Abstract
Cell-mediated immunity directed against human papillomavirus 16 (HPV-16) antigens was studied in 16 patients affected with classic vulvar intra-epithelial neoplasia (VIN), also known as bowenoid papulosis (BP). Ten patients had blood lymphocyte proliferative T cell responses directed against E6/2 (14–34) and/or E6/4 (45–68) peptides, which were identified in the present study as immunodominant among HPV-16 E6 and E7 large peptides. Ex vivo enzyme-linked immunospot–interferon (IFN)-γ assay was positive in three patients who had proliferative responses. Twelve months later, proliferative T cell responses remained detectable in only six women and the immunodominant antigens remained the E6/2 (14–34) and E6/4 (45–68) peptides. The latter large fragments of peptides contained many epitopes able to bind to at least seven human leucocyte antigen (HLA) class I molecules and were strong binders to seven HLA-DR class II molecules. In order to build a therapeutic anti-HPV-16 vaccine, E6/2 (14–34) and E6/4 (45–68) fragments thus appear to be good candidates to increase HPV-specific effector T lymphocyte responses and clear classic VIN (BP) disease lesions.
Keywords: HPV-16 infection, peptidic vaccination, T lymphocytes, tumour immunity, classic VIN
Introduction
The premalignant lesions of vulvar intra-epithelial neoplasia (VIN) involve the mucosal and/or cutaneous epithelium of the vulva. VIN may be human papillomavirus (HPV)-related classic VIN or -unrelated VIN. The former is by far the most frequent vulvar cancer precursor. It occurs in adult women and tends to be multi-focal. It is caused by high-risk HPV (HR-HPV) types, essentially type 16, and histologically is made of poorly to undifferentiated basal cells and/or highly atypical squamous epithelial cells [1]. The involvement of the entire thickness of the epithelium defines grade 3 of the disease. The disease progresses towards invasion in about 3% of treated patients and 9% of untreated patients, according to a review of more than 3000 cases [2]. Classic VIN can also regress spontaneously [3] in young women presenting with multi-focal pigmented papular lesions. Previously, we studied a patient who presented with multi-focal classic VIN and showed complete clearance of viral lesions 8 months after disease onset and 2 months after electrocoagulation of less than 50% of the classic VIN lesions [4]. Immunohistochemical study of her initial vulvar biopsy revealed a marked dermal infiltrate containing a majority of CD4+ T lymphocytes and an epidermal infiltrate made up of both CD4+ and CD8+ T cells. She also showed a proliferating response against one peptide from E6 protein and a high-frequency anti-E6 and anti-E7 effector blood T cells by ex vivo enzyme-linked immunospot–interferon-γ (ELISPOT–IFN-γ) assay just before clinical regression. Such a study of blood cellular immune responses, together with the analysis of vulvar biopsies obtained simultaneously and correlated with clinical outcome, has not been reported previously. In an anti-HPV vaccine trial conducted by Davidson et al.[5], classic VIN lesions regressed completely in a patient following vaccination. Interestingly, immunostaining of vulvar biopsy prior to the vaccine showed a marked CD4+ and CD8+ T lymphocyte infiltrate of both epithelial and subepithelial sheets. It may be speculated whether the regression of these patient lesions could be related to a spontaneous regression. Therefore, the observation of a CD4+ and CD8+ infiltrate within subepithelial and epithelial sheets in the biopsy and the visualization of very strong blood anti-HPV T cell responses in patients with classic VIN could be predictive of spontaneous clinical outcome. It may also be thought that high numbers of blood CD4+ and CD8+ lymphocytes after therapeutic vaccination could allow clearance of HPV-16 lesions in classic VIN, assuming that anti-HPV vaccine-induced T effector cells could home into the HPV cutaneous and mucosal lesions. In the present study, we assessed cellular responses against HPV-16 E6 and E7 peptides in 16 patients presenting with classic VIN with the aim of mapping and characterizing the highest immunogenic regions from these proteins as potential candidates for a peptidic therapeutic vaccination.
We used HPV-16 classic VIN as the model of HPV-16 grade 3 cervical intra-epithelial neoplasia (CIN3), despite numerous discrepancies between the diseases, such as: difference in the keratinization of the epithelium; lower number of Langerhans cells in the cervical epithelium; presence of a cervical transformation zone where the glandular epithelium is being replaced by squamous epithelium; and evolution of bowenoid papulosis (BP) towards invasive carcinoma less frequent than in CIN3 [3,6]. Nevertheless, cellular immunity plays a key role in the defence against all HPV-induced infections or lesions by destroying HPV-infected or -transformed keratinocytes. Indeed, the incidence of HPV infections and diseases increases significantly with CD4+ T cell impairment in immunosuppressed individuals, such as transplanted or human immunodeficiency virus (HIV)-infected patients [7–10]. In asymptomatic HPV-16 infections, most women resolve their infection spontaneously without clinical disease [11] concomitantly with blood anti-HPV-16 T helper type 1 (Th1) CD4+ T cell responses [12,13]. Similarly, regression of condyloma is associated with a dense epithelial cellular infiltrate made up of both CD4+ and CD8+ T lymphocytes [14], with a Th1 cytokine profile as measured by cytokine mRNAs in interferon (IFN)-treated condylomas [15,16]. Proliferative CD4+ T cell responses are also associated with spontaneously regressive CIN3 [17]. The evolution of CIN3 towards invasive cancers is featured by a decrease of CD4+ cellular infiltrate, an increase of CD8+ T lymphocytes [18–20], the appearance of suppressive T lymphocytes [21] and a loss of blood anti-HPV-16 CD4+ activity [22,23]. In high-grade CIN, positive intradermal reaction after intradermal injection of five HPV-16 E7 large peptides correlated with the spontaneous clearance of the lesions, which further indicates the presence and the very important role of HPV-specific CD4+ T lymphocytes [24].
Patients and methods
Clinical status of the patients at time of entry in the study
Sixteen consecutive classic VIN patients aged 24–67 years (mean 41 ± 9·6 years) (Table 1) entered the study. Classic VIN first symptoms had appeared from 6 to 168 months (mean 37 months ± 52 months) prior to inclusion (Table 1). Diagnosis was confirmed by standard pathological analysis. HPV-16 was isolated from the lesions of all patients. All except one were HIV-negative. Human leucocyte antigen (HLA) class I and class II antigens were determined in every case. At the time of study, 11 patients (nos 2, 3, 4, 5, 6, 8, 10, 11, 13, 14, 16) had suffered from recurrent lesions for more than 6 months and experienced numerous relapses despite multiple destructive treatments (cryotherapy, electrocoagulation or laser surgery), local topical therapy (5-fluorouracil, imiquimod) or systemic immunotherapy using IFN-α. The five remaining patients (nos 1, 7, 9, 12, 15) were previously untreated. Six of the 16 patients (nos 2, 5, 6, 12, 13, 15) had been affected with high-grade CIN treated by surgery or laser surgery and one patient suffered from anal intra-epithelial neoplasia (AIN3) 18 months previously (no. 11).
Table 1.
Clinical and laboratory data at time of inclusion in the study.
| Patient | Age (years) | HLA | HIV status | Time elapsed from classic VIN clinical onset | Previous treatments | Previous cervical HPV disease | Size of vulvar lesions |
|---|---|---|---|---|---|---|---|
| 1 | 23 | A2/33,B7/55 | Negative | 6 months | None | None known | 10 cm2 |
| DR1501/1601 | |||||||
| 2 | 28 | A2/32, B7/44 | Negative | 37 months | Cryotherapy | High grade CIN | 1 cm2 |
| DR1501/0801 | |||||||
| 3 | 67 | A2/29, B44 | Negative | 13 months | Surgery | None known | 0 |
| DR1501/0701 | |||||||
| 4 | 64 | A25/66, B7/55 | Negative | 8 months | Surgery | None known | 0 |
| DR1501/1501 | |||||||
| 5 | 41 | A1/2, B7/44 | Negative | 160 months | Efudix, interferon | High grade CIN | 0·5 cm2 |
| DR1501/0404 or 0423 | |||||||
| 6 | 34 | A3/28, B7/8 | Negative | 54 months | Electrocoagulation | High grade CIN | 0 |
| DR1501/0311 | |||||||
| 7 | 37 | A28/29, B44/51 | Negative | 7 months | None | None known | 2 cm2 |
| DR1301/1101 | |||||||
| 8 | 53 | A2/23, B35/62 | Negative | 11 months | Electrocoagulation | None known | 0 |
| DR0701/0701 | |||||||
| 9 | 34 | A2/9, B13/15 | Negative | 6 months | None | None known | 0·5 cm2 |
| DR0701/0701 | |||||||
| 10 | 39 | A2/9, B17/40 | Negative | 19 months | Laser vaporization | None known | 4·09 cm2 |
| DR0701/0411 | |||||||
| 11 | 50 | A2/29, B7/37 | Negative | 18 months | None | High grade CIN | 4·5 cm2 |
| DR0701/1101 | |||||||
| 12 | 27 | A1/2, B8 | Negative | 4 months | None | Low grade CIN | 3 cm2 |
| DR1501/0301 | |||||||
| 13 | 34 | A1/2, B13/51 | Negative | 168 months | Laser vaporization | High grade CIN | 20 cm2 |
| DR0701/1101 | Interferon, 5-FU | ||||||
| 14 | 42 | A3/29, B7/38 | Negative | 6 months | Laser vaporization | None known | 2 cm2 |
| DR1001/1101 | |||||||
| 15 | 38 | A9/35, B44 | Positive | 18 months | Imiquimod | Low grade CIN | 1 cm2 |
| DR0301/1101 | |||||||
| 16 | 49 | A11/19, B13/22 | Negative | 54 months | Laser vaporization | None known | 0·5 cm2 |
| DR0301/0701 | Surgery |
HLA, human leucocyte antigen; HIV, human immunodeficiency virus; HPV, human papillomavirus; VIN, vulvar intra-epithelial neoplasia.
Four patients (nos 3, 4, 6, 8) had no detectable vulvar lesion after a recent treatment. The lesion surfaces in the other 12 patients ranged between 0·5 and 20 cm2 (mean 4·1 cm2 ± 2·6 cm2).
Blood samples
In accordance with the Ethics Committee of Cochin hospital, 150 ml blood samples were collected the day of entry in the study in every patient after informed consent. In most cases, blood samples were collected further every 6 months for 12 or 18 months. Peripheral blood mononuclear cells (PBMC) were isolated by centrifugation through lymphocyte separation medium (Pharmacia, Uppsala, Sweden) and either used immediately or frozen with 10% dimethylsulphoxide (DMSO) and stored at −180°C in liquid nitrogen.
HPV-16 typing
HPV-16 typing was performed by polymerase chain reaction (PCR) with DNA extracted from keratinocytes followed by restriction mapping of the amplified products. Multiplex PCR was performed using specific E6 HPV-16 and HPV-18 primers, as described previously [25]. HeLa and SiHa cell lines were used as negative and positive controls, respectively. After 40 cycles of amplification, products were analysed on 5% polyacrylamide gels. When a HPV DNA band was detected, the amplified product was digested with restriction enzymes. The appropriate restriction pattern of amplified products, together with its size, confers virtually 100% specificity on the PCR reaction.
Synthetic peptides
Eighteen overlapping peptides (15-mer to 24-mer) spanning the entire length of the E6 and E7 proteins (Table 2) were synthesized by Neosystem (Strasbourg, France). Twelve short peptides (8–10 amino acids) included into E6/2 (14–34) and E6/4 (45–68) large peptides selected on the basis of the presence of known motifs of binding to different HLA class I molecules were synthesized by Chiron Mimotopes (Emeryville, CA, USA).
Table 2.
Synthetic large peptides from human papillomavirus-16 E6 and E7 proteins.
| E6/1 (1–22) | MGQKRTAMFQDPQERPRKLPQL |
| E6/2 (14–34) | ERPRKLPQLCTELQTTIHDII |
| E6/3 (30–50) | IHDIILECVYCKQQLLRREVY |
| E6/4 (45–68) | LRREVYDFAFRDLCIVYRDGNPYA |
| E6/5 (61–80) | YRDGNPYAVCDKCLKFYSKI |
| E6/6 (76–95) | FYSKISEYRHYCYRLYGTTL |
| E6/7 (91–110) | YGTTLEQQYNKPLCDLLIRC |
| E6/8 (105–126) | DLLIRCINCQKPLCPEEKQRHL |
| E6/9 (121–140) | EKQRHLDKKQRFHNIRGRWT |
| E6/10 (135–158) | IRGRWTGRCMSCCRSSRTRRETQL |
| E7/1 (1–20) | MGGDTPTLHEYMLDLQPETT |
| E7/2 (7–27) | TLHEYMLDLQPETTDLYCYEQ |
| E7/3 (21–40) | DLYCYEQLNDSSEEEDEIDG |
| E7/4 (35–55) | EDEIDGPAGQAEPDRAHYNIV |
| E7/5 (43–57) | GQAEPDRAHYNIVTF |
| E7/6 (60–74) | KCDSTLRLCVQSTHV |
| E7/7 (65–87) | LRLCVQSTHVDIRTLEDLLMGTL |
| E7/8 (78–98) | TLEDLLMGTLGIVCPICSQKP |
T cell proliferation assay
PBMC (2 × 105/200 µl) were cultured in 96-well round-bottomed microtitre plates in complete medium with individual antigenic peptides in triplicate. After 5 days of culture, 1 µCi of [3H]-TdR (NEN, Paris, France) was added to each well for 18 h. Cells were harvested using an automatic cell harvester (Skatron, Sterling, VA, USA) and [3H]-thymidine incorporation was quantified by scintillation counting. Proliferative responses with a stimulation index [SI = counts per minute (cpm) in the presence of antigen/cpm in control media which must be higher than 500 cpm] above 3 were scored as positive.
Ex vivo ELISPOT assay for single cell IFN-γ release
ELISPOT–IFN-γ assays were performed as described previously [26]. Briefly, nitrocellulose plates (Multi-Screen HA; Millipore, Bedford, MA, USA) were coated overnight at +4°C with 0·1 µg of mouse anti-human IFN-γ monoclonal antibody (mAb) (Genzyme, Russelheim, Germany). Plates were washed with phosphate-buffered saline (PBS) and blocked with RPMI-1640–glutamax medium (Gibco BRL, Paisley, Scotland, UK) supplemented with penicillin (100 UI/ml), streptomycin (100 µg/ml), non-essential amino acids (1%), HEPES buffer (10 mM) (Gibco BRL), sodium pyruvate (1 mM) (ICN, Costa Mesa, CA, USA) and 10% heat-inactivated human antibody serum (complete medium) for 2 h at 37°C. PBMC (4 × 105 cells/well in 100 µl) were incubated in duplicate with 5 µM of each peptide in complete medium with 50 UI/ml interleukin (IL)-2 (Boehringer, Mannheim, Germany) for 48 h. Plates were washed and 100 µl of polyclonal rabbit anti-human IFN-γ antibodies (Genzyme) diluted 1:250 were added. After overnight incubation at +4°C, plates were washed and 100 µl of polyclonal biotin-conjugated goat anti-rabbit IgG antibodies (Boehringer) diluted 1:500 were added for 2 h at 37°C. The plates were washed and incubated with alkaline phosphatase-labelled extravidin (diluted 1/5000; Sigma-Aldrich Chimie SARL, Lyon, France) for 1 h. Chromogenic alkaline phosphatase substrate (Bio-Rad Laboratories, Hercules, CA, USA) was added to the wells to develop spots. Blue spots were counted with an automatic microscope (Zeiss Apparatus; Carl Zeiss, Göttingen, Germany). Negative controls were PBMC incubated in complete medium alone. Positive controls were obtained by activating PBMC with 50 ng/ml phorbol myristate acetate and 500 ng/ml ionomycin (Sigma-Aldrich Chimie SARL) (2000 cells/well). Only large spots with fuzzy borders were scored as IFN-γ-spot-forming cells (SFC). Responses were considered significant when the mean number of SFC by 106 cells in two experimental wells was superior to the highest either mean number of SFC in the negative control (PBMC alone) plus 3 standard deviations or number of SFC in the negative control (PBMC alone) plus 25 SFC/106 cells.
HLA class I and peptide interactions
HLA molecules were purified from human Epstein–Barr virus (EBV)-transformed cell lines by using affinity columns coupled to various immunoglobulins (Igs), as described previously [27,28]. After denaturation in urea plus NaOH, HLA heavy chains and β2m were separated from endogenous peptides then incubated with different concentrations of exogenous peptides (10−4–10−10 M) and β2m. Reassembled HLA/peptide complexes were trapped in microtitration plate wells coated with anti-HLA monoclonal Igs, as described in Bourgault et al.[27]. Correctly folded HLA complexes were revealed with alkaline phosphatase-coupled antiβ2m Igs (M28) with 4-methyl-umbelliferyl phosphate as a substrate (M-8883; Sigma-Aldrich Chimie SARL). Fluorescence was measured at 360/460 nm in a Microfluor reader (Victor 1420; Wallac, Turku, Finland). Results were expressed as the lowest peptide concentrations yielding a significant binding (20% of maximal fluorescence).
HLA-DR-specific binding assays
Purification of HLA-DR molecules and peptide binding assays were performed as described previously [29,30]. These assays are specific for the HLA-DR molecules predominant in the European and North American populations, which are also frequent globally. Briefly, HLA-DR molecules purified from EBV homozygous cell lines by affinity chromatography were incubated with various concentrations of the competitor peptides and an appropriate biotinylated reference peptide. Unlabelled forms of the biotinylated peptides were used as reference peptides to assess the validity of each experiment. Their sequences and inhibitory concentration (IC50) values were as follows: HA 306–318 (PKYVKQNTLKLAT) for DRB1*0101 (6 nM); DRB1*0401 (30 nM), DRB1*1101 (17 nM) and DRB5*0101 (8 nM), YKL (AAYAAAKAAALAA) for DRB1*0701 (42 nM); A3152–166 (EAEQLRAYLDGTGVE) for DRB1*1501 (28 nM); MT 2–16 (AKTIAYDEEARRGLE) for DRB1*0301 (660 nM); B1 21–36 (TERVRLVTRHIYNREE) for DRB1*1301 (268 nM); LOL 191–210 (ESWGAVWRIDTPDKLTGPFT) for DRB3*0101 (9 nM); and E2/E168 (AGDLLAIETDKATI) for DRB4*0101 (3 nM). The peptide concentration that prevented binding of 50% of the labelled peptide (IC50) was evaluated. Data were expressed as relative affinity: ratios of the IC50 of the peptide by the IC50 of the reference peptide, which binds the HLA II molecule strongly.
Results
T cell proliferative responses
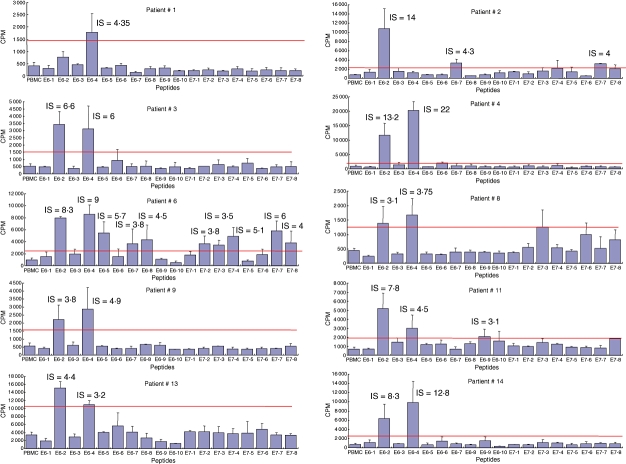
Proliferation assays using E6 and E7 large peptides covering both whole proteins performed at entry into the study showed that blood T lymphocytes from 10 patients (nos 1, 2, 3, 4, 6, 8, 9, 11, 13, 14) proliferated in the presence of one to 10 peptides (Fig. 1). The strongest responses in eight patients (nos 3, 4, 6, 8, 9, 11, 13, 14) were directed against both peptides E6/2 (aa 14–34) and E6/4 (aa 45–68), whereas T cells in patient 1 proliferated against peptide E6/4 and in patient 2 against E6/2 only, respectively (Fig. 1). SI of these strongest proliferative responses ranged from 3·1–22. Peptide E6/7 (aa 91–110) stimulated blood T lymphocytes from two patients (nos 2 and 6, SI = 3·8 and 4·3, respectively). One patient each displayed responses against peptide E6/5 (aa 61–80) (patient no. 6), peptide E6/8 (aa 105–126) (patient no. 6) and peptide E6/9 (aa 121–140) (patient no. 11). Finally, no response could be detected against peptides E6/1, E6/3, E6/6 and E6/10.
Fig. 1.
Proliferative responses against E6 and E7 long peptides in the responder patients. After [3H]-thymidine incorporation, proliferative responses with a stimulation index [SI = counts per minute (cpm) in the presence of antigen/cpm in control media] above 3 were scored as positive.
Only two patients (nos 2 and 6) had proliferative responses against E7 peptides. E7/7 (aa 65–87) was the better immunogenic peptide, recognized by two patients (with SI of 4 and 6), peptides E7/2 (7–27), E7/3 (21–40), E7/4 (35–55) and E7/8 (78–98) being recognized by only one patient. Peptides E7/1, E7/5 and E7/6 yielded no detectable response.
Ex vivo ELISPOT–IFN-γ assays
This assay was performed with E6 and E7 large peptides at entry into the study (Fig. 2). Numerous blood cells from patient 1 recognized three HPV-16 long peptides: E6/4, E7/2 and E7/3 with mean 270, 65 and 430 SFC/106 PBMCs. In patient 13 the recognized peptides were E6/7, E6/8, E7/1, E7/2, E7/3 and E7/8, with a mean of 43, 50, 38, 34, 33 and 30 SFC/106 PBMCs. These two patients both had large lesions (10 and 20 cm2, respectively). Nevertheless, their clinical outcome was different. The first patient experienced a complete and durable disappearance of the lesions 2 months after entry into the study following the electrocoagulation of less than 50% of the classic VIN lesion, whereas chronic and extensive lesions persisted in the second patient despite laser surgery.
Fig. 2.
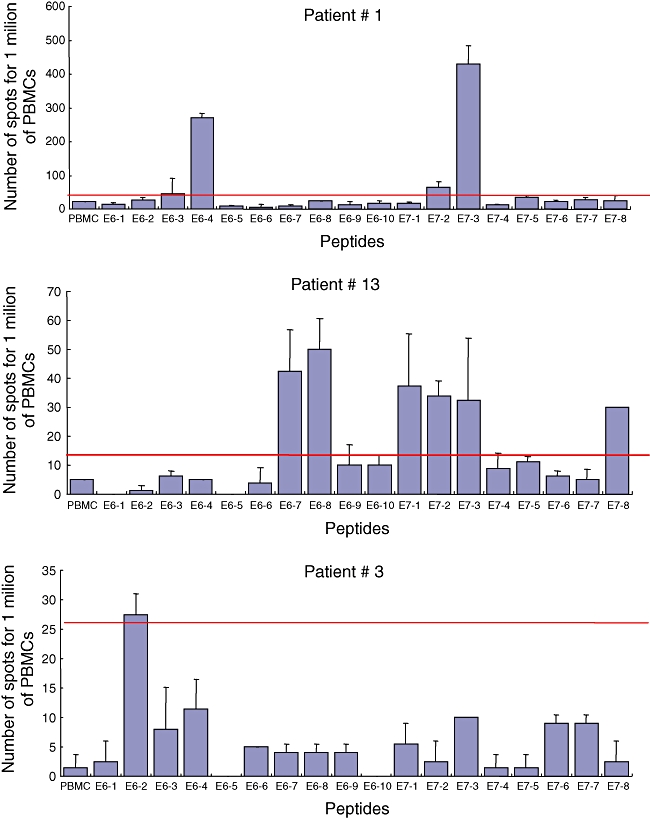
Ex vivo enzyme-linked immunospot–interferon-γ (ELISPOT–IFN-γ) responses in the three responder patients. Ex vivo ELISPOT–IFN-γ was performed as described in Materials and methods using peripheral blood mononuclear cells (PBMC) (4·105 cells/well) incubated with different peptides from E6 and E7 proteins (Table 2). The number of spots was calculated per million of PBMC.
With regard to the other women tested with smaller or no detectable lesions, a single patient (no. 3, without lesion) had weak blood T cell responses against peptide E6/2, with mean 28 specific SFC/106 PBMCs. All three patients with a positive ELISPOT–IFN-γ assay exhibited proliferative responses directed against the same or other E6 or E7 peptides.
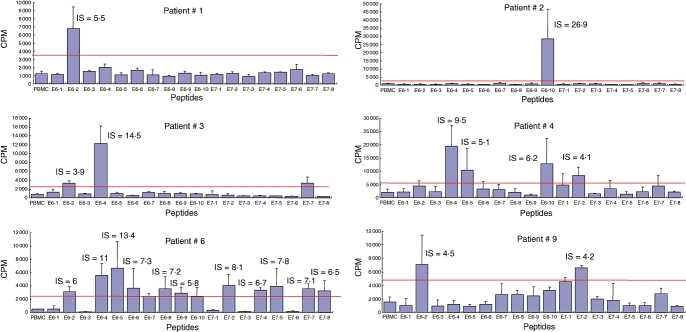
Outcome of the proliferative responses
T cells from six (nos 1, 2, 3, 4, 6, 9) of the 10 patients with initial proliferative responses still responded 12 months later, one (no. 8) lost detectable responses and three patients (nos 11, 13, 14) were lost of sight (Fig. 3). In the six responder patients, the recognized specificities were different from those observed initially, with a broadening of peptide recognition concomitant with a change on the recognition level of some specificity. E6/2 (14–34) and E6/4 (45–68) peptides were always the two that were recognized most strongly by four (nos 1, 3, 6, 9) and three (nos 3, 4, 6) patients, respectively. Four of these patients (nos 1, 3, 4, 9) received destructive treatment and remained free of vulvar lesions 1 year later, patient 2 had persistent lesions without improvement despite imiquimod therapy and patient 6 relapsed 12 months after the inclusion in the study.
Fig. 3.
Outcome of the proliferative responses 12 months after entry into the study in initial responder women. After [3H]-thymidine incorporation, proliferative responses with a stimulation index [SI = counts per minute (cpm) in the presence of antigen/cpm in control media] above 3 were scored as positive.
Outcome of the ELISPOT–IFN-γ responses
Patient 1, who had cleared more than 50% of her lesions spontaneously, had no detectable ex-vivo blood T cell effector cells 12 months later (data not shown). The two patients with a low initial ex-vivo ELISPOT–IFN-γ response (nos 3 and 13) also had no detectable circulating effector cells 12 months later, despite the persistence of the lesions in patient 13 (data not shown).
Peptides E6/2 (14–34) and E6/4 (45–68) contain several peptides that bind to HLA class II and class I molecules
In contrast to HLA class I molecules, class II molecules accommodate peptides of various sizes. We therefore submitted the whole E6/2 and E6/4 peptides directly to HLA-DR-specific binding assays, as these molecules are involved frequently in T cell epitope presentation. E6/2 (14–34) peptide bound to three of 10 HLA-DR molecules (Table 3). At least one of these three HLA-DR molecules, DR3, DR7, DR15, was shared by all except one responder studied. E6/4 (45–68) peptide bound to six of the 10 HLA class II molecules, DR1, DR4, DR7, DR11, DR15, DRB5, all shared by our patients.
Table 3.
Human leucocyte antigen (HLA) class II molecule binding activity of E6/2 (14–34) and E6/4 (45–64) peptides.
| Relative binding activity |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | DR1 | DR3 | DR4 | DR7 | DR11 | DR13 | DR15 | DRB3 | DRB4 | DRB5 | Number HLA II |
| E6/2 (14–34) | 632 | 8 | > 333 | 5 | > 577 | > 30 | 71 | 169 | 1167 | > 1208 | 3 |
| E6/4 (45–68) | 9 | 167 | 58 | 51 | 7 | > 30 | 6 | 293 | >3593 | 8 | 6 |
Peptide binding capacity was investigated by competitive enzyme-linked immunosorbent assays. Data are expressed as relative activity [ratio of the inhibitory concentration (IC50) of the peptide by the IC50 of the reference peptide. which is a high binder to the HLA class II molecule]. Relative activities inferior to 100 correspond to active peptides. Means were calculated from at least three independent experiments.
The HLA class I molecules binding of 12 short synthetic peptides (8–10-mers) included into E6/2 (14–34) and E6/4 (45–68) large peptides was tested against seven supertypes of HLA class I molecules (Table 4). Every short peptide was able to bind to at least one HLA class I molecule. Binding affinities ranged between 10−4 M (low HLA binders) and 10−9 M (high binders).
Table 4.
Binding of human papillomavirus-16 short E6 peptides included into E6/2 (14–34) and E6/4 (45–68) to human leucocyte antigen (HLA) class I molecules.
| Peptides |
HLA molecule |
|||||||
|---|---|---|---|---|---|---|---|---|
| Position | Sequence | A1/B18 | A2 | A3/11 | A24 | B7/35/51 | B27 | B44 |
| 15–22 | RPRKLPQL | 10−9 | ||||||
| 18–26 | KLPQLCTEL | 10−4 | ||||||
| 19–26 | LPQLCTEL | 10−5 | ||||||
| 21–30 | QLCTELQTTI | 10−5 | ||||||
| 24–33 | TELQTTIHDI | 10−5 | ||||||
| 26–34 | LQTTIHDII | 10−5 | ||||||
| 46–55 | RREVYDFAFR | 10−7 | ||||||
| 49–57 | VYDFAFRDL | 10−9 | ||||||
| 50–57 | YDFAFRDL | 10−6 | ||||||
| 52–60 | FAFRDLCIV | 10−4 | 10−6 | |||||
| 54–61 | FRDLCIVY | 10−4 | 10−4 | 10−4 | ||||
| 59–67 | IVYRDGNPY | 10−6 | ||||||
Binding capacities of peptides to immunopurified HLA class I molecules are expressed in M and have been evaluated from at least three independent experiments.
Discussion
Specific blood T CD8+ and CD4+ cells play an essential role in the defence against HPV, as observed previously in immunodeficient patients who are more susceptible to HPV persistent infections [9]. The high frequency (62%) of proliferative responses observed in classic VIN patients in the present study is in accordance with previous reports of CIN3 [22]. In contrast, other groups found far fewer proliferative responses (approximately 20%) in CIN3 [31–33]. These discrepancies could be explained by the fact that some patients with CIN3 show spontaneous regressions that might be associated with strong proliferative responses, whereas in other women the evolution towards invasive cancer is concomitant with the loss of proliferative T lymphocyte activity [33]. The high-level proliferative responses observed in our study might reflect the fact that BP is an intra-epithelial vulvar and perineal cutaneous and mucosal disease that progresses exceptionally to invasive carcinoma. Indeed, the evolution of BP towards invasive carcinoma is present in fewer than 3–4% of patients [2,3], whereas CIN3 evolves towards invasion in about 15% of cases [6].
Among 18 large peptides of the proteins E6 and E7, two were recognized in proliferative assays as immunodominant by T cells from 10 of 16 women (62%) at entry into the present study, namely E6/2 (aa 14–34) and E6/4 (aa 45–68). Four other peptides, E6/7 (aa 91–110), E7/2 (aa 7–27), E7/3 (aa 21–40) and E7/7 (aa 65–87), were recognized by only 12% of the women in proliferative or ELISPOT–IFN-γ tests. The E6 and E7 protein regions implicated in T cell recognition during HPV infection have not yet been well defined because of the usually low frequency of anti-HPV blood T cell responses and of the difficulties in studying them.
In protein E6, some peptides, including or overlapping our peptides E6/2 (aa 14–34) and E6/4 (aa 45–68), have already been described as recognized preferentially by CD4+ T cells. Among them, peptide E6 42–57, that is restricted by the HLA-DR7 molecule, has already been identified [34]. Regions E6 1–31, 22–51 and 24–45 can be also immunogenic for CD4+ T cells, as shown in CIN or sexually active healthy women [35]. Region E6 42–71, which includes peptide E6/4 (aa 45–68), has also been described as a target of proliferative responses in CIN patients [35]. Another E6111–158 region was described previously as inducing proliferative responses in infected asymptomatic subjects or in patients with CIN3 [33,35], as well as E6127–141 peptide in healthy young women [36]. Similarly, peptides E7 43–77, E7 50–62 and E7 58–68, which are restricted by DR3, DR15 and DR17, respectively, were defined as epitopic peptides for CD4+ T cells [34,37,38], and E7 region 51–98, including our E7/7 (aa 65–87) peptide, is also very immunogenic for proliferating T lymphocytes [22,23,31].
The characterization of E6 and E7 HPV-16 epitopes and the HLA restriction of their recognition by CD8+ T lymphocytes are more precise: E6 29–38, E7 11–20, E7 82–90 and E7 86–93 epitopes are presented by HLA-A2 [39–41], E6 80–88 and E7 44–52 by HLA-B18 [27] and E6 49–57 by HLA-A24 [42]. In women who cleared HPV-16 infection, cytotoxic T lymphocyte (CTL) responses are directed against epitopes located preferentially in the N-terminal half of the E6 protein (region 16–40) [43]. In this fragment, the dominant epitope E6 29–37 is restricted by HLA-B48, E6 31–38 by HLA-B4002 and the subdominant epitope E6 52–61 by HLA-B35 [44]. The same group had also shown that peptide E6 33–42 61 is recognized by CD8+ T lymphocytes in association with HLA-A68, peptide E6 52–61 in association with HLA-B57 and -B35, peptide E6 75–83 in association with HLA-B62, peptide E7 7–15 in association with HLA-B48 and peptide E7 79–87 in association with HLA-B60 [44–46]. In addition, E7 7–15 is also able to bind HLA-A2 and -B8 to be recognized by CTL [40,47].
From the latter results, two hot-spots of CD8+ T cell epitopes in protein E6 may be located in the regions E6 29–38 and 52–61, and another in protein E7 (region E7 7–15) [44]. Nevertheless, poor immunogenicity of E7 protein has been observed in many studies during both HPV-16 infection and after peptidic vaccination using long peptides spanning both E6 and E7 [48–49], such as those used in our study.
In this study we show that nearly the same regions of E6 protein (E6 14–34 and E6 45–68) are recognized by T lymphocytes from 10 of 16 patients presenting with classic VIN (PB). We have not characterized fully the nature of proliferative effector cells by CD4+ or CD8+ depletion experiments, except in patient 2, in whom the proliferative responses involved CD4+ T lymphocytes (data not shown). These results are consistent with CD4+ T cell responses, as large E6 peptides are known to induce proliferative responses more than short peptides. However, our previous study with short-term cultures of patient 1's lymphocytes showed a CD8+ epitope included in peptide E6/4 (data not shown and [4]). Hence, CD8+ T cells may also be involved in the proliferative responses. In addition, we tested the binding of E6 and E7 short peptides included in E6/2 (aa 14–34) and E6/4 (aa 45–68) to seven different supertypes of HLA class I molecules and we showed that regions E6 14–34 and E6 45–68 include several peptides able to bind to several different HLA class I molecules with a very high affinity (10−6–10−9 M).
Hence, the epitopes E6/2 14–34 and E6/4 45–68 could be recognized strongly by CD4+ and/or CD8+ T lymphocytes and could be particularly relevant in the design of a peptide vaccination. It is worth noting that our patients had not progressed towards invasive cancer of the vulva at their entry into the study. We may hypothesize that the T cell responses that we observed were able to contain the tumour cells in the epithelium. Therefore, E6/2 14–34 and E6/4 45–68 peptides could play a major role in protection against invasive cancer by stimulating T lymphocytes.
Recently, Piersma et al.[50] have shown positive proliferative responses of tumour-infiltrating lymphocytes against HPV-16 and HPV-18 E6 and E7 peptides in 23 of 54 patients with invasive cervical cancer (42%) without preferential recognition of the immunodominant region. This observation does not conflict with our data, the loss of the recognition of immunodominant epitopes by invasive cancer infiltrating T lymphocytes being able to reflect the development of tumour cells which escape to the immune system.
We observed the preferential presence of certain HLA class II DR molecules in our responding patients, HLA-DR15 and HLA-DR7 in 50% of the responding women and DR11 in 30%. No such an association between HLA class II molecules, T anti-HPV T cell responses and classic VIN has been described previously. A significantly high frequency of DRB1* 0901 or DQB1*03032 was observed in HPV-16-positive CIN3/invasive cervical carcinoma patients in Japan and China [51–53]. An increased risk of CIN3 has been associated with DRB1*1501 or DQA1*0102 in New Mexico [54]. Conversely, DRB1*1501 and DQB1*0602 haplotypes were shown recently to be protective against CIN2+, especially in individuals infected with oncogenic HPV in Canada [55]. In CIN1, DRB1*1301 was associated with an increased probability of regression [56] and DR B1*11, DR B1*15, DR B1*3 with persistence [57].
By studying the immunodominant E6 and E7 large peptides in HLA-DR-specific binding assays, we observed that E6/2 14–34 and E/4 45–68 peptides bound HLA-DR7, 11 and 15 (molecules shared by our patients) and to other HLA-DR such as DR1, DR3, DR4, DRB5. Nevertheless, it remains to be proven that HLA-DR molecules are the restricting element for proliferative CD4+ T cells. Indeed, HLA-DQ and -DP were described recently as proliferative response-restricting elements during HPV-16 infection [58,59].
The present study shows that following the disappearance of the lesions, either spontaneously or after destructive treatment, proliferative responses can persist at least for 1 year with a broadening of peptide recognition concomitant with a loss of some specificities and acquisition of others. This observation can be related to an immunospreading of the cellular immune response following deliverance of new HPV antigens in the blood after destruction of the lesions or to recirculation of effector T lymphocytes from the epithelium to the blood.
Using ELISPOT–IFN-γ assay, ex-vivo frequencies of specific anti-E6 or E7 peptides T lymphocytes were stronger in the present study in the two patients with large clinical lesions of classic VIN compared to the patients with smaller or no detectable lesions who had low blood T cell responses. In a previous study, six of nine patients with classic VIN had ex-vivo frequencies of specific anti-E6 or E7 peptides; CD8+ T lymphocytes comprised between 21 and 1360 SFC/106 CD4-depleted T lymphocytes [60]. However, no clinical correlation was reported in the latter study. Our results may be the consequence of better contact between T lymphocytes and a large area of HPV-16-infected keratinocytes, generating better ex-vivo T cell responses. After treatment and disappearance of the lesions in our patients, ex-vivo T cell responses became undetectable by ELIPSPOT–IFN-γ assay.
In conclusion, we have defined two immunodominant regions in HPV-16 E6 protein. They were recognized by blood effector T cells in 10 of 16 patients with classic VIN and they contained epitopic peptides able to bind to 13 different HLA class I and class II molecules. These immunodominant regions could be included in a peptidic vaccine in order to bypass the major histocompatibility complex barrier restriction for building a therapeutic anti-HPV-16 vaccine usable in previously HPV-16-infected women.
Acknowledgments
This work was supported by Association pour la Recherche sur le Cancer, Ligue Nationale Contre le Cancer and Délégation à la Recherche Clinique, Assistance Publique-Hôpitaux de Paris (CRC96160). The French Society for Dermatology offered some valuable help in the form of grants. We thank Sophie Caillat Zucman for HLA typing. This study is dedicated to the memory of Jean Gérard Guillet.
Disclosure
None.
References
- 1.Bruchim I, Gotlieb WH, Mahmud S, Tunitsky E, Grzywacz K, Ferenczy A. HPV-related vulvar intraepithelial neoplasia: outcome of different management modalities. Int J Gynaecol Obstet. 2007;99:23–7. doi: 10.1016/j.ijgo.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 2.van Seters M, van Beurden M, de Craen AJ. Is the assumed natural history of vulvar intraepithelial neoplasia III based on enough evidence? A systematic review of 3322 published patients. Gynecol Oncol. 2005;97:645–51. doi: 10.1016/j.ygyno.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Jones RW, Rowan D, MStewart AW. Vulvar intraepithelial neoplasia: aspects of the natural history and outcome in 405 women. Obstet Gynecol. 2005;106:1319–26. doi: 10.1097/01.AOG.0000187301.76283.7f. [DOI] [PubMed] [Google Scholar]
- 4.Bourgault Villada I, Moyal Barracco M, Ziol M, et al. Spontaneous regression of grade 3 vulvar intraepithelial neoplasia associated with human papillomavirus-16-specific CD4(+) and CD8(+) T-cell responses. Cancer Res. 2004;64:8761–6. doi: 10.1158/0008-5472.CAN-04-2455. [DOI] [PubMed] [Google Scholar]
- 5.Davidson EJ, Boswell CM, Sehr P, et al. Immunological and clinical responses in women with vulval intraepithelial neoplasia vaccinated with a vaccinia virus encoding human papillomavirus 16/18 oncoproteins. Cancer Res. 2003;63:6032–41. [PubMed] [Google Scholar]
- 6.Ostor AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol. 1993;12:186–92. [PubMed] [Google Scholar]
- 7.Arends MJ, Benton EC, McLaren KM, Stark LA, Hunter J, ABird CC. Renal allograft recipients with high susceptibility to cutaneous malignancy have an increased prevalence of human papillomavirus DNA in skin tumours and a greater risk of anogenital malignancy. Br J Cancer. 1997;75:722–8. doi: 10.1038/bjc.1997.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirgwin KD, Feldman J, Augenbraun M, Landesman S, Minkoff H. Incidence of venereal warts in human immunodeficiency virus-infected and uninfected women. J Infect Dis. 1995;172:235–8. doi: 10.1093/infdis/172.1.235. [DOI] [PubMed] [Google Scholar]
- 9.Sun XW, Kuhn L, Ellerbrock TV, Chiasson MA, Bush TJ, Wright TC., Jr. Human papillomavirus infection in women infected with the human immunodeficiency virus. N Engl J Med. 1997;337:1343–9. doi: 10.1056/NEJM199711063371903. [DOI] [PubMed] [Google Scholar]
- 10.Williams AB, Darragh TM, Vranizan K, Ochia C, Moss AR, Palefsky JM. Anal and cervical human papillomavirus infection and risk of anal and cervical epithelial abnormalities in human immunodeficiency virus-infected women. Obstet Gynecol. 1994;83:205–11. [PubMed] [Google Scholar]
- 11.Bulkmans NW, Berkhof J, Bulk S, et al. High-risk HPV type-specific clearance rates in cervical screening. Br J Cancer. 2007;96:1419–24. doi: 10.1038/sj.bjc.6603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong A, van der Burg SH, Kwappenberg KM, et al. Frequent detection of human papillomavirus 16 E2-specific T-helper immunity in healthy subjects. Cancer Res. 2002;62:472–9. [PubMed] [Google Scholar]
- 13.Welters MJ, de Jong A, van den Eeden SJ, et al. Frequent display of human papillomavirus type 16 E6-specific memory T-helper cells in the healthy population as witness of previous viral encounter. Cancer Res. 2003;63:636–41. [PubMed] [Google Scholar]
- 14.Coleman N, Birley HD, Renton AM, et al. Immunological events in regressing genital warts. Am J Clin Pathol. 1994;102:768–74. doi: 10.1093/ajcp/102.6.768. [DOI] [PubMed] [Google Scholar]
- 15.Arany I, Tyring SK. Status of local cellular immunity in interferon-responsive and -nonresponsive human papillomavirus-associated lesions. Sex Transm Dis. 1996;23:475–80. doi: 10.1097/00007435-199611000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Arany I, Tyring SK. Activation of local cell-mediated immunity in interferon-responsive patients with human papillomavirus-associated lesions. J Interferon Cytokine Res. 1996;16:453–60. doi: 10.1089/jir.1996.16.453. [DOI] [PubMed] [Google Scholar]
- 17.Kadish AS, Timmins P, Wang Y, et al. Regression of cervical intraepithelial neoplasia and loss of human papillomavirus (HPV) infection is associated with cell-mediated immune responses to an HPV type 16 E7 peptide. Cancer Epidemiol Biomarkers Prev. 2002;11:483–8. [PubMed] [Google Scholar]
- 18.Ferguson A, Moore M, Fox H. Expression of MHC products and leucocyte differentiation antigens in gynaecological neoplasms: an immunohistological analysis of the tumour cells and infiltrating leucocytes. Br J Cancer. 1985;52:551–63. doi: 10.1038/bjc.1985.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietl JA, Horny HP, Buchholz F. Lymphoreticular cells in invasive carcinoma of the uterine cervix: an immunohistological study. Int J Gynaecol Obstet. 1991;34:179–82. doi: 10.1016/0020-7292(91)90235-w. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh AK, Moore M. Tumour-infiltrating lymphocytes in cervical carcinoma. Eur J Cancer. 1992;28A:1910–16. doi: 10.1016/0959-8049(92)90034-y. [DOI] [PubMed] [Google Scholar]
- 21.van der Burg SH, Piersma SJ, de Jong A, et al. Association of cervical cancer with the presence of CD4+ regulatory T cells specific for human papillomavirus antigens. Proc Natl Acad Sci USA. 2007;104:12087–92. doi: 10.1073/pnas.0704672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luxton JC, Rowe AJ, Cridland JC, Coletart T, Wilson P, Shepherd PS. Proliferative T cell responses to the human papillomavirus type 16 E7 protein in women with cervical dysplasia and cervical carcinoma and in healthy individuals. J Gen Virol. 1996;77:1585–93. doi: 10.1099/0022-1317-77-7-1585. [DOI] [PubMed] [Google Scholar]
- 23.de Gruijl TD, Bontkes HJ, Walboomers JM, et al. Differential T helper cell responses to human papillomavirus type 16 E7 related to viral clearance or persistence in patients with cervical neoplasia: a longitudinal study. Cancer Res. 1998;58:1700–6. [PubMed] [Google Scholar]
- 24.Hopfl R, Heim K, Christensen N, et al. Spontaneous regression of CIN and delayed-type hypersensitivity to HPV-16 oncoprotein E7. Lancet. 2000;356:1985–6. doi: 10.1016/S0140-6736(00)03315-8. [DOI] [PubMed] [Google Scholar]
- 25.Jullian EH, Dhellemmes C, Saglio O, Chavinie J, Pompidou A. Improved detection of human papillomavirus types 16 and 18 in cervical scrapes by a multiplex polymerase chain reaction: a 4% prevalence among 120 French women with normal cytology. Lab Invest. 1993;68:242–7. [PubMed] [Google Scholar]
- 26.Gahery-Segard H, Pialoux G, Figueiredo S, et al. Long-term specific immune responses induced in humans by a human immunodeficiency virus type 1 lipopeptide vaccine: characterization of CD8+-T-cell epitopes recognized. J Virol. 2003;77:11220–31. doi: 10.1128/JVI.77.20.11220-11231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourgault Villada I, Beneton N, Bony C, et al. Identification in humans of HPV-16 E6 and E7 protein epitopes recognized by cytolytic T lymphocytes in association with HLA-B18 and determination of the HLA-B18-specific binding motif. Eur J Immunol. 2000;30:2281–9. doi: 10.1002/1521-4141(2000)30:8<2281::AID-IMMU2281>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 28.Connan F, Hlavac F, Hoebeke J, Guillet JG, Choppin J. A simple assay for detection of peptides promoting the assembly of HLA class I molecules. Eur J Immunol. 1994;24:777–80. doi: 10.1002/eji.1830240344. [DOI] [PubMed] [Google Scholar]
- 29.Texier C, Pouvelle S, Busson M, et al. HLA-DR restricted peptide candidates for bee venom immunotherapy. J Immunol. 2000;164:3177–84. doi: 10.4049/jimmunol.164.6.3177. [DOI] [PubMed] [Google Scholar]
- 30.Texier C, Pouvelle-Moratille S, Busson M, Charron D, Menez A, Maillere B. Complementarity and redundancy of the binding specificity of HLA-DRB1, -DRB3, -DRB4 and -DRB5 molecules. Eur J Immunol. 2001;31:1837–46. doi: 10.1002/1521-4141(200106)31:6<1837::aid-immu1837>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa M, Stites DP, Farhat S, et al. T-cell proliferative response to human papillomavirus type 16 peptides: relationship to cervical intraepithelial neoplasia. Clin Diagn Lab Immunol. 1996;3:205–10. doi: 10.1128/cdli.3.2.205-210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele JC, Mann CH, Rookes S, et al. T-cell responses to human papillomavirus type 16 among women with different grades of cervical neoplasia. Br J Cancer. 2005;93:248–59. doi: 10.1038/sj.bjc.6602679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukui T, Hildesheim A, Schiffman MH, et al. Interleukin 2 production in vitro by peripheral lymphocytes in response to human papillomavirus-derived peptides: correlation with cervical pathology. Cancer Res. 1996;56:3967–74. [PubMed] [Google Scholar]
- 34.Strang G, Hickling JK, McIndoe GA, et al. Human T cell responses to human papillomavirus type 16 L1 and E6 synthetic peptides: identification of T cell determinants, HLA-DR restriction and virus type specificity. J Gen Virol. 1990;71:423–31. doi: 10.1099/0022-1317-71-2-423. [DOI] [PubMed] [Google Scholar]
- 35.Kadish AS, Ho GY, Burk RD, et al. Lymphoproliferative responses to human papillomavirus (HPV) type 16 proteins E6 and E7: outcome of HPV infection and associated neoplasia. J Natl Cancer Inst. 1997;89:1285–93. doi: 10.1093/jnci/89.17.1285. [DOI] [PubMed] [Google Scholar]
- 36.Gallagher KM, Man S. Identification of HLA-DR1- and HLA-DR15-restricted human papillomavirus type 16 (HPV16) and HPV18 E6 epitopes recognized by CD4+ T cells from healthy young women. J Gen Virol. 2007;88:1470–8. doi: 10.1099/vir.0.82558-0. [DOI] [PubMed] [Google Scholar]
- 37.van der Burg SH, Ressing ME, Kwappenberg KM, et al. Natural T-helper immunity against human papillomavirus type 16 (HPV16) E7-derived peptide epitopes in patients with HPV16-positive cervical lesions: identification of 3 human leukocyte antigen class II-restricted epitopes. Int J Cancer. 2001;91:612–18. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1119>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Santin AD, Bellone S, Gupta S, Nakagawa M. A novel CD4 T-cell epitope described from one of the cervical cancer patients vaccinated with HPV 16 or 18 E7-pulsed dendritic cells. Cancer Immunol Immunother. 2009;58:301–8. doi: 10.1007/s00262-008-0525-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans M, Borysiewicz LK, Evans AS, et al. Antigen processing defects in cervical carcinomas limit the presentation of a CTL epitope from human papillomavirus 16 E6. J Immunol. 2001;167:5420–8. doi: 10.4049/jimmunol.167.9.5420. [DOI] [PubMed] [Google Scholar]
- 40.Ressing ME, Sette A, Brandt RM, et al. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J Immunol. 1995;154:5934–43. [PubMed] [Google Scholar]
- 41.Ressing ME, van Driel WJ, Celis E, et al. Occasional memory cytotoxic T-cell responses of patients with human papillomavirus type 16-positive cervical lesions against a human leukocyte antigen-A *0201-restricted E7-encoded epitope. Cancer Res. 1996;56:582–8. [PubMed] [Google Scholar]
- 42.Morishima S, Akatsuka Y, Nawa A, et al. Identification of an HLA-A24-restricted cytotoxic T lymphocyte epitope from human papillomavirus type-16 E6: the combined effects of bortezomib and interferon-gamma on the presentation of a cryptic epitope. Int J Cancer. 2007;120:594–604. doi: 10.1002/ijc.22312. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa M, Kim KH, Moscicki AB. Patterns of CD8 T-cell epitopes within the human papillomavirus type 16 (HPV 16) E6 protein among young women whose HPV 16 infection has become undetectable. Clin Diagn Lab Immunol. 2005;12:1003–5. doi: 10.1128/CDLI.12.8.1003-1005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa M, Kim KH, Gillam TM, Moscicki AB. HLA class I binding promiscuity of the CD8 T-cell epitopes of human papillomavirus type 16 E6 protein. J Virol. 2007;81:1412–23. doi: 10.1128/JVI.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa M, Kim KH, Moscicki AB. Different methods of identifying new antigenic epitopes of human papillomavirus type 16 E6 and E7 proteins. Clin Diagn Lab Immunol. 2004;11:889–96. doi: 10.1128/CDLI.11.5.889-896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Moscicki AB, Tsang L, Brockman A, Nakagawa M. Memory T cells specific for novel human papillomavirus type 16 (HPV16) E6 epitopes in women whose HPV16 infection has become undetectable. Clin Vaccine Immunol. 2008;15:937–45. doi: 10.1128/CVI.00404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oerke S, Hohn H, Zehbe I, et al. Naturally processed and HLA-B8-presented HPV16 E7 epitope recognized by T cells from patients with cervical cancer. Int J Cancer. 2005;114:766–78. doi: 10.1002/ijc.20794. [DOI] [PubMed] [Google Scholar]
- 48.Kenter GG, Welters MJ, Valentijn AR, et al. Phase I immunotherapeutic trial with long peptides spanning the E6 and E7 sequences of high-risk human papillomavirus 16 in end-stage cervical cancer patients shows low toxicity and robust immunogenicity. Clin Cancer Res. 2008;14:169–77. doi: 10.1158/1078-0432.CCR-07-1881. [DOI] [PubMed] [Google Scholar]
- 49.Welters MJ, Kenter GG, Piersma SJ, et al. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin Cancer Res. 2008;14:178–87. doi: 10.1158/1078-0432.CCR-07-1880. [DOI] [PubMed] [Google Scholar]
- 50.Piersma SJ, Jordanova ES, van Poelgeest MI, et al. High number of intraepithelial CD8+ tumor-infiltrating lymphocytes is associated with the absence of lymph node metastases in patients with large early-stage cervical cancer. Cancer Res. 2007;67:354–61. doi: 10.1158/0008-5472.CAN-06-3388. [DOI] [PubMed] [Google Scholar]
- 51.Chan PK, Cheung TH, Lin CK, et al. Association between HLA-DRB1 polymorphism, high-risk HPV infection and cervical neoplasia in southern Chinese. J Med Virol. 2007;79:970–6. doi: 10.1002/jmv.20805. [DOI] [PubMed] [Google Scholar]
- 52.Lie AK, Skarsvag S, Haugen OA, et al. Association between the HLA DQB1*0301 gene and human papillomavirus infection in high-grade cervical intraepithelial neoplasia. Int J Gynecol Pathol. 1999;18:206–10. doi: 10.1097/00004347-199907000-00003. [DOI] [PubMed] [Google Scholar]
- 53.Saito M, Okubo M, Hirata R, Takeda S, Maeda H. Association of human leukocyte antigen and T cell message with human papillomavirus 16-positive cervical neoplasia in Japanese women. Int J Gynecol Cancer. 2007;17:1314–21. doi: 10.1111/j.1525-1438.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- 54.Schiff MA, Apple RJ, Lin P, Nelson JL, Wheeler CM, Becker TM. HLA alleles and risk of cervical intraepithelial neoplasia among southwestern American Indian women. Hum Immunol. 2005;66:1050–6. doi: 10.1016/j.humimm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Ades S, Koushik A, Duarte-Franco E, et al. Selected class I and class II HLA alleles and haplotypes and risk of high-grade cervical intraepithelial neoplasia. Int J Cancer. 2008 doi: 10.1002/ijc.23459. [DOI] [PubMed] [Google Scholar]
- 56.Sastre-Garau X, Cartier I, Jourdan-Da Silva N, De Cremoux P, Lepage V, Charron D. Regression of low-grade cervical intraepithelial neoplasia in patients with HLA-DRB1*13 genotype. Obstet Gynecol. 2004;104:751–5. doi: 10.1097/01.AOG.0000139834.84628.61. [DOI] [PubMed] [Google Scholar]
- 57.Krul EJ, Schipper RF, Schreuder GM, Fleuren GJ, Kenter GG, Melief CJ. HLA and susceptibility to cervical neoplasia. Hum Immunol. 1999;60:337–42. doi: 10.1016/s0198-8859(98)00127-x. [DOI] [PubMed] [Google Scholar]
- 58.Peng S, Trimble C, Wu L, et al. HLA-DQB1*02-restricted HPV-16 E7 peptide-specific CD4+ T-cell immune responses correlate with regression of HPV-16-associated high-grade squamous intraepithelial lesions. Clin Cancer Res. 2007;13:2479–87. doi: 10.1158/1078-0432.CCR-06-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piersma SJ, Welters MJ, van der Hulst JM, et al. Human papilloma virus specific T cells infiltrating cervical cancer and draining lymph nodes show remarkably frequent use of HLA-DQ and -DP as a restriction element. Int J Cancer. 2008;122:486–94. doi: 10.1002/ijc.23162. [DOI] [PubMed] [Google Scholar]
- 60.Todd RW, Roberts S, Mann CH, Luesley DM, Gallimore PH, Steele JC. Human papillomavirus (HPV) type 16-specific CD8+ T cell responses in women with high grade vulvar intraepithelial neoplasia. Int J Cancer. 2004;108:857–62. doi: 10.1002/ijc.11645. [DOI] [PubMed] [Google Scholar]