Abstract
The mechanisms that regulate angiogenesis in hypoxia or hypoxic microenvironment are modulated by several pro- and antiangiogenic factors. Hypoxia-inducible factors (HIFs) have been established as the basic and major inducers of angiogenesis, but understanding the role of interacting proteins is becoming increasingly important to elucidate the angiogenic processes of a hypoxic response. In particular, with regard to wound healing and the novel therapies for vascular disorders such as ischemic brain and heart attack, it is essential to gain insights in the formation and regulation of HIF transcriptional machineries related to angiogenesis. Further, identification of alternative ways of inhibiting tumor growth by disrupting the growth-triggering mechanisms of increasing vascular supply via angiogenesis depends on the knowledge of how tumor cells develop their own vasculature. Here, we review our findings on the interactions of basic HIFs, HIF-1α and HIF-2α, with their regulatory binding proteins, histone deacetylase 7 (HDAC7) and translation initiation factor 6 (Int6), respectively. The present results and discussion revealed new regulatory interactions of HIF-related mechanisms.
Keywords: angiogenic proteins; anoxia; eukaryotic initiation factors; histone deacetylases; hypoxia-ischemia, brain; neovascularization, pathologic
Hypoxia and angiogenesis (vascular physiology and pathology)
Hypoxia means a reduction in the physiological oxygen level. It is caused by vascular and pulmonary diseases or by triggering of cancerous-tissue growth and leads to cellular dysfunction. There are 3 types of hypoxia, namely, chronic hypoxia, acute hypoxia, and hypoxia with reperfusion. Chronic hypoxia (oxygen tension, 2-3%), in particular, causes uncontrolled proliferation of cells under limited oxygen diffusion (Harris, 2002). In contrast, acute hypoxia, and particularly that with reperfusion, causes tumor vasculature-specific temporary closure of aberrant, irregular blood vessels with a following reversal of blood flow (Brown and Giaccia, 1998; Harris, 2002). In that case, even if the physiological oxygen level is restored, the increased blood flow supplies excessive oxygen; this results in increased free-radical concentration, tissue damage, and concomitant activation of stress-response genes-a condition known as "reoxygenation injury" (Prabhakar, 2001)-in the tissue. In chronic hypoxia, because of the relatively long duration of hypoxic conditions, normal cells/tissues cannot survive; however, tumor cells are able to continue proliferation within the hypoxic area, because these cells undergo genetic and adaptive changes leading to development of resistance against hypoxic conditions. One of the most well-studied and predominant hypoxic responses is the induction of angiogenic and growth factors, which lead to the formation and growth of new blood vessels (Figure 1). Blood vessels can grow in typically 2 ways: (1) vasculogenesis, in which they are derived from endothelial progenitors and (2) angiogenesis and arteriogenesis, which involves sprouting and collateral growth of mural cells, forming collateral bridges between arterial networks. When tissues grow beyond the physiological oxygen diffusion limit, the relative hypoxia triggers expansion of vascular beds by inducing angiogenic factors in the cells of the vascular beds, which are physiologically oxygenated by simple diffusion of oxygen. One of the angiogenic factors, VEGF, has been reported to be the most remarkable one stimulating angiogenesis in a strictly dose-dependent manner (Ferrara et al., 2003). Other factors such as angiopoietin-2/angiopoietin-1 (Graham et al., 1998; Phelan et al., 1998; Kietzmann et al., 1999), Tie2 (Kuwabara et al., 1995), PDGF (Negus et al., 1998; Wykoff et al., 2000), basic fibroblast growth factor (bFGF) (Sakuda et al., 1992), and monocyte chemoattractant protein 1 (MCP-1) (Phillips et al., 1995) have also been reported as indispensable factors responsible not only for increasing vascular permeability, endothelial sprouting, maintenance, differentiation and remodeling but also cell proliferation, migration, enhancement of endothelial assembly, and lumen acquisition (Figure 1). In hypoxia, angiogenesis is modulated because of the concomitant inflammation and by several factors secreted from immune cells, because leukocyte subtypes produce a myriad of angiogenic factors, various interleukins such as TGF-β1 and MCP-1, and proteinases (Vacca et al., 1998; Norrby, 2002). Thus, hypoxia provides an important environment not only for angiogenesis but also for related phenomena in the hypoxic or surrounding area; this implies that hypoxia is more than simply a regulator of angiogenesis (Paleolog, 2004).
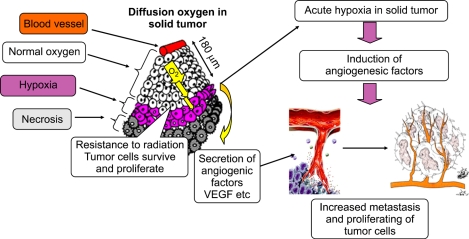
Figure 1.
Hypoxia and tumor angiogenesis: Hypoxia occurs in chronic and acute vascular diseases and tumor formation. It is toxic to normal cells, but cancer cells can survive and continue to proliferate in hypoxia. Human tumors grew like cords around blood vessels, and tumor cells located > 180 µm from the blood vessels were observed to become necrotic. Hypoxia induces angiogenesis during tumor growth; after tumor growth progresses, an oxygen gradient develops from the oxygen source to the periphery of the tumor. Cells lacking oxygen and nutrition because of their distance from the blood supply become necrotic, whereas those closer to the blood supply begin sensing hypoxia and secrete angiogenic factors. As a result, angiogenesis occurs and the tumor develops its own vasculature, independent of the original tissue.
Regulation of HIFs in hypoxia
Hypoxia inducible factors (HIFs) are transcriptional complexes, which are heterodimers consisting of HIF-α and HIF-β (aryl hydrocarbon receptor nuclear translocator [ARNT]) subunits. They belong to the Per/ARNT/Sim (PAS) subfamily of the basic helixloop-helix (bHLH) family of transcription factors. HIF-α subunits are the main hypoxia sensors; they are initially regulated by protein degradation, which ensures a quick response to oxygen reduction within 2 min (Jewell et al., 2001), compared with transcriptional regulation which produces a delayed response due to RNA processing and cytosolic translation. Constitutively expressed cytosolic HIF-α subunits are hydroxylated via two prolyl residues comprising Pro402 and Pro564 (Hon et al., 2002) in human HIF-1α, and Pro405 and Pro531 in human HIF-2α (Kondo et al., 2003). Under normoxic conditions, hydroxylation of the proline residues triggers protein degradation via ubiquitin-proteasome systems (Jaakkola et al., 2001; Ivan et al., 2001; Masson et al., 2001; Yu et al., 2001), a prerequisite for interaction with the von Hippel-Lindau (VHL) E3 ubiquitin ligase complex that contains elongin B, elongin C, Cul2, and Rbx1 (Kaelin, 2002) which only recognizes hydroxylated prolins as a target (Kaelin, 2002) (Figure 2). In hypoxia, the HIF heterodimer is formed by the association of HIF-α subunits with their binding partner HIF-1β forming an activated transcriptional core complex. HIF-1β proteins are constitutively expressed and not controlled by oxygen tensions within the cells/tissues, and they are involved in additional hypoxia-independent transcriptional activities related to dioxin response and estrogen metabolism (Whitlock 1999; Brunnberg et al., 2003). In addition to the protein degradation signaling, HIF-1α proteins are downregulated by an other oxygen-dependent inhibitor, factor inhibiting HIF-1 (FIH-1), which is an asparaginyl hydroxylase that hydroxylates an asparagine residue in the C-terminal transactivation domain (TAD), preventing the interaction of the proteins with the transcriptional coactivator p300 (Koivunen et al., 2004). Among the 3 HIF-α isoforms (Figure 2), HIF-1α and HIF-2α are apparently closely homologous and recognize hypoxia-responsive elements (HREs) in the promoter regions of 546 and 143 genes, respectively (Tian et al., 1997; Wiesener et al., 1998; Pugh as well as Ratcliffe, 2003; Mole et al., 2009). In contrast, HIF-3α is involved in negative regulation of hypoxic response, as an inhibitory PAS domain protein (Makino et al.,, 2001; Pugh and Ratcliffe, 2003).
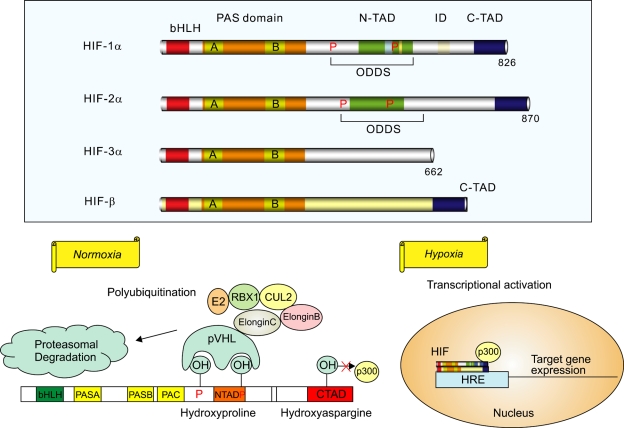
Figure 2.
HIF pathway: Upper panel: Known HIF-α isoforms and their cofactor HIF-β. HIF-1β and HIF-2α appear closely related, whereas HIF-3α seems to be involved in negative regulation of hypoxia response. Lower panel: Schematic diagram of HIF-1α regulation. In normoxia, the proline residues are hydroxylated and recognized by pVHL, which targets HIF-1α for degradation. pVHL is part of a large complex that comprises elongin B, elongin C, CUL2, RBX1, and a ubiquitin-conjugating enzyme (E2). The asparagine residue in the C-TAD of HIF-1α is an oxygen-dependent hydroxylation-regulated binding site for p300, which is inactive in its hydroxylated state. In hypoxia, prolyl hydroxylase cannot modify HIF-1α, and the protein remains stable. The stabilized HIF-1α is translocated to the nucleus, where it interacts with cofactors HIF-1β and p300 and transcribes hypoxia-related target genes. HIF, hypoxia-inducible factor; bHLH, basic helix-loop-helix; PAS, Per-ARNT-SIM; N-TAD, N-terminal transactivation domain; C-TAD, C-terminal transactivation domain; ID, inhibitory domain; ODDs, oxygen-dependent degradation domains; pVHL, von Hippel-Lindau protein; CUL2, cullin 2; RBX1, RING-box protein 1; E2, ubiquitin-conjugating enzyme; HRE, hypoxia-responsive element.
Recent studies on HIF-1α or HIF-2α inactivation in mice models (knockout mice) revealed different characteristics and functions of the 2 isoforms (Rosenberger et al., 2002; Holmquist-Mengelbier et al., 2006). The HIF-2α-knockout mice developed a syndrome of multiple-organ deficiency and biochemical abnormalities and showed altered gene expression patterns. Histological and ultrastructural analyses revealed retinopathy, hepatic steatosis, cardiac hypertrophy, skeletal myopathy, hypocellular bone marrow, azoospermia, and mitochondrial abnormalities in these mice. Analyses of serum and urine metabolites revealed hypoglycemia, lactic acidosis, altered Krebs cycle, and dysregulated fatty acid oxidation (Scortegagna et al., 2003). In contrast, the HIF-1α-knockout mice mainly showed abnormal vascular development and embryonic lethality (Ryan et al., 1998; Kotch et al., 1999), and superisingly, VEGF mRNA levels were raised in Hif-1 α-/- mice embryos. These distinct phenotypes observed in the HIF-1α- and HIF-2α-knockout mice indicate differences in tissue specificity and temporal expression patterns of the 2 isoforms, implying that substitution of one with the other is not possible. Raval et al. (2005) have suggested that HIF-1α and HIF-2α have distinct transcriptional targets. This is because in clear cell renal carcinomas (CCRCs), the usual predominance of HIF-1α expression in non-neoplastic renal tubules (Rosenberger et al., 2002) is altered strikingly in favor of HIF-2α expression in neoplastic epithelial cells; moreover, selective activation or suppression of HIFs has been reported. In mouse embryonic fibroblasts, HIF-2α is transcriptionally inactive and remains in the cytoplasm (Park et al., 2003), while there is evidence that a dose-dependent repressor restricts the activity of nuclear HIF-2α in embryonic stem cells (Hu et al., 2006). Furthermore, genetic manipulation in CCRC cells indicates that activation of HIF-2α, but not HIF-1α, promotes tumor growth (Kondo et al., 2002; Raval et al., 2005). This parallels patterns of transcriptional selectivity in CCRCs, by which HIF-2α drives the expression of genes encoding prosurvival factors such as VEGF, TGF-α, and cyclin D1, whereas HIF-1α drives the expression of genes encoding proapoptotic factors such as BCL2/adenovirus E1B-interacting protein 1, NIP3 (BNIP3) (Raval et al., 2005). In addition, HIF-1α primarily regulates transcriptional activation of VEGF in response to hypoxia and insulin-like growth factor-I (IGF-I), compared with HIF-2α in MCF-7 cells; in contrast, in renal carcinoma cells that constitutively express HIF-1α and HIF-2α because of loss of VHL function, it was observed that high basal VEGF, glucose transporter 1 (GLUT1), urokinase-type plasminogen activator receptor, and plasminogen activator inhibitor 1 expression was predominantly dependent on HIF-2α activity (Carroll and Ashcroft, 2006).
Here, we address the question whether distinct transcriptional responses of HIF-1α and HIF-2α might be integrated in a way that supports a particular type of physiological adaptation to hypoxia. Further, superimposed on target gene-specific patterns of transcriptional selectivity, there might be additional levels of control that limit or enhance HIF-2α activity in certain contexts. Thus, in our study, we focused on novel HIF-1α- and HIF-2α-binding proteins and discovered histone deacetylase 7 (HDAC7) and translation initiation factor 6 (Int6)/subunit of eukaryotic translation initiation factor 3 (eIF3e), respectively, by using the yeast two-hybrid system. Here, we demonstrate and discuss the regulation of both the HIF-α isoforms discussed above by the novel binding proteins, focusing on the interaction of HDAC7 with HIF-1α and that of Int6 with HIF-2α.
Role of HDAC7 in HIF-1α regulation
In our study, we used the yeast two-hybrid system to identify novel proteins that interact with the inhibitory domain (ID) of HIF-1α and may affect HIF-1α transcriptional activity. We identified HDAC7 as a HIF-1α-interacting protein, which is a transcriptional repressor belonging to the mammalian class II HDAC family. Class II HDAC family members include HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10. HDAC4, HDAC5, and HDAC7 are closely homologous to each other, whereas HDAC6, HDAC9, and HDAC10 share only the conserved HDAC domain (Kao et al., 2000; de Ruijter et al., 2003). HDAC7 plays a role in the repression of gene transcription by associating with HDAC3 in vivo via recruitment of corepressors-nuclear receptor corepressor (N-CoR) and silencing mediator for retinoic acid receptor (RAR) as well as thyroid hormone receptor (TR) (SMRT) in the nucleus. HDAC7, like 2 other class II HDAC family members HDAC4 and HDAC5, requires HDAC3 for enzymatic activity (Fischle et al., 2001, 2002; Guenther et al., 2001; Yang et al., 2002). HDAC4, HDAC5, and HDAC7 contain a highly homologous conserved catalytic domain (HDAC domain) in the C-terminal region (Figure 3). The N-terminal region and the C-terminal tail of HDAC7 are less homologous to the corresponding regions of HDAC4 and HDAC5. HDAC4, HDAC5, and HDAC7 also contain N-terminal nuclear localization signal sequences and C-terminal signal-responsive nuclear export signal sequences (Figure 3) (Bertos et al., 2001; McKinsey et al., 2001). In addition, the 3 proteins are known to shuttle between the cytoplasm and nucleus in a process regulated by calcium/calmodulin-dependent protein kinase (McKinsey et al., 2000; Kao et al., 2001). By studying clinical samples, Ouaissi et al. (2008) recently reported that HDAC7 expression is significantly associated with adenocarcinomas of the pancreas from clinical samples. In our study, we showed that HDAC7 forms a complex with HIF-1α and p300 in the nucleus under hypoxic conditions, resulting in enhanced transcription of HIF-1α target genes (VEGF and Glut-1). In contrast, HDAC4 and HDAC5 do not bind to HIF-1α. Moreover, HDAC7 translocates from the cytoplasm to the nucleus under hypoxic conditions. We propose that the HDAC domain of HDAC7 may be masked through HDAC7-HIF-1α interaction, causing interference in the binding of HIF-1α to N-CoR/SMRT corepressor under hypoxic conditions. Alternately, the ID of HIF-1α may act as a regulator of TAD function. Binding of HDAC7 to HIF-1α may lead to a conformational change within the ID of HIF-1α, resulting in facilitated binding of coactivators such as cyclic AMP response element-binding protein (CREB)-binding protein (CBP) and p300 and increased transcriptional activity under hypoxic conditions. HDAC7 regulates the transcriptional activity of HIF-1α, but not that of HIF-2α (Kato et al., 2004). Previous reports suggest that HDAC7 silencing in endothelial cells alters cell morphology, migration, and capacity to form capillary tube-like structures in vitro but does not affect cell adhesion, proliferation, or apoptosis. HDAC7 has been identified to function as a key modulator of endothelial cell migration and hence angiogenesis, at least in part, by regulating PDGF-B/PDGFR-β gene expression (Mottet et al., 2007).
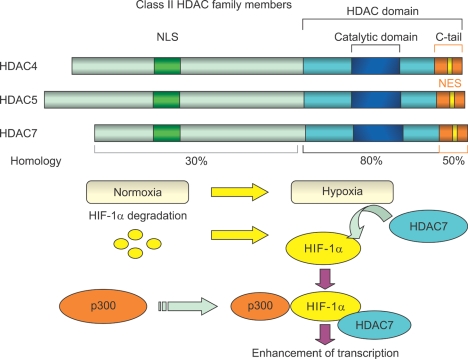
Figure 3.
Class II HDAC family members and interaction of HDAC7 with HIF-1α in its activated state in hypoxia: HDAC4, HDAC5, and HDAC7 contain a highly homologous conserved catalytic domain (HDAC domain) in the C-terminal region. The N-terminal region and the C-terminal tail of HDAC7 are less homologous to the corresponding regions of HDAC4 and HDAC5. HDAC4, HDAC5, and HDAC7 also contain N-terminal NLS sequences and C-terminal signal-responsive NES sequences. In addition, the 3 proteins are known to shuttle between the cytoplasm and nucleus in a process regulated by calcium/calmodulin-dependent protein kinase. In hypoxia, HDAC7 forms a complex with HIF-1α and p300 in the nucleus, resulting in enhanced transcription of HIF-1α target genes. HDAC, histone deacetylase; NLS, nuclear localization signal; NES, nuclear export signal; HIF, hypoxia-inducible factor.
Role of Int6 in HIF-2α regulation
To distinguish between HIF-1α and HIF-2α activities and identify HIF-2α-specific binding proteins, we performed yeast two-hybrid analysis and identified the tumor suppressor Int6/eIF3e/p48 as a novel HIF-2α-binding protein (Chen et al., 2007). The Int6 gene was first described as a tumor-suppressor gene during screening of mouse mammary tumor virus (MMTV) integration sites and was recognized as the insertional mutagenesis target of MMTV, which is critical for murine breast tumor formation (Marchetti et al., 1995). MMTV insertion into mouse Int6-coding DNA sequences appears to create a C-terminal truncated protein (Int6-ΔC; Figure 4), which functions as a dominant-negative mutant. Overexpression of these truncated proteins can transform cells in culture, and injection of these transformed cells into nude mice can induce tumor formation (Rasmussen et al., 2001; Mayeur and Hershey, 2002). Human Int6 may also be indirectly involved in proteolysis via at least 3 proteins that have been identified as Int6-binding proteins, namely, Tax (Desbois et al., 1996), Ret finger protein (Morris-Desbois et al., 1999), and p56 (Guo and Sen, 2000) (Figure 4). Tax, a transactivator of human T-cell leukemia virus type I, binds to Int6 and stimulates the activity of the proteasome (Desbois et al., 1996; Asano et al., 1997). Ret finger protein contains a RING finger motif, which is widely present among ubiquitin ligases (Jackson and Eldridge, 2002), and localizes to promyelocytic leukemia gene product nuclear bodies, which are rich in proteasomes (Desbois et al., 1996). p56 was first identified as one of the molecules whose expression is induced by interferons; it contains 2 ubiquitin-like domains, suggesting that it can also bind to the proteasome (Guo and Sen, 2000). The Int6 protein has been independently identified as a subunit (eIF3e) of eIF3. In addition, this protein can interact with 2 other multisubunit complexes: constitutive photomorphogenesis (COP9) signalosome (CSN) and proteasome (von Arnim and Chamovitz, 2003). Taken together, diverse involvements of Int6 in protein expression and proteolytic control make its exact role in tumorigenesis complex to understand, and therefore, exact mechanisms are not fully understood thus far. During our research, we further analyzed the specific interaction between Int6 and HIF-2α and observed that the N-terminal region of Int6 specifically binds to the C-terminal ID of HIF-2α, but not to that of HIF-1α or HIF-3α. Binding of Int6 induced HIF-2α instability in MCF-7 and HeLa cells, whereas that of the dominant-negative mutant Int6-ΔC with deleted C-terminal proteasome/Int6/Nip1/thyroid receptor-interacting protein 15 (TRIP-15) (PINT) domain (von Arnim and Chamovitz, 2003) induced stable HIF-2α expression, even under normoxic conditions. HIF-2α degradation could be completely inhibited by the proteasome inhibitors MG132 and MG101, but Int6-induced HIF-2α degradation was still observed in 786-O cells lacking endogenous pVHL expression. These results imply that Int6 is able to bind to HIF-2α and induce its instability through the ubiquitin-proteasome pathway, independent of pVHL. Further, the results suggest which pathway (pVHL and/or Int6) is critical for the regulation of HIF-2α stability and that HIF-2α degradation is regulated independent from each other. Indeed, the yeast two-hybrid binding assays demonstrated that HIF-2α had different binding sites for pVHL and Int6 and that the binding was non-competitive. The association of pVHL with HIF-2α triggered ubiquitin-dependent HIF-2α degradation in a hypoxia-dependent manner. However, HIF-2α degradation by Int6 was independent of oxygen concentration. A luciferase assay performed using the HRE promoter plasmid showed that Int6-ΔC expression increased the stability and transcriptional activity of HIF-2α. Moreover, by using siRNA against HIF-2α (Chen et al., 2007), we found that the silencing of endogenous Int6 was sufficient to induce HIF-2α expression even under normoxic conditions. This normoxic stabilization of HIF-2α enhanced the expression of angiogenic factors such as angiopoietin, fetal liver kinase 1, Tie-2, and VEGF. We also compared HIF-2α stabilization with HIF-1α overexpression and found HIF-1α to be the less potent inducer of angiogenesis, making HIF-2α the major candidate for treatment of vascular diseases. Taken together, our present results suggest that novel regulation of HIF-2α by Int6 plays a critical role in the regulation of vascular remodeling and development.
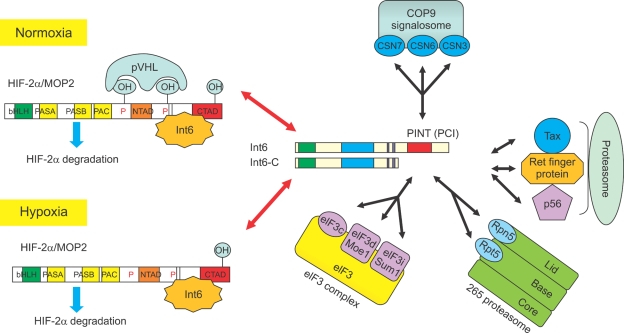
Figure 4.
Multiple functions of Int6: Left panel: Schematic diagram of Int6-HIF-2α interaction leading to oxygen-independent HIF-2α degradation. The N-terminal region of Int6 specifically binds to the C-terminal inhibitory domain of HIF-2α. Binding of Int6 induced HIF-2α instability, whereas that of the dominant-negative mutant Int6-ΔC with deleted C-terminal PINT domain induced stable HIF-2α expression, even under normoxic conditions. Right panel: Known function of Int6, which may also be indirectly involved in proteolysis via at least 3 Int6-binding proteins, namely, Tax, Ret finger protein, and p56. Int6 has a C-terminal PCI domain, which has also been found in the subunits of the following 3 complexes: proteasome regulatory lid, CSN, and eIF3 complex. HIF, hypoxia-inducible factor; MOP2, member of PAS protein 2; pVHL, von Hippel-Lindau protein; bHLH, basic helix-loop-helix; PAS, Per-ARNT-SIM; N-TAD, N-terminal transactivation domain; C-TAD, C-terminal transactivation domain; Int6, translation initiation factor 6; CSN, constitutive photomorphogenesis (COP9) signalosome; PINT, proteasome/Int6/Nip1/thyroid receptor-interacting protein 15 (TRIP-15); PCI, proteasome/CSN/eIF3; eIF3, eukaryotic translation initiation factor 3; Moe1, microtubule over extended; Sum1, suppressor of uncontrolled mitosis 1; Tax, transactivator x; Ret, receptor tyrosine kinase; Rpn5, regulatory particle non-ATPase 5; Rpt5, regulatory particle, ATPase like.
Conclusion
Angiogenesis is a strongly regulated process, influenced by the microenvironment and modulated by a multitude of pro- and antiangiogenic factors. A thorough understanding of the angiogenic process may lead to the identification of novel therapies for ischemic vascular diseases as well as diseases characterized by excessive angiogenesis, such as rheumatoid arthritis, psoriasis, or tumors. Hypoxia is a highly endangering condition for cells and responses must be quick and effective. One of these responses is angiogenesis and hypoxia is known to be a potent inducer of VEGF via HIF-related transcription in vitro. However, extended research revealed that cofactors are essential to ensure maximum efficiency of the transcriptional machinery related to changes in oxygen availability within cells/tissues, and the roles of different HIFs in eliciting hypoxic responses seem to be more divergent as originally assumed. In our research, we could establish direct involvement of HDAC7 as a binding partner of HIF-1α and that it upregulates the HIF-1α transcriptional activity. On the other hand, angiogenesis can be detrimental when not restricted to the absolutely necessary level and is miss-leaded by tumor cells. Therefore, angiogenic factors must be highly active but also under strict control. The instant HIF-1α protein degradation in normoxia is a basic control mechanism. In our studies, we demonstrated an alternative alternative mechanism involving HIF-2α. Int6 induces HIF-2α degradation in a hypoxia-independent manner after direct interaction. In addition, silencing of Int6 produces a potent, physiological inducer of angiogenesis (data not shown) and might be useful in the treatment of diseases related to insufficient blood supply. Further, the newly discovered binding proteins-HDAC7 for HIF-1α and Int6 for HIF-2α-support the assumption that the 2 HIF-α isoforms play distinct roles in eliciting hypoxia-related responses. HIF-2α can be considered as one of the master switches for inducing angiogenic factors, at least in specific cells, in the time windows that we used in our experiments. However, complete understanding of the complex interactions involved in hypoxic responses and their regulatory finetuning remains a subject of further investigation.
Abbreviations
- ARNT
aryl hydrocarbon receptor nuclear translocator
- bFGF
basic fibroblast growth factor
- bHLH
basic helix-loop-helix
- BNIP3
BCL2/adenovirus E1B 19-kDa interacting protein 3
- CBP
cyclic AMP response element-binding protein (CREB)-binding protein
- CCRC
clear cell renal carcinoma
- CSN
constitutive photomorphogenesis (COP9) signalosome
- eIF3
eukaryotic translation initiation factor 3
- FIH-1
factor inhibiting HIF-1
- GLUT1
glucose transporter 1
- HDAC
histone deacetylase
- HIF
hypoxia-inducible factor
- HRE
hypoxia-responsive element
- ID
inhibitory domain
- IGF-1
insulin-like growth factor-1
- Int6
translation initiation factor 6
- MCP-1
monocyte chemoattractant protein 1
- MMTV
mouse mammary tumor virus
- N-CoR
nuclear receptor corepressor
- PAS
Per/ARNT/SIM
- PINT
proteasome/Int6/Nip1/thyroid receptor-interacting protein 15 (TRIP-15)
- SMRT
silencing mediator for retinoic acid receptor (RAR) and thyroid hormone receptor (TR)
- TAD
transactivation domain
- VHL
von Hippel-Lindau
References
- 1.Asano K, Merrick WC, Hershey JW. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- 2.Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: Structure, function, and regulation. Biochem Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 3.Brown JM, Giaccia AJ. The unique physiology of solid tumors: Opportunities (and problems) for cancer therapy. Cancer Res. 1998;58:1408–1416. [PubMed] [Google Scholar]
- 4.Brunnberg S, Pettersson K, Rydin E, Matthews J, Hanberg A, Pongratz I. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc Natl Acad Sci USA. 2003;100:6517–6522. doi: 10.1073/pnas.1136688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll VA, Ashcroft M. Role of hypoxia-inducible factor (HIF)-1α versus HIF-2α in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: Implications for targeting the HIF pathway. Cancer Res. 2006;66:6264–6270. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Uchida K, Endler A, Shibasaki F. Mammalian tumor suppressor Int6 specifically targets hypoxia inducible factor 2α for degradation by hypoxia- and pVHL-independent regulation. J Biol Chem. 2007;282:12707–12716. doi: 10.1074/jbc.M700423200. [DOI] [PubMed] [Google Scholar]
- 7.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 9.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 10.Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 11.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 12.Graham CH, Fitzpatrick TE, McCrae KR. Hypoxia stimulates urokinase receptor expression through a heme protein-dependent pathway. Blood. 1998;91:3300–3307. [PubMed] [Google Scholar]
- 13.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo J, Sen GC. Characterization of the interaction between the interferon-induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumor virus. J Virol. 2000;74:1892–1899. doi: 10.1128/jvi.74.4.1892-1899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 16.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, Poellinger L, Pahlman S. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 18.Hu CJ, Iyer S, Sataur A, Covello KL, Chodosh LA, Simon MC. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in stem cells. Mol Cell Biol. 2006;26:3514–3526. doi: 10.1128/MCB.26.9.3514-3526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 20.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, von Kriegsheim A, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 21.Jackson PK, Eldridge AG. The SCF ubiquitin ligase: An extended look. Mol Cell. 2002;9:923–925. doi: 10.1016/s1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- 22.Jewell UR, Kvietikova I, Scheid A, Bauer C, Wenger RH, Gassmann M. Induction of HIF-1α in response to hypoxia is instantaneous. FASEB J. 2001;15:1312–1314. [PubMed] [Google Scholar]
- 23.Kaelin WG., Jr Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 24.Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 25.Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, Tamamizu-Kato S, Shibasaki F. Histone deacetylase 7 associates with hypoxia-inducible factor 1α and increases transcriptional activity. J Biol Chem. 2004;279:41966–41974. doi: 10.1074/jbc.M406320200. [DOI] [PubMed] [Google Scholar]
- 27.Kietzmann T, Roth U, Jungermann K. Induction of the plasminogen activator inhibitor-1 gene expression by mild hypoxia via a hypoxia response element binding the hypoxia-inducible factor-1 in rat hepatocytes. Blood. 1999;94:4177–4185. [PubMed] [Google Scholar]
- 28.Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 29.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 30.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotch LE, Iyer NV, Laughner E, Semenza GL. Defective vascularization of HIF-1α-null embryos is not associated with VEGF deficiency but with mesenchymal cell death. Dev Biol. 1999;209:254–267. doi: 10.1006/dbio.1999.9253. [DOI] [PubMed] [Google Scholar]
- 32.Kuwabara K, Ogawa S, Matsumoto M, Koga S, Clauss M, Pinsky DJ, Lyn P, Leavy J, Witte L, Joseph-Silverstein J, FurieMB, Torcia G, Cozzolino F, Kamada T, Stern DM. Hypoxia-mediated induction of acidic/basic fibroblast growth factor and platelet-derived growth factor in mononuclear phagocytes stimulates growth of hypoxic endothelial cells. Proc Natl Acad Sci USA. 1995;92:4606–4610. doi: 10.1073/pnas.92.10.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 34.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayeur GL, Hershey JW. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e) FEBS Lett. 2002;514:49–54. doi: 10.1016/s0014-5793(02)02307-4. [DOI] [PubMed] [Google Scholar]
- 37.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyteenhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris-Desbois C, Bochard V, Reynaud C, Jalinot P. Interaction between the Ret finger protein and the int-6 gene product and co-localisation into nuclear bodies. J Cell Sci. 1999;112:3331–3342. doi: 10.1242/jcs.112.19.3331. [DOI] [PubMed] [Google Scholar]
- 41.Mottet D, Bellahcene A, Pirotte S, Waltergny D, Deroanne C, Lamour V, Lidereau R, Castronovo V. Histone deacetylase 7 silencing alters endothelial cell migration, a key step in angiogenesis. Circ Res. 2007;101:1237–1246. doi: 10.1161/CIRCRESAHA.107.149377. [DOI] [PubMed] [Google Scholar]
- 42.Negus RP, Turner L, Burke F, Balkwill FR. Hypoxia down-regulates MCP-1 expression: Implications for macrophage distribution in tumors. J Leukoc Biol. 1998;63:758–765. doi: 10.1002/jlb.63.6.758. [DOI] [PubMed] [Google Scholar]
- 43.Norrby K. Mast cells and angiogenesis. APMIS. 2002;110:355–371. doi: 10.1034/j.1600-0463.2002.100501.x. [DOI] [PubMed] [Google Scholar]
- 44.Ouaissi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, Payan MJ, Dahan L, Pirro N, Seitz JF, Mas E, Lombardo D, Ouaissi A. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol. 2008;15:2318–2328. doi: 10.1245/s10434-008-9940-z. [DOI] [PubMed] [Google Scholar]
- 45.Paleolog E. Hypoxia: Not merely a regulator of angiogenesis? Arthritis Res Ther. 2004;6:75–77. doi: 10.1186/ar1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park SK, Dadak AM, Haase VH, Fontana L, Giaccia AJ, Johnson RS. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1α (HIF-1α): Role of cytoplasmic trapping of HIF-2α. Mol Cell Biol. 2003;23:4959–4971. doi: 10.1128/MCB.23.14.4959-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phelan MW, Forman LW, Perrine SP, Faller DV. Hypoxia increases thrombospondin-1 transcript and protein in cultured endothelial cells. J Lab Clin Med. 1998;132:519–529. doi: 10.1016/s0022-2143(98)90131-7. [DOI] [PubMed] [Google Scholar]
- 48.Phillips PG, Birnby LM, Narendran A. Hypoxia induces capillary network formation in cultured bovine pulmonary microvessel endothelial cells. Am J Physiol. 1995;268:L789–L800. doi: 10.1152/ajplung.1995.268.5.L789. [DOI] [PubMed] [Google Scholar]
- 49.Prabhakar NR. Oxygen sensing during intermittent hypoxia: Cellular and molecular mechanisms. J Appl Physiol. 2001;90:1986–1994. doi: 10.1152/jappl.2001.90.5.1986. [DOI] [PubMed] [Google Scholar]
- 50.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen SB, Kordon E, Callahan R, Smith GH. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene. 2001;20:5291–5301. doi: 10.1038/sj.onc.1204624. [DOI] [PubMed] [Google Scholar]
- 52.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenberger C, Mandriota S, Jurgensen JS, Wiesener MS, Horstrup JH, Frei U, Ratcliffe PJ, Maxwell PH, Bachmann S, Eckardt KU. Expression of hypoxia-inducible factor-1α and -2α in hypoxic and ischemic rat kidneys. J Am Soc Nephrol. 2002;13:1721–1732. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- 54.Ryan HE, Lo J, Johnson RS. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakuda H, Nakashima Y, Kuriyama S, Sueishi K. Media conditioned by smooth muscle cells cultured in a variety of hypoxic environments stimulates in vitro angiogenesis. A relationship to transforming growth factor-beta 1. Am J Pathol. 1992;141:1507–1516. [PMC free article] [PubMed] [Google Scholar]
- 56.Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, Marck BT, Matsumoto AM, Shelton JM, Richardson JA, Bennett MJ, Garcia JA. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat Genet. 2003;35:331–340. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- 57.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 58.Vacca A, Ribatti D, Iurlaro M, Albini A, Minischetti M, Bussolino F, Pellegrino A, Ria R, Rusnati M, Presta M, Vincenti V, Persico MG, Dammacco F. Human lymphoblastoid cells produce extracellular matrix-degrading enzymes and induce endothelial cell proliferation, migration, morphogenesis, and angiogenesis. Int J Clin Lab Res. 1998;28:55–68. doi: 10.1007/s005990050018. [DOI] [PubMed] [Google Scholar]
- 59.von Arnim AG, Chamovitz DA. Protein homeostasis: A degrading role for Int6/eIF3e. Curr Biol. 2003;13:R323–R325. doi: 10.1016/s0960-9822(03)00238-0. [DOI] [PubMed] [Google Scholar]
- 60.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 61.Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks KL, Wood SM, Gatter KC, Harris AL, Pugh CW, Ratcliffe PJ, Maxwell PH. Induction of endothelial PAS domain protein-1 by hypoxia: Characterization and comparison with hypoxia-inducible factor-1α. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- 62.Wykoff CC, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Identification of novel hypoxia dependent and independent target genes of the von Hippel-Lindau (VHL) tumour suppressor by mRNA differential expression profiling. Oncogene. 2000;19:6297–6305. doi: 10.1038/sj.onc.1204012. [DOI] [PubMed] [Google Scholar]
- 63.Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 64.Yu F, White SB, Zhao Q, Lee FS. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]