time into synthetic DNA (Figure 2). In early in vitro studies, the oligonucleotides with adducts at known sites were acted upon with purified polymerases (Figure 2A) and repair proteins, which gave results that helped predict the biological relevance of a lesion and helped define the cellular repair systems that might protect against it. A second step involved the use of shuttle vectors that were globally modified by a DNA-damaging agent (Figure 2B). Chemical or enzymatic tools allowed the mapping of some (but not all) lesion sites along a stretch of DNA. The damage spectrum was then compared with the spectrum of mutations that arose when the modified vector was replicated within cells. Often multiple types of mutation were observed at a single site and it was impossible to ascertain if a single lesion gave rise to multiple mutations at, for example, a guanine site or whether there were several distinct guanine adducts each of which had its own signature and singular mutation. Nevertheless, this approach was and continues to be a cornerstone of mutation research.
Fig. 2.
(A) Methods to evaluate the biological relevance of DNA damage. The ability of a DNA lesion (lollipop structure) to block polymerases in vitro and cause mispairing during DNA synthesis can be evaluated in a system in which a template containing the lesion is primed with a complementary oligonucleotide that terminates to the 3′ side of the lesion. DNA synthesis may result in incorporation of non-complementary bases or in truncated products, which can be evaluated on sequencing gels. The same in vitro constructs, in double-stranded or single-stranded form, can also be used as substrates for DNA repair reactions using purified DNA repair proteins or cellular extracts. (B) To determine the mutagenic properties of the full population of adducts that form from treatment of DNA with a mutagen, a plasmid or viral vector is treated with the damaging agent. Replication of the vector in cells results in repair of some adducts but those that evade repair can possibly be converted into mutations. Sequencing the genomes of progeny can generate the mutational spectrum, which indicates the types and frequencies of specific mutations along the DNA sequence being studied. In parallel, one can map the locations of some of the DNA adducts by using enzymatic or chemical probes. The corresponding damage spectrum is often compared with the mutational spectrum in order to formulate hypotheses with regard to which DNA adduct might have caused specific mutations. (C) The most sophisticated system for analysis of mutagenesis involves chemical or enzymatic synthesis of an oligonucleotide that contains a candidate for mutagenesis (often the candidate is nominated based on the data from experiments shown in part B). The oligonucleotide is inserted into the genome of a virus or plasmid, which is later replicated within cells, either intra- or extra-chromosomally. Progeny are analyzed to determine the type, amount and genetic requirements for mutagenesis by the lesion. In parallel, the reduction in viable progeny is determined as an estimate of the extent to which each lesion inhibits replication of the genome.
The fusion of chemistry and biology, termed ‘chemical biology’, gave rise to a more advanced technology in which synthetic oligonucleotides containing well-characterized single DNA lesions were genetically engineered into the genomes of viruses or plasmids, which could be introduced into bacterial or mammalian cells (Figure 2C). Within the cell, the lesion would encounter the host repair and replication systems much in the same way that the lesion would be treated if it had formed endogenously. Lethal endpoints could be measured as a decrease in viral or plasmid progeny. Mutagenic outcomes could be determined by interrogating the vector genomes in the vicinity of the genomic site that originally contained the adduct. The relative importance of various DNA repair and polymerase systems to deal with or process the adduct could be determined by introduction of the vector into cell strains with known defects in repair or replication. In time, the quantitative and qualitative features of mutagenesis and toxicity of a wide array of DNA-damaging agents were profiled by this new technology.
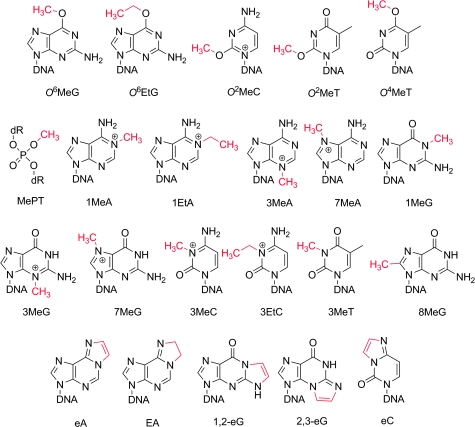
This review examines in detail the application of a variety of experimental systems, primarily the use of site-specifically modified vector genomes, to categorize the mutagenic and toxic properties of DNA alkylating agents. Such agents are common environmental carcinogens, some are formed endogenously and cause spontaneous DNA damage and some have found use as cancer chemotherapeutic agents. The paper specifically reviews current knowledge of the biological properties of each of the lesions formed by low molecular weight alkylating agents. The structures of the relevant lesions are shown in Figure 3. By compiling data on lesion mutagenicity, genotoxicity and repairability, we develop a biological ‘fingerprint’ for each lesion (Table I). It is noteworthy that some lesions have mutagenicities at or approaching 100%, whereas others display comparatively weak mutagenic properties; however, it must be kept in mind that a lesion with a mutagenicity of only 0.1% creates mutations at a rate that is five orders of magnitude greater than the basal or spontaneous rate of mutagenesis. In the review, exocyclic monoadducts are covered first, followed by adducts in which endocyclic atoms are the points of attachment to the alkyl residue. The final sections of the review cover small cyclic adducts. To keep the manuscript of a manageable size, we have limited our attention to adducts of one or two carbon residues, avoiding larger adducts and some of the lipid-derived adducts that have been reviewed elsewhere (6).
Fig. 3.
Structures of DNA alkylation lesions.
Table I.
Mutagenicity, genotoxicity and repairability of DNA alkylation lesions
| Lesion | Mutagenic specificity | Genotoxicity | Repaired by prokaryotic enzymes/systems | Repaired by eukaryotic enzymes/systems |
| O6MeG | G → A | Toxic in presence of MMR | Ada, Ogt | MGMT |
| UvrABC (NER) | NER | |||
| MMR | MMR | |||
| O6EtG | G → A | Toxic in Escherichia coli | Not repaired by Ada or Ogt | MGMT |
| NER | ||||
| MMR | ||||
| O2MeC | Possibly toxic | |||
| O2MeT | Possibly toxic | |||
| O4MeT | T → C | Toxic, but less than O6MeG and O6EtG in E.coli | Ada, Ogt | NER |
| T → A in MMR-deficient cells | MGMT (minimal) | |||
| MePT | Not known | Not known | Ada | |
| 1MeA | A → T | Mutagenic and toxic to E.coli in the absence of AlkB | AlkB | AAG |
| ABH2, ABH3 | ||||
| 1EtA | Mutagenic and toxic to E.coli in the absence of AlkB | AlkB | ABH2, ABH3 | |
| 3MeA | A → T | Highly toxic | AlkA | AAG/MPG |
| Tag | ||||
| UvrA | ||||
| 7MeA | Fapy-7MeA | |||
| A → G | ||||
| 1MeG | G → T | Mutagenic and toxic to E.coli in the absence of AlkB | AlkB | AAG |
| G → A | ||||
| G → C | ||||
| 3MeG | Possibly toxic | AlkA | ||
| Tag | ||||
| 7MeG | Fapy-7MeG | Toxic via formation of AP sites and Fapy-7MeG | AlkA | AAG/ |
| G → C | Fpg | MPG | ||
| G → T | hOGG1 | |||
| 3MeC | C → T | Toxic | AlkB | AAG |
| C → A | ABH2, ABH3 | |||
| 3EtC | C → T | Toxic | AlkB | |
| C → A | ||||
| 3MeT | AlkB-deficient cell | Strong block to replication | Weak substrate for AlkB | FTO |
| T → A | ||||
| T → C | ||||
| 8MeG | G → C | AlkA | Not repaired by known enzymes | |
| eA | AlkB-deficient cell | Mutagenic and toxic to E.coli in the absence of AlkB | AlkA | AAG |
| A → T | ||||
| A → G | AlkB | |||
| A → C | ||||
| EA | A → T (weak) | Toxic to E.coli in the absence of AlkB | AlkB | AAG |
| A → C (weak) | ||||
| A → G (weak) | Weak substrate for AlkA | |||
| 1,2-eG | G → T | Mutagenic and causes frameshift | AAG, MUG | |
| G → C | ||||
| 2,3-eG | G → A | Mutagenic | AlkA | |
| eC | C → A | Mutagenic and toxic to E.coli in the absence of AlkB | AlkB | hTDG |
| C → T | dsUDG |
1,2-eG, 1,N2-Ethenoguanine; 2,3-eG, 3,N2-ethenoguanine; Fpg, formamidopyrimidine-DNA-glycosylase; FTO, Fat mass and obesity associated protein; O2MeC, O2-methylcytosine; O2MeT, O2-methylthymine.
O6-methylguanine and O6-ethylguanine
O6-methylguanine (O6MeG), which causes G → A transitions (7), is the primary mutagenic lesion under most conditions of alkylation damage to the genome (8). O6MeG is formed from both endogenous (9,10) and exogenous sources (11), and studies have correlated its persistence to organ-specific tumorigenicity in rats (12). O6-ethylguanine (O6EtG) is the major mutagenic lesion formed by ethylating agents (13) and also primarily causes G → A transitions (14).
Escherichia coli has two O6MeG-DNA methyltransferases that can repair the adduct—the constitutive Ogt protein and the inducible Ada protein, which directly reverse methylation damage by transferring the alkyl group to one of the internal cysteine residues on each repair protein. This transfer irreversibly inactivates the repair proteins, making the non-enzymatic stoichiometric reaction ‘suicidal’ (15). Ada is part of the adaptive response, which was discovered when E.coli treated with a low dose of a methylating agent acquired resistance to the mutagenicity and toxicity of subsequent higher doses (16). The alkyl groups from O6AlkGua and O4AlkThy are transferred to Cys-321 at the C-terminus, whereas those from a third substrate, methylphosphotriester (MePT), is transferred to the N-terminus of Ada. It was initially believed that the methyl group from MePT was transferred to Cys-69 on the protein (17) but recent evidence identifies Cys-38 as the acceptor residue (18). Methylation of Cys-38 of Ada converts it to a transcriptional activator of the genes encoding the ‘adaptive response’ to alkylating agents, namely, ada, alkA, alkB and aidB. This is the most nucleophilic of all available cysteine residues in Ada since it is not part of a network of hydrogen bonds. Methylation at this site reduces the overall negative charge on Ada. Reduction in charge density is important for the role of Ada as a transcription factor as it enhances its interaction with negatively charged DNA by 1000-fold (19). The number of Ada molecules is estimated to rise from one to two molecules in an unadapted state to ∼3000 molecules in a fully adapted cell (20,21). It was initially found that Ada preferentially repairs O6MeG as compared with O4-methylthymine (O4MeT) (22) but recent evidence suggests that it repairs both lesions with equal efficiency (23).
The second DNA methyltransferase, Ogt, was discovered by deletion of the ada operon (24,25). Unlike Ada, Ogt is constitutively expressed in E.coli, shows a preference for repair of O4MeT and larger alkyl adducts and does not repair MePT (25). It is estimated that there are ∼30 molecules of Ogt in wild-type E.coli (21). The mammalian homolog of Ogt and Ada is O6-methylguanine-DNA methyltransferase (MGMT) (also referred to as AGT). This enzyme works in a similar suicidal fashion but is not inducible, and it shows a 35-fold higher preference for repairing O6MeG over O4MeT (23). Human MGMT can be silenced by epigenetic modifications (26). This silencing plays a dual role in carcinogenesis as tumors not expressing MGMT acquire a mutator phenotype but also become more susceptible to killing by alkylating agents (27).
Ogt is speculated to provide protection at low levels of sporadic exposure to alkylating agents, whereas the adaptive response becomes more important against higher chronic exposures or acute exposures that trigger the transcriptional switch of the adaptive response operon. In addition to the methyltransferases, the UvrABC nucleotide excision repair (NER) pathway can also repair O6MeG. Excision of O6MeG on duplex substrates has been shown to occur in vitro (28) and in vivo (29). When O6MeG is present in a single-stranded context in vivo, NER does not affect mutation frequency of the lesion; the mutation frequencies in E.coli uvrB+ada−ogt− cells are very similar to those found in uvrB−ada−ogt− cells (30). Interestingly, Chambers et al. (31) found a 40-fold decrease in the G → A transition caused by an O6MeG lesion introduced on a single-stranded ΦX174 genome in an NER-deficient (uvrA) cell strain versus wild-type. The authors suggest a shielding mechanism by which UvrA binds to the lesion and protects it from repair by Ada or Ogt, leading to elevated mutation frequencies. There is some evidence of the NER pathway playing a role in repair of O6MeG in Drosophila melanogaster (32) and of O6EtG in D.melanogaster (33) and mammalian cells (34).
The mismatch repair (MMR) pathway has also been implicated in the cellular response to O6MeG (35). O6MeG can be processed by post-replicative MMR in E.coli in a double-stranded context, but in a single-stranded context (a gapped plasmid), the mutation frequencies in wild-type and mutS− cells are the same (36). Using an M13 single-stranded system containing an O6MeG lesion, Rye et al. (37) have shown that dam− and mutH− strains display the same mutation frequency as wild-type, but mutS− and mutL− strains show a decrease. This result suggests that MMR proteins may aid in the repair of O6MeG in a cooperative fashion. Whereas early work suggested that O6EtG is not repaired by alkyltransferases or MMR in E.coli (38), more recent studies suggest that it is repaired by the same machinery that repairs O6MeG in mammalian cells (39). Nevertheless, in rat mammary cells, O6EtG is repaired 20 times faster than O6MeG by an unknown, MGMT-independent mechanism (40). In line with expectations based upon this finding, a G → A mutation is not seen as a frequent event at codon 12 of the H-ras gene in tumors initiated by N-ethyl-N-nitrosourea compared with tumors initiated by N-methyl-N-nitrosourea (MNU).
The toxicity of O6MeG has been established by several studies, and it appears that abortive MMR or inhibition of replication systems may play roles in converting the adduct into lethal intermediates. Evidence that O6MeG is potently toxic in mammalian cells comes from a number of studies, including those in MGMT knockout mice, which display hypersensitivity to the lethal effects of alkylating agents that generate O6MeG (41). There are two proposed mechanisms by which this lesion contributes to the toxicity generated by alkylating agents. The first suggests that the lesion reduces the efficiency of replication by polymerases. This phenomenon has been studied using in vitro systems. The rates of replication by T4 and T5 phage DNA polymerases and E.coli polymerase I decrease linearly with increasing proportion of O6MeG in the synthetic oligonucleotide used as a template (42). Also, human polymerase β, subcloned in an E.coli plasmid, is blocked by O6MeG present on a single-stranded DNA template (43). The second mechanism leading to toxicity is that of futile cycling of the MMR system at an O6MeG:T pair (44,45). The model proposes recognition of this base pair by the MMR enzymes, which results in the removal of the newly incorporated thymine from the nascent strand opposite the lesion. On re-replication, O6MeG preferentially pairs once again with an incoming thymine (7), reinitiating the repair and replication cycle. This persistent iteration of excision and synthesis is thought to result in a stabilized nick or small gap in one strand of DNA, which may activate damage signaling pathways (46). The recursive cycling mechanism is thought to be of practical significance in that it may explain the lethal effects of the anticancer drug, temozolomide (47). In E.coli, O6EtG is more toxic than O6MeG (38) but the mechanism underlying this differential toxicity is unknown.
O6MeG is known to be highly mutagenic. To study the mutations formed in vivo, Loechler et al. (7) constructed single-stranded M13mp8 DNA containing O6MeG at a specific position and transfected the same into E.coli. It was found that the predominant mutation generated by this lesion was a G → A transition. In wild-type E.coli, the lesion was weakly mutagenic, but challenging the Ada and Ogt repair systems of the cell by treatment with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG; which forms alkyl adducts in the host genome) resulted in a robust, dose-dependent demonstration of the mutagenic power of this adduct (7). This early study showed how significant even a few molecules per cell of a DNA repair protein could be as a protection against DNA damage. The ethyl homolog of O6MeG, O6EtG, introduced at a specific position in ΦX174 and transfected into E.coli produces higher mutation frequencies compared with O6MeG in the same system (48,49). O6MeG and O6EtG also have been site-specifically incorporated in Chinese hamster ovary cells and are shown to have a mutation frequency of 19 and 11%, respectively, in cells lacking O6-alkylguanine-DNA alkyltransferase (50).
A recent study used site-specific mutagenesis to generate single-stranded M13mp7 genomes containing O6MeG in all 16 possible permutations and combinations of nearest neighbor sequence contexts. These genomes were then introduced into E.coli mutants of different repair backgrounds and the mutation frequencies were determined by a novel and very sensitive assay. It was found that O6MeG went from being 10% mutagenic in repair-proficient cells to 100% mutagenic in repair-deficient cells (30). Moreover, it was found that DNA repair in vivo is sequence context dependent.
With regard to effects on gene expression, O6MeG can inhibit carbon-5 methylation of cytosines in 5′-deoxycytidine-deoxyguanosine-3′ (dCpG) motifs by interfering with the binding of 5-methylcytosine DNA methyltransferases; eventually, this interference with natural methylation can lead to genome hypomethylation. The pairing of O6MeG with thymine can also lead to DNA hypomethylation (51). By these mechanisms, the formation of this adduct could affect the epigenetic program of mammalian cells.
O4-methylthymine
O4MeT is one of the mutagenic lesions formed concurrently with O6MeG when DNA is exposed to alkylating agents that react with DNA by an SN1 mechanism. O4MeT is formed at a much lower level than O6MeG, for example, the methylated thymine was detected at a level 126 times lower than that of O6MeG in calf thymus DNA treated with MNU (52). Although it is not an abundant lesion, O4MeT can be very mutagenic. Using site-specific mutagenesis tools, it was shown that O4MeT incorporated in single-stranded M13mp19 had a mutation frequency of 12% in repair-proficient E.coli. O6MeG gave a mutation frequency of <2% in the same repair-proficient system. Pre-treatment with MNNG to deplete or occupy endogenous repair enzymes doubled this mutation frequency (53). Similar results were obtained using double-stranded and gapped plasmids in E.coli (mutation frequency of 45% for O4MeT versus 6% for O6MeG), leading to the conclusion that O4MeT is much more mutagenic than O6MeG (38) on a mole-per-mole basis under normal conditions of DNA repair proficiency in cells. O4MeT mimics cytosine in structure and generates an overwhelming majority of T → C transitions (54); it can also cause a small number of T → A transversions in MMR-deficient cells (39). O4MeT has been examined as a site-specific adduct in mammalian vectors and again appears to be more mutagenic than O6MeG in both repair-proficient and repair-deficient backgrounds (39,55). In E.coli, O4MeT is toxic but less so than O6MeG and O6EtG (38). O4MeT has been shown to be toxic to mammalian cells deficient in NER capability (56), suggesting a role for this repair pathway in the cellular defense against this adduct.
In E.coli, O4MeT is repaired by the same alkyltransferases that repair O6MeG. The Ogt protein from E.coli seems to have a preference for repair of O4MeT over O6MeG (25). Ada repairs O6MeG and O4MeT with equal efficiency but human and rat alkyltransferases show a preference for O6MeG repair (23,57). Studies in mammalian cells have shown that the mutation frequency of O4MeT does not vary significantly in the presence or absence of alkyltransferase, indicating that it is probably not repaired by MGMT (39,55). In fact, mammalian alkyltransferases may actually inhibit repair of O4MeT by the NER pathway by binding to and shielding the lesion, as evidenced in E.coli in studies using plasmids expressing human and mouse methyltransferases (58). A study done in human cell lines using site-specifically modified plasmids containing O4MeT shows that repair is not influenced by the levels of alkyltransferase and that NER seems to be the most significant repair system for this lesion (56). With regard to repair by MMR, one in vitro study found that E.coli MutS (a DNA MMR binding protein) does not bind to oligonucleotide duplexes containing a site-specifically incorporated O4MeT:A base pair (35), whereas another shows that hMutSα, a protein of the MMR pathway in humans, recognizes and binds to a O4MeT:A base pair quite well but very poorly to an O4MeT:G base pair (59).
O2-methylcytosine and O2-methylthymine
O2-methylcytosine (O2MeC) and O2-methylthymine (O2MeT) are minor reaction products formed by treatment of DNA with alkylating agents such as MNU or MNNG. Both lesions are repaired in vitro by E.coli AlkA (60). O2MeC and O2MeT are predicted to interfere with minor groove contacts; yet, there have been very few studies of these modifications, making the lesions good candidates for future study.
Methylphosphotriester
Methylation damage can occur on the DNA sugar-phosphate backbone to form MePT. The physical accessibility and negative charge of the phosphate oxygens make them a favorable site for chemical reaction. When double-stranded DNA is treated with MNU, 17% of the total methylation occurs on the backbone to yield MePT (13). These adducts react with water and other nucleophiles much faster than the common diester form of phosphate linking adjacent nucleosides, leading to facile cleavage of the backbone. Of the two diastereomers formed, only the Sp-MePT is repaired by the Cys-38 residue in the N-terminal domain of Ada. This selective repair results because the oxygen atom on the phosphate in the Sp diastereomer is only 3.5Å away from the acceptor cysteine residue versus 5Å in the Rp configuration (18). As discussed earlier, N-terminal domain of Ada has an inherent electrostatic switch that works in a methylation-dependent fashion to modulate its affinity for DNA and ability to act as a transcription activator. There is no known homolog of N-terminal domain of Ada in eukaryotes, thus making the repair of MePT in mammalian cells uncertain.
In vivo studies using wild-type Ada and truncated Ada (lacking MePT repair capability) transfected into HeLa cells showed the same extent of resistance to the cytotoxic effects of alkylating agents, similar sister chromatid exchange induction, as well as host-cell reactivation of adenovirus (61). This observation suggests that MePT may not have cytotoxic effects in cells. The role of MePT seems to be a chemosensor for detection of methylation damage and induction of the adaptive response in E.coli, but their role, if any, in eukaryotes is unknown.
N1-methyladenine and N1-ethyladenine
N1-methyladenine (1MeA) is formed by alkylating agents mainly in single-stranded DNA and has been detected in vitro (62,63,64,65,66,67,68) and in vivo (64,69,70,71,72). SN2 agents, such as methylmethanesulfonate (MMS) and the naturally occurring methyl halides, can generate 1MeA (15); similarly, the ethyl homolog, N1-ethyladenine (1EtA), is formed by ethylating agents both in vitro and in vivo (64). The preference for formation in single-stranded DNA is owed to location of the N1 atom of adenine at a site usually protected by base pairing in double-stranded DNA (73). 1MeA is cytotoxic because it disturbs DNA replication (15). 1-Methyldeoxyadenosine is known to be unstable due to a base-catalyzed Dimroth rearrangement, a complex mechanism, the net result of which is the migration of the N1 methyl group to the exocyclic N6 position of adenine (74).
A specialized DNA repair system protects cells from N1-substituted DNA lesions. The AlkB enzyme of the adaptive response repairs 1MeA both in vitro and in vivo (75) in an oxidative reaction that liberates formaldehyde from the methylated base, affording complete reversal of the damage. The role of AlkB in the repair of 1MeA seems to be the prevention of genotoxicity, because this very toxic adduct is only weakly mutagenic in cells. AlkB and its human homologs, ABH2 and ABH3, also repair 1EtA residues in DNA, with the release of acetaldehyde as the repair product (76). Studies of 1MeA in vivo reveal that the lesion severely blocks DNA replication, but the replication block can be partially overcome by the induction of SOS bypass polymerases. The 1MeA blockade is completely removed in AlkB-proficient cells (75), underscoring the physiological relevance of the AlkB system for countering the toxicity of this base. While very toxic, as indicated above, 1MeA is at best weakly mutagenic. To the extent that it is mutagenic, 1MeA induces A to T mutations, which are enhanced following induction of the SOS polymerases. The base composition for A versus T was, respectively, 99 versus 0.61% in SOS−/AlkB− cells, 99.7 versus 0.06% in SOS−/AlkB+ cells, and 98.6 versus 1.0% in SOS+/AlkB− cells (75).
While the AlkB protein can repair the 1EtA lesion, it cannot repair 3-ethyladenine damage, which parallels AlkB's activity on 1MeA but not on 3-methyladenine (76). AlkB repairs 1EtA somewhat less well than 1MeA.
N3-methyladenine
N3-methyladenine (3MeA) can be formed in DNA by methylating agents as well as non-enzymatically by intracellular SAM. In a mammalian cell, SAM or some other methylating agent reacts with DNA to generate an estimated 600 3MeA per day (77). The half-life of 3MeA in vivo is estimated to be between 4–24 h (78). While 3MeA is not particularly mutagenic, it is a cytotoxic DNA lesion by virtue of its ability to block replication or by virtue of its ability to give rise to a chemically or enzymatically generated abasic/apurinic site (AP site). With regard to replication inhibition, it is thought that the methyl at the N3 position of purines sterically interferes with the required contact between the polymerase and minor groove on DNA (79). This property makes it essential for the cell to have in place defenses against this form of damage. 3MeA-DNA-glycosylases have evolved in both prokaryotic and eukaryotic systems to afford the efficient repair of this lesion. The prokaryotic system includes the highly selective and constitutive 3-methyladenine-DNA glycosylase I (TAG) protein and the inducible AlkA glycosylase with a broader specificity. The eukaryotic system is comprised of human 3MeA-DNA-glycosylase (AAG) and N-methylpurine-DNA glycosylase (MPG). AlkA and TAG repair 3MeA with equal efficiency on double-stranded DNA, but AlkA is 10- to 20-fold more efficient on single-stranded DNA (80). There is also evidence that UvrA, an ATPase and DNA-binding protein of the NER pathway, may be able to mitigate the cytotoxic effects of this lesion. One study used a neutral DNA equilibrium binding agent, Me-lex [N-methylpyrrolecarboxamide dipeptide (lex) modified with an O-methyl sulfonate ester functionality], to introduce selectively 3MeA lesions in the minor groove of DNA. It was shown that this agent shows increasing toxicity to E.coli mutants lacking one base excision repair (BER) enzyme (AlkA), two BER enzymes (AlkA and TAG) or both BER and NER repair capabilities (AlkA, TAG and UvrA), in that order (81).
3MeA is not considered to be a seriously promutagenic lesion based upon work done in bacterial and in yeast systems. In 3MeA-DNA-glycosylase I (tag)-deficient E.coli mutants, treatment with MNU leads to a 5-fold increase in mutation frequency only under SOS-induced conditions. Furthermore, in repair-proficient cells, removal of 3MeA from the DNA does not show a significant difference in mutagenesis in SOS-induced versus SOS-uninduced cells (82). To study the mutational profile of 3MeA in eukaryotic cells, the p53 gene cDNA on a yeast expression vector was treated with Me-lex in vitro and transfected into a yeast strain containing the p53-dependent reporter ADE2 gene. The results show that Me-lex is a weak mutagen compared with MNU but that it induces A → T transversions as the most common genetic change (40% of all mutations) (83). Mutagenicity increased 2- to 3-fold in 3MeA-glycosylase-deficient strains, which suggests that the lesion driving the mutations is 3MeA (84). Interestingly, the methylated adenines in Me-lex-treated DNA give rise to mutations in a strictly sequence-specific manner.
The cytotoxicity of 3MeA is well established in the literature. In vitro studies showing chain termination one nucleotide 3′ to adenines in methylated DNA templates pointed to 3MeA as a strong block to DNA replication. 3MeA in DNA has also been shown to be toxic in E.coli (81). Using Me-lex in combination with 3-methyladenine-DNA-glycosylase-proficient and -deficient cell lines, Engelward et al. (85) showed that 3MeA can cause p53 induction, S phase arrest, sister chromatid exchange, chromosome aberrations and apoptosis in mammalian cells. As with N7-methylguanine (7MeG), enhanced repair of 3MeA by DNA-glycosylases of the BER pathway can lead to a flood of AP sites that can also contribute to mutations and lethality (86).
N7-methyladenine
N7-methyladenine (7MeA) is a minor lesion formed at a level 40-fold below that of 7MeG, which is typically the most abundant lesion in alkylated DNA (87). Like 7MeG, 7MeA possesses a cationic imidazole ring, which facilitates depurination and, alternatively, can favor hydrolysis of the five-membered ring to form the formamidopyrimidine (Fapy) derivative, Fapy-7MeA; this latter hydrolysis reaction is especially favored for the 7MeA in RNA (88), which has a stabilized glycosidic bond as compared with DNA. The half-life of 7MeA in DNA in vivo is only 2–3 hours, which is similar to its half-life in vitro at pH 7.2, 37°C (89). Fapy-7MeA is a mutagenic lesion displaying A → G transitions in single-stranded M13mp18 DNA transfected into SOS-induced E.coli (87,90). In these studies, dimethylsulfate-treated DNA was compared before and after treatment with alkali, which hydrolyzed the imidazole rings of N7-methylated adenines and guanines, forming the Fapy derivatives. Dimethylsulfate- and alkali-treated DNA was 60-fold more mutagenic than DNA treated with dimethylsulfate alone and showed mutations primarily at A:T sites.
N1-methylguanine
N1-methylguanine (1MeG) has been found both in vitro (67) and in vivo (91). With regard to biological relevance, the AlkB protein can repair 1MeG both in vitro and in vivo (75,92). The glycosylase AAG, which repairs 3MeA and a range of other lesions, is also active against 1MeG in vitro (93) but the in vivo relevance of AAG against this adduct has not been established as yet. 1MeG is a very strong block to replication, which can be partially overcome when the DNA lesion is partially repaired by AlkB; lesion bypass of 1MeG in vivo increases 8-fold from 2% in AlkB− cells to 16% in AlkB+ cells. Similarly, AlkB causes a reduction in the mutagenicity of 1MeG from a very high frequency of 80% in AlkB− cells to 4% in AlkB+ cells. Taken together, these data indicate that AlkB is a powerful protection against the mutagenic activity of this dangerous alkylated base. The mutational fingerprint of 1MeG reveals G → T (57% of all progeny), G → A (17%) and G → C (6%) mutations. In many instances, the induction of the SOS bypass polymerases results in increased bypass of a given lesion at the expense of reduced fidelity at the site of damage; however, the SOS polymerases are somewhat anti-mutagenic when they bypass this modified base (75).
N3-methylguanine
N3-methylguanine (3MeG) is thought to block replication in the same way as 3MeA does, but it is formed in DNA at a 15-fold lower level. The half-life of 3MeG in vivo has been shown to be 3–4 hours (89). It has been shown that E.coli alkA mutants are sensitive to alkylating agents even though they express Tag (94), which repairs 3MeA (a known cytotoxic lesion) as efficiently as AlkA on double-stranded DNA (80). This result suggests that 3MeG contributes to the toxic effects of alkylation seen in these cells.
Using cell extracts from adapted E.coli, it was shown that the AlkA protein can repair 3MeG present on methylated DNA in vitro. The same study also shows persistence of this adduct in unadapted E.coli 30 min after exposure to MNNG (95). A second in vitro study has shown that TAG also repairs 3MeG present on a synthetic G:C-rich double-stranded DNA sequence, albeit with an efficiency of only 1/70th that of AlkA (96).
N7-methylguanine and its degradation products
The N7 atom of guanine is the most chemically vulnerable site to attack by alkylating electrophiles as it has the highest negative electrostatic potential of all the other atoms within the DNA bases (97). This property also makes it a highly reactive ligand for metal ions such as platinum (98). When double-stranded DNA is treated with MMS or MNNG, 82 and 67% of the methylation occurs on the N7 position of guanine, respectively (13). Within the cell, 7MeG is produced at the rate of 4000 residues/human genome/day by the non-enzymatic reaction of SAM with DNA (77), and its steady-state level in repair-proficient cells is estimated to be 3000 bases (99). 7MeG has been detected in human DNA at the level of a few adducts per 107 bases (100). 7MeG by itself does not have any major mutagenic or cytotoxic effects. However, methylation at the N7 position destabilizes the N-glycosidic bond leading to spontaneous depurination of this lesion (101) and the resulting AP sites are toxic. AP sites can also be formed during repair of 7MeG by N-alkylpurine DNA-glycosylases, which are part of the BER pathway. Although not examined directly in the context of alkylation, the mutagenic and toxic properties of AP sites have been thoroughly investigated (86).
In addition to its role as a source of AP sites, 7MeG can manifest toxicity by converting to its imidazole ring-opened form. Hydrolysis of the imidazole ring of 7MeG forms 2,6-diamino-4-hydroxy-5N-methyl-formamidopyrimidine (Fapy-7MeG). Whereas this lesion does not cause misparing with deoxyadenosine monophosphate (dAMP) or deoxythymidine monophosphate (dTMP), in vitro experiments using E.coli DNA polymerase I and poly[ 5′-deoxyguanosine-deoxycytidine-3′ (dGpC)] templates (102) or Klenow fragment and M13mp18 template DNA (103) show that Fapy-7MeG blocks DNA chain elongation. Fapy-7MeG lesions present on M13mp18 phage template DNA also lead to a 2- to 3-fold increase in G → C and G → T transversions when transfected into SOS-induced E.coli (87). However, DNA polymerase I preferentially incorporates deoxycytidine monophosphate (dCMP) opposite Fapy-7MeG and a Fapy-7MeG:C pair is extended most efficiently compared with other possibilities. This property makes Fapy-7MeG a lesion with weak mutagenic potential (88).
In E.coli, AlkA is known to excise 7MeG from methylated DNA (95). In humans, this reaction is carried out by AAG/MPG (104). There exist specific DNA-glycosylases in E.coli [formamidopyrimidine-DNA-glycosylase (Fpg)] and mammalian cells [human 7,8-dihydro-8-oxoguanine DNA glycosylase (hOGG1)] (105) that remove Fapy-7MeG lesions. Escherichia coli Fpg repairs 7MeG very efficiently, with a Km in the nanomolar range (88). It has been shown in a mammalian cell line by site-directed mutagenesis that over-expression of MPG sensitizes cells to alkylation damage by converting 7MeG into toxic AP sites, which lead to strand breaks. 7MeG by itself is not toxic to cells, nor is over-expression of MPG, but in combination, they can overwhelm the cell with AP sites leading to cytotoxicity. Rinne et al. (106) propose that these two aspects combined with appropriate delivery systems could be exploited for the selective targeting of tumor cells, thereby reducing the peripheral effects of DNA damage by drugs.
N3-methylcytosine and N3-ethylcytosine
N3-methylcytosine (3MeC) is formed by SN2 agents such as MMS and the naturally occurring methyl halides (15) preferentially in single-stranded DNA. It has been detected both in vitro (62,64–68,107,108) and in vivo (64,70,71,91,108). The corresponding ethyl homolog, N3-ethylcytosine (3EtC), is formed by ethylating agents in single-stranded DNA and also has been detected in vitro (64,65) and in vivo (64,109). As with the 1-alkyladenines, these lesions probably exist only or predominantly in single-stranded DNA because this site of modification is normally protected by base pairing (73). 3MeC stalls DNA synthesis and is likely to be toxic (15).
In E.coli, the AlkB protein has good activity against 3MeC and 3EtC both in vitro and in vivo (62,63,75). The appreciable mutagenesis and toxicity of the 3-alkylcytosines in vivo is decimated by AlkB, although a portion of the toxicity can also be overcome by induction of the SOS bypass polymerases. With regard to mutagenic potential, if a cell has no AlkB and uninduced SOS bypass polymerases, 3MeC and 3EtC are 30% mutagenic, with the predominant mutations being C → T and C → A. Basal expression of AlkB of a few molecules per cell abrogates the mutagenicity of 3MeC and 3EtC, whereas expression of SOS bypass polymerases in the absence of AlkB increases the mutagenicity of both lesions to a striking 70%. Although investigations involving replication past 1MeA and 1MeG, which similarly have a blocked Watson–Crick hydrogen bonding face, by DNA polymerases in vitro are lacking, it is known from in vitro studies that 3MeC inhibits replication by DNA polymerase I and does not cause mutation (107,110,111). However, some adduct bypass occurs with the incorporation of dAMP and dTMP opposite 3MeC (107). Therefore, the rules for misreplication of the 3-alkylcytosine lesions are the same both in vitro and in vivo, although the replicative system in cells is capable of a much higher mutation rate than is achieved in vitro (75,107).
N3-methylthymine
N3-methylthymine (3MeT) has been found both in vitro (64,65,67,68,112) and in vivo (112,113) and is formed through the reaction of DNA with SN2 alkylating agents such as MMS. This adduct is a very weak substrate for AlkB, and it is a strong block to replication in vivo, which can be only slightly overcome by SOS bypass polymerase induction (75). Recently, fat mass obesity associated protein (FTO) has been shown as a 2-oxoglutarate-dependent demethylase for nucleic acid (114,115). FTO can efficiently repair 3MeT in single-stranded DNA but not in double-stranded DNA; it also shows strong activity on the demethylation of 3-methyluracil in single-stranded RNA (114,116). While there are numerous epidemiological studies associating the FTO gene with obesity, the biological basis for metabolic effects of this gene are still under investigation.
From the standpoint of its potential to induce genetic change, 3MeT is ∼60% mutagenic in SOS−/AlkB− cells, providing mostly T → A (47%) and T → C (9%) mutations. Studies performed in vitro also show that 3MeT is a strong block to the Klenow fragment of DNA polymerase I, which slightly increases deoxythymidine triphosphate (dTTP) incorporation on a poly(dC-d3MeT) template (117); interestingly, T is exclusively incorporated opposite the analogous 3-ethyldeoxythymidine adduct in one study (118), whereas A is exclusively incorporated in another (119).
8-Methylguanine
C8-alkylated DNA bases exist but have not been reported extensively in the literature. Recent studies have suggested that carbon-centered radicals can be a source of C8-alkylated lesions. 8-Methylguanine (8MeG) was shown to be produced in vitro in RNA (120) and DNA (121) by methyl radicals generated by oxidation of 1,2-dimethylhydrazine and methylhydrazine, respectively. Proof of in vivo DNA alkylation by carbon-centered radicals was given by Netto et al. (122) who detected 8MeG in DNA isolated from the liver and colon of rats administered 1,2-dimethylhydrazine. Other studies have shown that this lesion can also be produced in vitro and in vivo by genotoxic agents such as tert-butylhydroperoxide, diazoquinones and arenediazonium ions (123). These findings are significant as they suggest a possible contribution of 8MeG in the carcinogenic effects of these agents, especially 1,2-dimethylhydrazine, which induces adenocarcinomas of the colon in rodents.
Site-specific studies using 8MedG-containing oligonucleotides prepared by phosphoramidite synthesis have explored the mutagenicity and toxicity of this lesion. It was found that 8MeG on the template strand blocks in vitro extension of DNA by mammalian polymerase α, but not by the E.coli Klenow fragment (124). The products from the primer extension reaction were then analyzed for mutations. 8MeG was found to direct exo− Klenow fragment-based incorporation of dCMP most of the time (77%) but also paired occasionally with deoxyguanosine monophosphate (dGMP) (1.1%) and dAMP (0.41%). Similar numbers were obtained for extension assays with mammalian polymerase α. Replication with the Klenow fragment also introduced small amounts of one (0.38%) and two (0.81%) base-pair deletions (124). These numbers mirror the thermodynamic stability of the 8MeG:deoxynucleoside monophosphate base (dNMP) pair, decreasing in the order dCMP > dGMP > dAMP >> dTMP. 1,2-Dimethylhydrazine induces both O6MeG and 8MeG in similar amounts in the DNA of rats (122). However, the mutation frequencies of 8MeG are two orders of magnitude less than those of O6MeG (125). Therefore, we may conclude that 8MeG is a weakly mutagenic lesion that in principle can contribute to G → C transversions in cells.
The repair of 8MeG has been studied in vitro by Gasparutto et al. (123). In this study, the authors incorporated 8MeG site specifically into oligonucleotides and probed the ability of bacterial, yeast and mammalian glycosylases to repair this lesion. Of the extensive list of enzymes evaluated, only AlkA was able to excise 8MeG. Human MPG did not repair 8MeG, nor did any of the glycosylases involved in repair of oxidative damage (Fpg, Nth of E.coli; Ntg1, Ntg2, Ogg1 of Saccharomyces cerevisiae and human Ogg1) (123).
8MeG has been shown to stabilize the Z-conformation of DNA in short oligonucleotides even in low salt concentrations. This property may be relevant in vivo as Z-DNA is thought to have a role in the regulation of DNA supercoiling (126). This lesion is also used as a chemical modification to stabilize quadruplex structures of G-rich sequences of DNA, which are proposed to have a role in telomeric DNA stability and in repression of transcription at the c-myc promoter (127). The wide range of potential biological activities of this lesion makes it a prime target for future investigations.
1,N6-ethenoadenine and 1,N6-ethanoadenine
The formation of 1,N6-ethenoadenine (eA) results from the reaction of adenine with products of unsaturated lipid peroxidation (128–131). This bifunctional DNA lesion arises endogenously under normal physiological conditions in both rodents and humans (132,133). Of great toxicological concern is the observation that eA is induced by common industrial agent vinyl chloride and its metabolites, such as chloroacetaldehyde. eA also occurs in chronically inflamed human and rodent tissues (134). Oxidative stress associated with inflammation is increasingly being linked to neurological disease, cancer promotion and accelerated aging (135).
In duplex DNA, eA can be repaired in vitro by glycosylases of the BER pathway (93,136). Mammalian cells can also repair etheno lesions by this route in vivo (137,138). Indeed, the BER enzyme AAG and its homologs are likely to be the primary vehicles of repair of eA in the duplex genomes of eukaryotes. In contrast, the in vivo repair of etheno adducts in E.coli was not clearly understood until recently; for example, one early study showed that neither BER nor NER figures prominently in etheno lesion repair (139). Early genetic studies on the mutagenicity of eA in E.coli reinforced this conundrum (140). The eA adduct was neither toxic nor mutagenic despite the fact that the base lacks any structural possibility of Watson–Crick complementarity. The issues raised in these studies were resolved in 2005 when biochemical studies provided the possibility that the direct reversal enzyme, AlkB, may play a significant role in the defense of cells against this type of bifunctional DNA damage. These biochemical studies showed that AlkB and its human homolog ABH3 can efficiently repair eA in vitro (141,142). AlkB uses a unique iron-mediated biochemical reaction involving α-ketoglutarate as a cofactor to putatively epoxidize the exocyclic double bond of eA. An epoxide may be hydrolyzed to a glycol with the glycol moiety being liberated as the dialdehyde, glyoxal. The direct reversal mechanism is also likely to be operative in vivo, as evidenced by genetic studies in which a single-stranded vector containing a single eA was replicated in AlkB-proficient and -deficient E.coli cells. In AlkB-deficient cells, eA is 35% mutagenic, yielding 25% A → T, 5% A → G and 5% A → C mutations. SOS induction causes an increased incorporation of dAMP opposite to eA (141).
1,N6-ethanoadenine (EA) is the chemically reduced form of eA and forms through the reaction of adenine with the antitumor drug bis-chloroethylnitrosourea. EA can be weakly repaired by the E.coli enzyme AlkA (143) and the corresponding human enzyme AAG (93,144), which suggested that BER is a means of repair of this adduct. Recent work, however, by Frick et al. shows that the direct reversal repair enzyme AlkB easily alleviates the toxicity of EA in E.coli in vivo (145). In an AlkB-proficient cell, EA is almost non-toxic (i.e. easily bypassed) and not significantly mutagenic. However, in AlkB-deficient cells, EA is extremely toxic, showing an 86% reduction in replication. The adduct is weakly mutagenic causing A → C (2%), A → G (1%) and A → T (1%) mutations (145).
1,N2-ethenoguanine and 3,N2-ethenoguanine
1,N2-ethenoguanine (1,2-eG) and its isomer 3,N2-ethenoguanine (2,3-eG) are cyclic DNA adducts formed, as with eA, by reagents such as chloroacetylaldehyde (146) or 4-hydroperoxy-2-nonenal (147). Significantly, the former has been found in the liver DNA of rodents exposed to vinyl chloride (146). 1,2-eG can moderately block DNA polymerase and cause G → T and G → C base substitutions, as well as frameshift mutations (148). It can be repaired by mammalian uracil-DNA glycosylase (MUG) and AAG (93,149). In recent work, 1,2-eG was shown to be repaired, albeit weakly, by BER using a truncated form of the AAG enzyme (93). The AlkA protein can release 2,3-eG from DNA (150). The glycosidic bond of 2,3-eG is extremely labile, a property that has made assessment of biological significance of this modified base a difficult task (146). Nevertheless, Loeb and colleagues successfully determined the mutation frequency of the lesion to be ∼13% in E.coli, where it primarily induces G → A transitions.
3,N4-ethenocytosine
3,N4-ethenocytosine (eC) is produced from the same precursors and by the same pathways that generate eA in DNA (128–130,151). As with eA, the BER pathway ([human thymine-DNA-glycosylase (hTDG)] in human and [double-stranded uracil-DNA-glycosylase (dsUDG)] in E.coli) is an established strategy used by nature to suppress the biological effects of this adduct (137,151). The cellular defense network against eC additionally involves the AlkB pathway, at least in E.coli., which should be mechanistically similar to that of eA repair by AlkB (141). In E.coli, AlkB has a modest effect on eC toxicity but reduces the mutation rate of the adduct by about two-thirds from 82% in AlkB-deficient cells to 37% in AlkB-proficient hosts, implying incomplete conversion to cytosine prior to polymerase traversal. The mutations of eC in AlkB-deficient and -proficient cells are C → A and C → T, which are of approximately equal abundance in each cellular background.
Perspective
Thirty years ago, when Carcinogenesis was a new Journal, the field of cancer research looked very different from the way it looks today. The field was richly populated by chemists who identified carcinogens and studied the molecular transformations whereby those agents damaged DNA. The work described in this review started shortly after the Journal began when the complexities of DNA adduction confounded attempts to relate specific types of DNA damage with genetic changes that, presumably, attend the conversion of a normal cell into a fully malignant one. From that time to the present, much has been learned. Many oncogenes and tumor-suppressor genes have been discovered and placed like footsteps on the path between normalcy and malignancy (152). More recently, linkages have been made between the genetic events of oncogene activation and tumor-suppressor gene inactivation and parallel disruptions in biochemical networks. These studies are revealing the secrets of how cancer cells obtain the energy and the raw materials to finance their growth into a tumor (153,154). One revelation to come out of the last few decades is that the number of mutations in cancers is far in excess of the number one would expect on the basis of normal replication errors or perhaps even the enhanced rate of replication errors that occur when a polymerase tries to copy past a mutagenic DNA lesion such as those described in the manuscript. While it seems likely that genetic changes induced by carcinogens are an important step in the early stage of malignant transformation, it now seems clear that we need to find other chemical or biochemical events that underpin the ‘mutator phenotype’ of tumors (155). Answers may come from studies of virally induced diseases, such as human immunodeficiency virus and hepatitis, where recent work has discovered enzymatic DNA-targeted base deamination systems that cause a high density of mutations within a genome (156). Answers might also come from the field of immunology where enzymes such as activation-induced deaminase cause, once again, a high density of mutations in a localized stretch of DNA (157,158).
One of the most important contributions of work on the chemical biology of mutagenesis has been the collateral impact of this field on the nearby field of DNA repair. It is now common for workers in the repair field to use oligonucleotides with single lesions, originally made for studies of mutagenesis, to characterize the detailed biochemical mechanisms by which repair enzymes or complexes reverse the damage. Moreover, studies of mutagenesis done using cells that are defective in a specific repair enzyme (141) or that express specialized polymerases (159) have provided high-quality data that have established the physiological relevance of specific enzymes as protectors from damage or as the vehicles by which damage is processed into events with disastrous consequences for the cell and organism.
Looking ahead, there is much to do. To give one example, the process of inflammation is clearly associated with cancer development (135). The range of DNA damages created by inflammation-generated reactive oxygen and nitrogen species is vast, and the task will be a large one to determine how each of these lesions contributes to the biological endpoints downstream of an inflammatory event. As a second example, workers will soon develop modified versions of the tools described herein to probe what may become a new field … RNA repair. Some mRNA species are so long that it takes a day to transcribe them (160–162). These important molecules not only need to have their informational integrity protected but also the energy used in their synthesis is large and would be wasted if a single lesion, for example, an eA residue, made them unreadable. Finally, while this review focuses on only one class of lesion, the small alkylated bases, it illustrates how much can be learned about the chemical rules of mutagenesis. Ten years ago, studies of the mutagenic properties of 5-hydroxycytosine (163), which induces C to T transitions, were the starting point for a novel application in the development of anti-viral agents (164). It is expected that additional examples of this nature, in which basic studies of mutagenesis drive clinical development, will help propel this field into a robust future.
Funding
National Institutes of Health (CA080024, CA26731, ES02109).
Acknowledgments
We thank James C. Delaney and Bogdan Fedeles for editorial and artistic contributions.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AAG
human 3-methyladenine-DNA glycosylase
- AP site
apurinic site
- BER
base excision repair
- dAMP
deoxyadenosine monophosphate
- dCMP
deoxycytidine monophosphate
- dGMP
deoxyguanosine monophosphate
- dNMP
deoxynucleoside monophosphate
- dTTP
deoxythymidine triphosphate
- EA
1,N6-ethanoadenine
- eA
1,N6-ethenoadenine
- eC
3,N4-ethenocytosine
- 1EtA
N1-ethyladenine
- 3EtC
N3-ethylcytosine
- Fapy
formamidopyrimidine
- 1MeA
N1-methyladenine
- 3MeA
N3-methyladenine
- 7MeA
N7-methyladenine
- 3MeC
N3-methylcytosine
- 1MeG
N1-methylguanine
- 3MeG
N3-methylguanine
- 7MeG
N7-methylguanine
- 8MeG
8-methylguanine
- MePT
methylphosphotriester
- 3MeT
N3-methylthymine
- MGMT
O6-methylguanine-DNA methyltransferase
- MMR
mismatch repair
- MMS
methylmethanesulfonate
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- MNU
N-methyl-N-nitrosourea
- MGP
N-methylpurine-DNA glycosylase
- NER
nucleotide excision repair
- O6EtG
O6-ethylguanine
- O6MeG
O6-methylguanine
- O4MeT
O4-methylthymine
- SAM
S-adenosylmethionine
- TAG
3-methyladenine-DNA glycosylase I
References
- 1.Loeb LA, et al. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 2008;68:6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conticello S. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu AK, et al. Site-specifically modified oligodeoxynucleotides as probes for the structural and biological effects of DNA-damaging agents. Chem. Res. Toxicol. 1988;1:1–18. doi: 10.1021/tx00001a001. [DOI] [PubMed] [Google Scholar]
- 4.Groopman JD, et al. Protective interventions to prevent aflatoxin-induced carcinogenesis in developing countries. Annu. Rev. Public Health. 2008;29:187–203. doi: 10.1146/annurev.publhealth.29.020907.090859. [DOI] [PubMed] [Google Scholar]
- 5.Essigmann JM, et al. Interactions of aflatoxin B1 and alkylating agents with DNA: structural and functional studies. Cold Spring Harb. Symp. Quant. Biol. 1983;47:327–337. doi: 10.1101/sqb.1983.047.01.038. [DOI] [PubMed] [Google Scholar]
- 6.West JD, et al. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem. Res. Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 7.Loechler EL, et al. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc. Natl Acad. Sci. USA. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl T, et al. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 9.Taverna P, et al. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J. Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuker DEG, et al. Nitrosated glycine derivatives as a potential source of O6-methylguanine in DNA. Cancer Res. 1997;57:366–369. [PubMed] [Google Scholar]
- 11.Loveless A. Possible relevance of O6-alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 12.Margison GP, et al. Chemical carcinogenesis in the nervous system. Preferential accumulation of O6-methylguanine in rat brain deoxyribonucleic acid during repetitive administration of N-methyl-N-nitrosourea. Biochem. J. 1975;148:521–530. doi: 10.1042/bj1480521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 14.Engelbergs J, et al. Role of DNA repair in carcinogen-induced ras mutation. Mutat. Res. 2000;450:139–153. doi: 10.1016/s0027-5107(00)00021-x. [DOI] [PubMed] [Google Scholar]
- 15.Sedgwick B. Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 16.Samson L, et al. A new pathway for DNA repair in Escherichia coli. Nature. 1977;267:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- 17.Sedgwick B, et al. Functional domains and methyl acceptor sites of the Escherichia coli Ada protein. J. Biol. Chem. 1988;263:4430–4433. [PubMed] [Google Scholar]
- 18.He C, et al. A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli Ada. Mol. Cell. 2005;20:117–129. doi: 10.1016/j.molcel.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Myers LC, et al. Metal dependence of transcriptional switching in Escherichia coli Ada. J. Biol. Chem. 1995;270:6664–6670. doi: 10.1074/jbc.270.12.6664. [DOI] [PubMed] [Google Scholar]
- 20.Mitra S, et al. O6-methylguanine-DNA methyltransferase in wild-type and ada mutants of Escherichia coli. J. Bacteriol. 1982;152:534–537. doi: 10.1128/jb.152.1.534-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebeck GW, et al. Characterization of the major DNA repair methyltransferase activity in unadapted Escherichia coli and identification of a similar activity in Salmonella typhimurium. J. Bacteriol. 1989;171:4563–4568. doi: 10.1128/jb.171.9.4563-4568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassanfar M, et al. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine. Suggestive evidence for O4-methylthymine repair by eukaryotic methyltransferases. J. Biol. Chem. 1991;266:2767–2771. [PubMed] [Google Scholar]
- 23.Paalman SR, et al. Specificity of DNA repair methyltransferases determined by competitive inactivation with oligonucleotide substrates: evidence that Escherichia coli Ada repairs O6-methylguanine and O4-methylthymine with similar efficiency. Biochemistry. 1997;36:11118–11124. doi: 10.1021/bi970740t. [DOI] [PubMed] [Google Scholar]
- 24.Rebeck GW, et al. A second DNA methyltransferase repair enzyme in Escherichia coli. Proc. Natl Acad. Sci. USA. 1988;85:3039–3043. doi: 10.1073/pnas.85.9.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potter PM, et al. Characterisation and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E.coli. Nucleic Acids Res. 1987;15:9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esteller M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 27.Esteller M, et al. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 28.Voigt JM, et al. Repair of O6-methylguanine by ABC excinuclease of Escherichia coli in vitro. J. Biol. Chem. 1989;264:5172–5176. [PubMed] [Google Scholar]
- 29.Samson L, et al. Alternative pathways for the in vivo repair of O6-alkylguanine and O4-alkylthymine in Escherichia coli: the adaptive response and nucleotide excision repair. EMBO J. 1988;7:2261–2267. doi: 10.1002/j.1460-2075.1988.tb03066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaney JC, et al. Effect of sequence context on O6-methylguanine repair and replication in vivo. Biochemistry. 2001;40:14968–14975. doi: 10.1021/bi015578f. [DOI] [PubMed] [Google Scholar]
- 31.Chambers RW, et al. uvrA and recA mutations inhibit a site-specific transition produced by a single O6-methylguanine in gene G of bacteriophage ΦX174. Proc. Natl Acad. Sci. USA. 1985;82:7173–7177. doi: 10.1073/pnas.82.21.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nivard MJ, et al. Mutational spectra induced under distinct excision repair conditions by the 3 methylating agents N-methyl-N-nitrosourea, N-methyl-N’-nitro-N-nitrosoguanidine and N-nitrosodimethylamine in postmeiotic male germ cells of Drosophila. Mutat. Res. 1996;352:97–115. doi: 10.1016/0027-5107(96)00011-5. [DOI] [PubMed] [Google Scholar]
- 33.Tosal L, et al. In vivo repair of ENU-induced oxygen alkylation damage by the nucleotide excision repair mechanism in Drosophila melanogaster. Mol. Genet. Genomics. 2001;265:327–335. doi: 10.1007/s004380000419. [DOI] [PubMed] [Google Scholar]
- 34.Bronstein SM, et al. Efficient repair of O6-ethylguanine, but not O4-ethylthymine or O2-ethylthymine, is dependent upon O6-alkylguanine-DNA alkyltransferase and nucleotide excision repair activities in human cells. Cancer Res. 1992;52:2008–2011. [PubMed] [Google Scholar]
- 35.Rasmussen LJ, et al. The Escherichia coli MutS DNA mismatch binding protein specifically binds O6-methylguanine DNA lesions. Carcinogenesis. 1996;17:2085–2088. doi: 10.1093/carcin/17.9.2085. [DOI] [PubMed] [Google Scholar]
- 36.Pauly GT, et al. Mutagenesis in Escherichia coli by three O6-substituted guanines in double-stranded or gapped plasmids. Biochemistry. 1995;34:8924–8930. doi: 10.1021/bi00027a045. [DOI] [PubMed] [Google Scholar]
- 37.Rye PT, et al. Mismatch repair proteins collaborate with methyltransferases in the repair of O6-methylguanine. DNA Repair (Amst.) 2008;7:170–176. doi: 10.1016/j.dnarep.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pauly GT, et al. Comparison of mutagenesis by O6-methyl- and O6-ethylguanine and O4- methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis. 1998;19:457–461. doi: 10.1093/carcin/19.3.457. [DOI] [PubMed] [Google Scholar]
- 39.Pauly GT, et al. Mutagenesis by O6-methyl-, O6-ethyl-, and O6-benzylguanine and O4-methylthymine in human cells: effects of O6-alkylguanine-DNA alkyltransferase and mismatch repair. Chem. Res. Toxicol. 2001;14:894–900. doi: 10.1021/tx010032f. [DOI] [PubMed] [Google Scholar]
- 40.Engelbergs J, et al. Fast repair of O6-ethylguanine, but not O6-methylguanine, in transcribed genes prevents mutation of H-ras in rat mammary tumorigenesis induced by ethylnitrosourea in place of methylnitrosourea. Proc. Natl Acad. Sci. USA. 1998;95:1635–1640. doi: 10.1073/pnas.95.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glassner BJ, et al. DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis. 1999;14:339–347. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- 42.Snow ET, et al. Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J. Biol. Chem. 1984;259:8095–8100. [PubMed] [Google Scholar]
- 43.Abbotts J, et al. Expression of human DNA polymerase β in Escherichia coli and characterization of the recombinant enzyme. Biochemistry. 1988;27:901–909. doi: 10.1021/bi00403a010. [DOI] [PubMed] [Google Scholar]
- 44.Karran P, et al. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 45.Goldmacher VS, et al. Isolation and partial characterization of human cell mutants differing in sensitivity to killing and mutation by methylnitrosourea and N-methyl-N’-nitro-N-nitrosoguanidine. J. Biol. Chem. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 46.York SJ, et al. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. J. Biol. Chem. 2006;281:22674–22683. doi: 10.1074/jbc.M603667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newlands ES, et al. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat. Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 48.Chambers RW. Site-specific mutagenesis in cells with normal DNA repair systems: transitions produced from DNA carrying a single O6-alkylguanine. Nucleic Acids Res. 1991;19:2485–2488. doi: 10.1093/nar/19.9.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambers RW. Site-directed mutagenesis in single cells: transitions produced by DNA carrying a single O6-alkylguanine residue. Mutat. Res. 1993;299:123–133. doi: 10.1016/0165-1218(93)90090-z. [DOI] [PubMed] [Google Scholar]
- 50.Ellison KS, et al. Site-specific mutagenesis by O6-alkylguanines located in the chromosomes of mammalian cells: influence of the mammalian O6-alkylguanine-DNA alkyltransferase. Proc. Natl Acad. Sci. USA. 1989;86:8620–8624. doi: 10.1073/pnas.86.22.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franco R, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 52.Dolan ME, et al. Extent of formation of O4-methylthymidine in calf thymus DNA methylated by N-methyl-N-nitrosourea and lack of repair of this product by rat liver O6-alkylguanine-DNA-alkyltransferase. Carcinogenesis. 1985;6:1611–1614. doi: 10.1093/carcin/6.11.1611. [DOI] [PubMed] [Google Scholar]
- 53.Dosanjh MK, et al. Comparative mutagenesis of O6-methylguanine and O4-methylthymine in Escherichia coli. Biochemistry. 1991;30:7027–7033. doi: 10.1021/bi00242a031. [DOI] [PubMed] [Google Scholar]
- 54.Preston BD, et al. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc. Natl Acad. Sci. USA. 1986;83:8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altshuler KB, et al. Intrachromosomal probes for mutagenesis by alkylated DNA bases replicated in mammalian cells: a comparison of the mutagenicities of O4-methylthymine and O6-methylguanine in cells with different DNA repair backgrounds. Chem. Res. Toxicol. 1996;9:980–987. doi: 10.1021/tx960062w. [DOI] [PubMed] [Google Scholar]
- 56.Klein JC, et al. Role of nucleotide excision repair in processing of O4-alkylthymines in human cells. J. Biol. Chem. 1994;269:25521–25528. [PubMed] [Google Scholar]
- 57.Zak P, et al. Repair of O6-methylguanine and O4-methylthymine by the human and rat O6-methylguanine-DNA methyltransferases. J. Biol. Chem. 1994;269:730–733. [PubMed] [Google Scholar]
- 58.Samson L, et al. Mammalian DNA repair methyltransferases shield O4MeT from nucleotide excision repair. Carcinogenesis. 1997;18:919–924. doi: 10.1093/carcin/18.5.919. [DOI] [PubMed] [Google Scholar]
- 59.Duckett DR, et al. Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl Acad. Sci. USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCarthy TV, et al. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishizaki K, et al. Expression of the truncated E. coli O6-methylguanine methyltransferase gene in repair-deficient human cells and restoration of cellular resistance to alkylating agents. Mutat. Res. 1987;184:121–128. doi: 10.1016/0167-8817(87)90068-x. [DOI] [PubMed] [Google Scholar]
- 62.Trewick SC, et al. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 63.Falnes PO, et al. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 64.Singer B, et al. Molecular Biology of Mutagens and Carcinogens. New York: Plenum; 1983. [Google Scholar]
- 65.Beranek DT, et al. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980;1:595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- 66.Gomes JD, et al. Reverse-phase high-performance liquid chromatography of chemically modified DNA. Anal. Biochem. 1983;129:387–391. doi: 10.1016/0003-2697(83)90566-3. [DOI] [PubMed] [Google Scholar]
- 67.Chang CJ, et al. Chemical modification of deoxyribonucleic acids: a direct study by carbon-13 nuclear magnetic resonance spectroscopy. J. Org. Chem. 1983;48:5151–5160. [Google Scholar]
- 68.Ashworth DJ, et al. Chemical modification of nucleic acids. Methylation of calf thymus DNA investigated by mass spectrometry and liquid chromatography. Biomed. Mass Spectrom. 1985;12:309–318. doi: 10.1002/bms.1200120703. [DOI] [PubMed] [Google Scholar]
- 69.Margison GP, et al. Methylated purines in the deoxyribonucleic acid of various Syrian-golden-hamster tissues after administration of a hepatocarcinogenic dose of dimethylnitrosamine. Biochem. J. 1976;157:627–634. doi: 10.1042/bj1570627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faustman EM, et al. A method for the rapid quantitation of methylated hepatic DNA-purines using high pressure liquid chromatography. J. Pharmacol. Methods. 1980;4:305–312. doi: 10.1016/0160-5402(80)90050-9. [DOI] [PubMed] [Google Scholar]
- 71.Beranek DT, et al. Correlation between specific DNA-methylation products and mutation induction at the HGPRT locus in Chinese hamster ovary cells. Mutat. Res. 1983;110:171–180. doi: 10.1016/0027-5107(83)90026-x. [DOI] [PubMed] [Google Scholar]
- 72.Faustman-Watts EM, et al. DNA-purine methylation in hepatic chromatin following exposure to dimethylnitrosamine or methylnitrosourea. Biochem. Pharmacol. 1984;33:585–590. doi: 10.1016/0006-2952(84)90312-5. [DOI] [PubMed] [Google Scholar]
- 73.Bodell WJ, et al. Influence of hydrogen bonding in DNA and polynucleotides on reaction of nitrogens and oxygens toward ethylnitrosourea. Biochemistry. 1979;18:2860–2863. doi: 10.1021/bi00580a029. [DOI] [PubMed] [Google Scholar]
- 74.Engel JD. Mechanism of the Dimroth rearrangement in adenosine. Biochem. Biophys. Res. Commun. 1975;64:581–586. doi: 10.1016/0006-291x(75)90361-7. [DOI] [PubMed] [Google Scholar]
- 75.Delaney JC, et al. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl Acad. Sci. USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duncan T, et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rydberg B, et al. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singer B. N-nitroso alkylating agents: formation and persistence of alkyl derivatives in mammalian nucleic acids as contributing factors in carcinogenesis. J. Natl Cancer Inst. 1979;62:1329–1339. [PubMed] [Google Scholar]
- 79.Fronza G, et al. The biological effects of N3-methyladenine. J. Cell. Biochem. 2004;91:250–257. doi: 10.1002/jcb.10698. [DOI] [PubMed] [Google Scholar]
- 80.Bjelland S, et al. Different efficiencies of the Tag and AlkA DNA glycosylases from Escherichia coli in the removal of 3-methyladenine from single-stranded DNA. FEBS Lett. 1996;397:127–129. doi: 10.1016/s0014-5793(96)01166-0. [DOI] [PubMed] [Google Scholar]
- 81.Shah D, et al. Evidence in Escherichia coli that N3-methyladenine lesions induced by a minor groove binding methyl sulfonate ester can be processed by both base and nucleotide excision repair. Biochemistry. 2001;40:1796–1803. doi: 10.1021/bi0024658. [DOI] [PubMed] [Google Scholar]
- 82.Chaudhuri I, et al. 3-Methyladenine mutagenesis under conditions of SOS induction in Escherichia coli. Carcinogenesis. 1991;12:2283–2289. doi: 10.1093/carcin/12.12.2283. [DOI] [PubMed] [Google Scholar]
- 83.Kelly JD, et al. Relationship between DNA methylation and mutational patterns induced by a sequence selective minor groove methylating agent. J. Biol. Chem. 1999;274:18327–18334. doi: 10.1074/jbc.274.26.18327. [DOI] [PubMed] [Google Scholar]
- 84.Monti P, et al. Influences of base excision repair defects on the lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent. J. Biol. Chem. 2002;277:28663–28668. doi: 10.1074/jbc.M203384200. [DOI] [PubMed] [Google Scholar]
- 85.Engelward BP, et al. A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. J. Biol. Chem. 1998;273:5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 86.Fortini P, et al. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Tudek B, et al. Mutagenic specificity of imidazole ring-opened 7-methylpurines in M13mp18 phage DNA. Acta Biochim. Pol. 1999;46:785–799. [PubMed] [Google Scholar]
- 88.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J. Biochem. Mol. Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 89.Lawley PD, et al. Removal of minor methylation products 7-methyladenine and 3-methylguanine from DNA of Escherichia coli treated with dimethyl sulphate. Chem. Biol. Interact. 1976;12:211–220. doi: 10.1016/0009-2797(76)90100-9. [DOI] [PubMed] [Google Scholar]
- 90.Tudek B, et al. Biological properties of imidazole ring-opened N7-methylguanine in M13mp18 phage DNA. Nucleic Acids Res. 1992;20:3079–3084. doi: 10.1093/nar/20.12.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Culp LA, et al. Methylated bases in DNA of animal origin. Arch. Biochem. Biophys. 1970;136:73–79. doi: 10.1016/0003-9861(70)90328-0. [DOI] [PubMed] [Google Scholar]
- 92.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee CY, et al. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Evensen G, et al. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982;296:773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- 95.Karran P, et al. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982;296:770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- 96.Bjelland S, et al. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pullman A, et al. Molecular electrostatic potential of the nucleic acids. Q. Rev. Biophys. 1981;14:289–380. doi: 10.1017/s0033583500002341. [DOI] [PubMed] [Google Scholar]
- 98.Jamieson ER, et al. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 99.Kunkel TA. The high cost of living. Trends Genet. 1999;15:93–94. doi: 10.1016/s0168-9525(98)01664-3. [DOI] [PubMed] [Google Scholar]
- 100.Szyfter K, et al. Tobacco smoke-associated N7-alkylguanine in DNA of larynx tissue and leucocytes. Carcinogenesis. 1996;17:501–506. doi: 10.1093/carcin/17.3.501. [DOI] [PubMed] [Google Scholar]
- 101.Saffhill R, et al. Mechanisms of carcinogenesis induced by alkylating agents. Biochim. Biophys. Acta. 1985;823:111–145. doi: 10.1016/0304-419x(85)90009-5. [DOI] [PubMed] [Google Scholar]
- 102.Boiteux S, et al. Imidazole open ring 7-methylguanine: an inhibitor of DNA synthesis. Biochem. Biophys. Res. Commun. 1983;110:552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- 103.O'Connor TR, et al. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988;16:5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O'Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Asagoshi K, et al. Enzymatic properties of Escherichia coli and human 7,8-dihydro-8-oxoguanine DNA glycosylases. Nucleic Acids Symp. Ser. 2000;44:11–12. doi: 10.1093/nass/44.1.11. [DOI] [PubMed] [Google Scholar]
- 106.Rinne ML, et al. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Res. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boiteux S, et al. Mutagenesis by alkylating agents: coding properties for DNA polymerase of poly (dC) template containing 3-methylcytosine. Biochimie. 1982;64:637–641. doi: 10.1016/s0300-9084(82)80103-x. [DOI] [PubMed] [Google Scholar]
- 108.Kawasaki H, et al. High-performance liquid chromatographic determination of 3-methylcytosine in deoxyribonucleic acid treated with carcinogenic methylating agents in vitro and in vivo. Chem. Pharm. Bull. 1985;33:1170–1174. doi: 10.1248/cpb.33.1170. [DOI] [PubMed] [Google Scholar]
- 109.Frei JV, et al. Alkylation of deoxyribonucleic acid in vivo in various organs of C57BL mice by the carcinogens N-methyl-N-nitrosourea, N-ethyl-N-nitrosourea and ethylmethanesulphonate in relation to induction of thymic lymphoma. Some applications of high-pressure liquid chromatography. Biochem. J. 1978;174:1031–1044. doi: 10.1042/bj1741031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abbott PJ, et al. DNA synthesis with methylated poly(dC-dG) templates. Evidence for a competitive nature to miscoding by O6-methylguanine. Biochim. Biophys. Acta. 1979;562:51–61. doi: 10.1016/0005-2787(79)90125-4. [DOI] [PubMed] [Google Scholar]
- 111.Saffhill R. Differences in the promutagenic nature of 3-methylcytosine as revealed by DNA and RNA polymerising enzymes. Carcinogenesis. 1984;5:691–693. doi: 10.1093/carcin/5.5.691. [DOI] [PubMed] [Google Scholar]
- 112.Den EL, et al. Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis. 1986;7:393–403. doi: 10.1093/carcin/7.3.393. [DOI] [PubMed] [Google Scholar]
- 113.Singer B, et al. Escherichia coli polymerase I can use O2-methyldeoxythymidine or O4-methyldeoxythymidine in place of deoxythymidine in primed poly(dA-dT).poly(dA-dT) synthesis. Proc. Natl Acad. Sci. USA. 1983;80:4884–4888. doi: 10.1073/pnas.80.16.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yi C, et al. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 2009;42:519–529. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jia G, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huff AC, et al. DNA damage at thymine N-3 abolishes base-pairing capacity during DNA synthesis. J. Biol. Chem. 1987;262:12843–12850. [PubMed] [Google Scholar]
- 118.Grevatt PC, et al. The role of N3-ethyldeoxythymidine in mutagenesis and cytotoxicity by ethylating agents. J. Biol. Chem. 1991;266:1269–1275. [PubMed] [Google Scholar]
- 119.Bhanot OS, et al. Incorporation of dA opposite N3-ethylthymidine terminates in vitro DNA synthesis. Biochemistry. 1990;29:10357–10364. doi: 10.1021/bi00497a010. [DOI] [PubMed] [Google Scholar]
- 120.Kang JO, et al. Methylation of RNA purine-bases by methyl radicals. Arch. Biochem. Biophys. 1993;306:178–182. doi: 10.1006/abbi.1993.1497. [DOI] [PubMed] [Google Scholar]
- 121.Augusto O, et al. Formation of 8-methylguanine as a result of DNA alkylation by methyl radicals generated during horseradish peroxidase-catalyzed oxidation of methylhydrazine. J. Biol. Chem. 1990;265:22093–22096. [PubMed] [Google Scholar]
- 122.Netto LE, et al. Identification of C8-methylguanine in the hydrolysates of DNA from rats administered 1,2-dimethylhydrazine. Evidence for in vivo DNA alkylation by methyl radicals. J. Biol. Chem. 1992;267:21524–21527. [PubMed] [Google Scholar]
- 123.Gasparutto D, et al. Excision of 8-methylguanine site-specifically incorporated into oligonucleotide substrates by the AlkA protein of Escherichia coli. DNA Repair. 2002;1:437–447. doi: 10.1016/s1568-7864(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 124.Kohda K, et al. Synthesis, miscoding specificity, and thermodynamic stability of oligodeoxynucleotide containing 8-methyl-2′-deoxyguanosine. Chem. Res. Toxicol. 1996;9:1278–1284. doi: 10.1021/tx9601059. [DOI] [PubMed] [Google Scholar]
- 125.Shibutani S. Quantitation of base substitutions and deletions induced by chemical mutagens during DNA synthesis in vitro. Chem. Res. Toxicol. 2002;6:625–629. doi: 10.1021/tx00035a006. [DOI] [PubMed] [Google Scholar]
- 126.Sugiyama H, et al. Synthesis, structure and thermodynamic properties of 8-methylguanine-containing oligonucleotides: Z-DNA under physiological salt conditions. Nucleic Acids Res. 1996;24:1272–1278. doi: 10.1093/nar/24.7.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu Y, et al. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.el Ghissassi F, et al. Formation of 1,N6-ethenoadenine and 3,N4-ethenocytosine by lipid peroxidation products and nucleic acid bases. Chem. Res. Toxicol. 1995;8:278–283. doi: 10.1021/tx00044a013. [DOI] [PubMed] [Google Scholar]
- 129.Chung FL, et al. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 130.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 131.Blair IA. Lipid hydroperoxide-mediated DNA damage. Exp. Gerontol. 2001;36:1473–1481. doi: 10.1016/s0531-5565(01)00133-4. [DOI] [PubMed] [Google Scholar]
- 132.Nair J, et al. 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytine in liver DNA from humans and untreated rodents detected by immunoaffinity/32P-postlabeling. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 133.Barbin A, et al. Endogenous deoxyribonucleic acid (DNA) damage in human tissues: a comparison of ethenobases with aldehydic DNA lesions. Cancer Epidemiol. Biomarkers Prev. 2003;12:1241–1247. [PubMed] [Google Scholar]
- 134.Barbin A. Etheno-adduct-forming chemicals: from mutagenicity testing to tumor mutation spectra. Mutat. Res. 2000;462:55–69. doi: 10.1016/s1383-5742(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 135.Hussain SP, et al. Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 136.Saparbaev M, et al. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Engelward BP, et al. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl Acad. Sci. USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ham AJ, et al. New immunoaffinity-LC-MS/MS methodology reveals that Aag null mice are deficient in their ability to clear 1,N6-etheno-deoxyadenosine DNA lesions from lung and liver in vivo. DNA Repair (Amst.) 2004;3:257–265. doi: 10.1016/j.dnarep.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 139.Pandya GA, et al. Escherichia coli responses to a single DNA adduct. J. Bacteriol. 2000;182:6598–6604. doi: 10.1128/jb.182.23.6598-6604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Basu AK, et al. Mutagenic and genotoxic effects of three vinyl chloride-induced DNA lesions: 1,N6-ethenoadenine, 3,N4-ethenocytosine, and 4-amino-5-(imidazol-2-yl)imidazole. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 141.Delaney JC, et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 142.Mishina Y, et al. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guliaev AB, et al. Chloroethylnitrosourea-derived ethano cytosine and adenine adducts are substrates for Escherichia coli glycosylases excising analogous etheno adducts. DNA Repair (Amst.) 2004;3:1311–1321. doi: 10.1016/j.dnarep.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 144.Guliaev AB, et al. Structural insights by molecular dynamics simulations into differential repair efficiency for ethano-A versus etheno-A adducts by the human alkylpurine-DNA N-glycosylase. Nucleic Acids Res. 2002;30:3778–3787. doi: 10.1093/nar/gkf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Frick LE, et al. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl Acad. Sci. USA. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cheng KC, et al. The vinyl chloride DNA derivative N2,3-ethenoguanine produces G –>A transitions in Escherichia coli. Proc. Natl Acad. Sci. USA. 1991;88:9974–9978. doi: 10.1073/pnas.88.22.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee SH, et al. 4-Hydroperoxy-2-nonenal-induced formation of 1,N2-etheno-2′-deoxyguanosine adducts. Chem. Res. Toxicol. 2005;18:780–786. doi: 10.1021/tx0497088. [DOI] [PubMed] [Google Scholar]
- 148.Langouet S, et al. Misincorporation of dNTPs opposite 1,N2-ethenoguanine and 5,6,7,9-tetrahydro-7-hydroxy-9-oxoimidazo[1,2-a]purine in oligonucleotides by Escherichia coli polymerases I exo- and II exo-, T7 polymerase exo-, human immunodeficiency virus-1 reverse transcriptase, and rat polymerase beta. Biochemistry. 1997;36:6069–6079. doi: 10.1021/bi962526v. [DOI] [PubMed] [Google Scholar]
- 149.Saparbaev M, et al. 1,N(2)-ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N-glycosylase. J. Biol. Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 150.Matijasevic Z, et al. Release of N2,3-ethenoguanine from chloroacetaldehyde-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Proc. Natl Acad. Sci. USA. 1992;89:9331–9334. doi: 10.1073/pnas.89.19.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saparbaev M, et al. 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hanahan D, et al. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 153.Zong WX, et al. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Loeb LA, et al. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- 156.Suspène R, et al. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Goodman MF, et al. AID-initiated purposeful mutations in immunoglobulin genes. In: Frederick WA, Tasuku H, editors. Advances in Immunology. Academic Press; 2007. pp. 127–155. [DOI] [PubMed] [Google Scholar]
- 158.Chelico L, et al. Stochastic properties of processive cytidine DNA deaminases AID and APOBEC3G. Phil. Trans. R. Soc. B. 2009;364:583–593. doi: 10.1098/rstb.2008.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Neeley WL, et al. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 160.Kabnick KS, et al. Determinants that contribute to cytoplasmic stability of human c-fos and ß-globin mRNAs are located at several sites in each mRNA. Mol. Cell. Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Morceau F, et al. GTP-mediated differentiation of the human K562 cell line: transient overexpression of GATA-1 and stabilization of the gamma-globin mRNA. Leukemia. 2000;14:1589–1597. doi: 10.1038/sj.leu.2401890. [DOI] [PubMed] [Google Scholar]
- 162.Yi X, et al. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999;19:3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kreutzer DA, et al. Oxidized, deaminated cytosines are a source of C–>T transitions in vivo. Proc. Natl Acad. Sci. USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Loeb LA, et al. Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res. Hum. Retroviruses. 2000;16:1–3. doi: 10.1089/088922200309539. [DOI] [PubMed] [Google Scholar]