Abstract
The bovine papillomavirus E5 protein is a 44-aa transmembrane protein that forms a stable complex with the cellular platelet-derived growth factor (PDGF) β receptor and induces constitutive tyrosine phosphorylation and activation of the receptor, resulting in cell transformation. The E5 protein does not resemble PDGF, but rather activates the receptor in a ligand-independent fashion, thus providing a unique system to examine activation of receptor tyrosine kinases. Here, we used a variety of approaches to explore the mechanism of receptor activation by the E5 protein. Chemical cross-linking experiments revealed that the E5 protein activated only a small fraction of the endogenous PDGF β receptor in transformed fibroblasts and suggested that this fraction was constitutively dimerized. Coimmunoprecipitation experiments using extracts of cells engineered to coexpress full-length and truncated PDGF β receptors confirmed that the E5 protein induced oligomerization of the receptor. Furthermore, in cells expressing the E5 protein, a kinase-active receptor was able to trans-phosphorylate a kinase-negative mutant receptor but was unable to catalyze intramolecular autophosphorylation. These results indicated that the E5 protein induced PDGF β receptor activation by forming a stable complex with the receptor, resulting in receptor dimerization and trans-phosphorylation.
The binding of ligands to the extracellular domain of receptor tyrosine kinases typically results in receptor dimerization (1–3). This allows trans-phosphorylation of receptor molecules in the dimeric complex, stimulation of the intrinsic tyrosine kinase activity of the receptor, and receptor autophosphorylation, thereby generating binding sites on the receptor for signal-transduction proteins (4). These events initiate a signaling cascade that ultimately culminates in a variety of downstream events, including cell proliferation (5).
The E5 protein of bovine papillomavirus type 1 is a 44-aa, homodimeric transmembrane protein that transforms cultured fibroblasts by activating the platelet-derived growth factor (PDGF) β receptor, a receptor tyrosine kinase (5–8). PDGF β receptor activated by the E5 protein displays increased tyrosine kinase activity in vitro, constitutive tyrosine phosphorylation, and constitutive binding to proteins involved in normal signaling by the PDGF receptor, including phosphatidylinositol 3′-kinase, phospholipase C-γ, and ras GTPase-activating protein (9). There is substantial genetic, biochemical, and physiological evidence that PDGF β receptor activation plays an essential role in E5-induced cell transformation (7–16).
Although both the E5 protein and PDGF activate the PDGF β receptor, they do not share significant structural similarity, and the mechanism by which the E5 protein activates the PDGF β receptor is not known. Like PDGF, the E5 protein forms a stable complex with the activated PDGF β receptor in transformed cells, but it does not form complexes with or activate other, even closely related receptor tyrosine kinases (9–13, 15–18). The E5 protein also can associate with a subunit of the vacuolar H+–ATPase (19), but the role of this interaction in transformation is not clear (20). Schlegel and colleagues (20) have reported E5 mutants that activate the PDGF β receptor without appearing to form a stable complex with it. However, our ongoing analysis strongly suggests that receptor activation and transformation of mouse C127 fibroblasts and Ba/F3 cells requires stable complex formation between the E5 protein and the PDGF β receptor (refs. 13, 16, and 21; unpublished data). Genetic and biochemical studies indicate that the E5 protein interacts with the transmembrane and juxtamembrane domain of the PDGF β receptor and that the extracellular ligand-binding domain of the receptor is not required for complex formation and receptor activation (9, 11, 15, 19, 21). Thus, the interaction between the E5 protein and the PDGF β receptor provides a unique system to investigate the mechanism by which small proteins activate receptor tyrosine kinases in a ligand-independent manner.
Because PDGF triggers receptor activation by inducing receptor dimerization, the E5 protein also may trigger activation by recruiting multiple PDGF β receptor molecules into a complex. We recently proposed (6) a model of the complex between a dimer of the E5 protein and the PDGF β receptor, involving direct interactions between the transmembrane and juxtamembrane domains of these two proteins. A central feature of this model is that the PDGF β receptor in the complex is itself dimeric. However, the oligomeric state of the PDGF β receptor in E5-transformed cells has not been examined. Here, we demonstrate that expression of the E5 protein induced oligomerization, most likely dimerization, of the PDGF β receptor. Moreover, trans-phosphorylation between PDGF β receptor molecules in the complex occurred. Thus, although the E5 protein and PDGF are structurally dissimilar and the interactions that drive binding of these two proteins to the receptor are entirely distinct, the impact on receptor structure and function is similar.
MATERIALS AND METHODS
Expression Vectors.
ΔPR, a human PDGF β receptor lacking amino acids 38–442 in the extracellular ligand-binding domain, was expressed from LXSN-CP84-ΔX, a retrovirus vector encoding G418 resistance (9). K634R is a full-length human PDGF β receptor containing a Lys-to-Arg substitution in the kinase domain that eliminates tyrosine kinase activity (22), which is expressed from the pBabe-Puro retrovirus vector which also encodes puromycin resistance (23). RVY is an empty retrovirus vector encoding hygromycin resistance, and RVY-BE5 and RVY-BE5-D33V express the wild-type E5 protein and a transformation-defective E5 protein, respectively (13, 14). Retrovirus stocks were generated as described (21).
Cell Culture.
Normal and E5-transformed C127 mouse fibroblasts and Ba/F3 cell derivatives were maintained as described (7, 9). Ba/F3 cells expressing ΔPR alone, ΔPR plus the E5 protein, and the E5 protein alone were described previously (9). Additional Ba/F3 derivatives expressing various combinations of foreign proteins were generated as described (21) by sequential infection and selection with either 1 mg/ml G418, 1,000 units/ml of hygromycin B, or 1 μg/ml of puromycin or the appropriate combination of drugs. In general, uncloned drug-resistant cell lines were analyzed, but in some cases in which cloned cells lines were used, similar results were obtained.
Protein Analysis.
Ba/F3-derived cells (5 × 107) were lysed in 1 ml of radioimmunoprecipitation assay (RIPA) buffer [20 mM morpholinepropanesulfonic acid (Mops; pH 7.0)/150 mM NaCl/1 mM EDTA/1% Nonidet P-40/1% deoxycholate/0.1% SDS] containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM Na3VO4, and 10 μg/ml leupeptin and aprotinin on ice, and clarified supernatant was collected. Extracts for coimmunoprecipitation were used immediately; otherwise, extracts were frozen at −70°C for future use.
For E5 immunoprecipitation, 8–10 μl of anti-E5 rabbit antiserum (αE5) was added to 1,000–1,500 μg of extract (9). For immunoprecipitation of full-length and truncated PDGF β receptor, 4–8 μl of anti-PDGF β receptor rabbit antiserum (αPR), raised against the 13-amino acid carboxyl-terminal peptide of PDGF β receptor, was added to 100–400 μg of extract (17). For specific immunoprecipitation of full-length PDGF β receptor, 8–10 μl of anti-human PDGF β receptor antibody [which we designate αPRex (catalog no. 1263–00, Genzyme)], an mAb recognizing the extracellular ligand-binding domain of human PDGF β receptor, was added to 600–1,000 μg of extract. After rotating at 4°C for 2 hr to overnight, antibody–antigen complexes were collected with protein A-Sepharose. Rabbit anti-mouse serum (Pierce) was added first in the case of the αPRex immunoprecipitations. The immunoprecipitate was washed and resuspended in 8–15 μl of 2× Laemmli sample buffer containing DTT and 3% 2-mercaptoethanol and boiled for 5 min. For phosphatase treatment, the washed immunoprecipitate was resuspended in 20 mM Tris (pH 7.4) and 0.1% 2-mercaptoethanol, 5.3 milliunits of protein tyrosine phosphatase 34-kDa fragment (Boehringer Mannheim cat. no. 1500775) was added, and the mixture was incubated at 37°C for 60 min before addition of sample buffer. The boiled samples were electrophoresed in 7.5% SDS/PAGE gels. The separated proteins were transferred to Immobilon-P (Millipore) and probed with αPR to detect total PDGF receptor or with mAb 4G10 (Upstate Biotechnology, Lake Placid, NY) to detect phosphotyrosine, as described (9, 21). To strip a filter for reprobing, it was incubated in 2% SDS, 62.5 mM Tris⋅HCl (pH 6.7), and 100 mM 2-mercaptoethanol at 70°C for 15 min with occasional agitation, washed three times with 10 mM Tris·HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40, and incubated in blocking solution at room temperature for at least 1 hr.
Chemical Cross-Linking.
Confluent RVY or RVY E5-transformed C127 cells were serum-starved for 1 day. Cells were treated with PDGF BB (25 ng/ml) for 1.5 hr at 4°C or left untreated. After lysis with EBC buffer (50 mM Tris, pH 8.0/120 mM NaCl/0.5% Nonidet P-40) containing 1 mM PMSF, 1 mM Na3VO4, 10 μg/ml leupeptin and aprotinin, the extracts were treated with 1 mM BS3 [bis(sulfosuccinimidyl) suberate] for 30 min at room temperature or left untreated. The cross-linking reaction was stopped by adding methylammonium chloride to a final concentration of 70 mM. αPR was used to immunoprecipitate the PDGF receptor, and after electrophoresis on 5% SDS/PAGE gels, either αPR or anti-phosphotyrosine antibody were used for immunoblotting.
RESULTS
Cross-Linking Experiments.
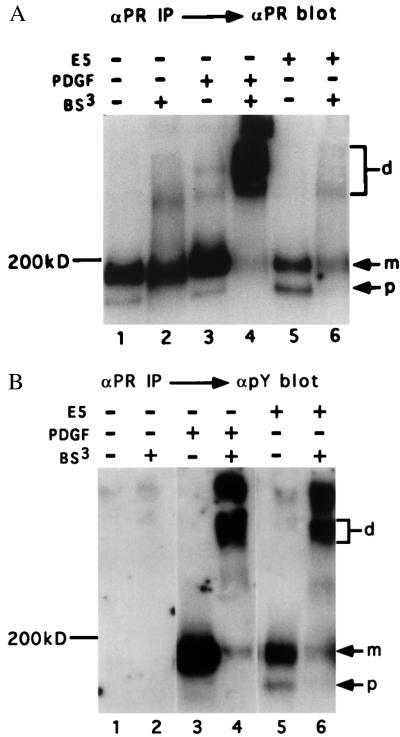
To assess the ability of the E5 protein to induce oligomerization of the endogenous PDGF β receptor in mouse fibroblasts, chemical cross-linking experiments were carried out. Normal mouse C127 cells or cells stably transformed by the E5 protein were treated with PDGF or left untreated. Cells then were lysed in detergent-containing buffer, and a portion of each cell extract was treated with the cross-linking agent, BS3. Total PDGF β receptor was immunoprecipitated and electrophoresed in a gel containing SDS to disrupt any noncovalent bonds. After transfer, the filter was probed with αPR to visualize total receptor (Fig. 1A) or with an antibody that recognizes phosphotyrosine to visualize activated receptor (Fig. 1B).
Figure 1.
Cross-linking analysis of PDGF β receptor in C127 cells. Extracts of PDGF-treated or untreated normal or E5-transformed C127 cells were prepared and treated with the chemical cross-linker BS3 or left untreated. Total PDGF β receptor was immunoprecipitated with αPR and detected by immunoblotting with αPR (A) or anti-phosphotyrosine antibody (B). In B, approximately 3-fold more extract was loaded in lanes 5 and 6 as in the other lanes to facilitate visualization of activated PDGF β receptor species. The position of monomeric mature (m) and precursor (p) PDGF β receptor is shown, as is the position of dimeric receptor (d).
In the absence of PDGF and cross-linker treatment, the PDGF β receptor migrated as a ≈200-kDa mature form and a 180-kDa precursor form with immature carbohydrates (Fig. 1A, lane 1). PDGF treatment resulted in a dramatic decrease in the mobility of the receptor in the sample exposed to BS3 (Fig. 1A, lane 4), but not in the sample unexposed to cross-linker (Fig. 1A, lane 3). A broad band that entered the resolving gel was observed and is thought to consist of dimers of ligand-activated receptor (24, 25). In addition, a significant amount of cross-linked receptor failed to enter the gel, which may represent higher order oligomers or dimers cross-linked to other cellular proteins, such as downstream substrates. In contrast, cross-linker treatment had little effect on the mobility of the receptor isolated from E5-transformed cells (lane 6). Thus, expression of the E5 protein did not appear to induce dimerization of a significant fraction of the PDGF β receptor.
PDGF β receptor also was examined by phosphotyrosine blotting (Fig. 1B). In the absence of cross-linker treatment, PDGF induced tyrosine phosphorylation of the mature cell-surface form of the PDGF β receptor (Fig. 1, lane 3), whereas the E5 protein induced tyrosine phosphorylation of both the mature and immature receptor forms (Fig. 1B, lane 5). As expected, treatment with cross-linker induced dimerization of the activated receptor in extracts of PDGF-treated cells (Fig. 1B, lane 4). Strikingly, cross-linker treatment converted a substantial fraction of the activated PDGF β receptor in E5-transformed cells into slower migrating species that comigrated with the cross-linked activated receptor in PDGF-treated cells (Fig. 1B, lane 6), suggesting that much of the E5-activated PDGF receptor was constitutively dimeric. However, because the majority of total PDGF β receptor in E5-transformed cells migrated as a monomer after cross-linking (Fig. 1A), the E5 protein appeared to induce activation of only a small fraction of the receptor.
Coimmunoprecipitation Experiments.
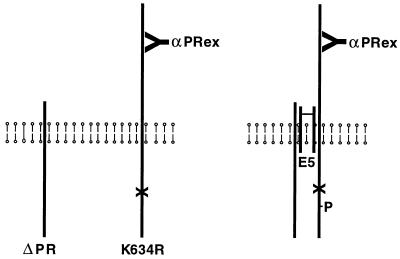
To obtain further evidence that the E5 protein induced oligomerization of the PDGF β receptor, we carried out experiments in murine Ba/F3 cells, which do not express endogenous PDGF β receptor. In these cells, complex formation can occur between the E5 protein and exogenous PDGF β receptor, resulting in tyrosine phosphorylation of the PDGF β receptor and interleukin-3-independent cell proliferation (9). We established stable Ba/F3 cell lines expressing various combinations of the E5 protein and two forms of the human PDGF β receptor: a full-length PDGF β receptor (K634R) containing a mutation that abolished enzymatic activity and a truncated version (ΔPR) lacking most of the extracellular ligand-binding domain but retaining a catalytically active kinase domain (Fig. 2). This truncation does not result in constitutive activation of the receptor (9).
Figure 2.
Schematic diagram of coimmunoprecipitation experiments. The vertical lines represent the full-length kinase-negative receptor (K634R), the truncated kinase-active receptor (ΔPR), and a dimer of the E5 protein traversing the cell membrane. αPRex antibody, which specifically recognizes the extracellular domain of the full-length receptor, also is shown. (Left) Receptors existing independently in cells in the absence of the E5 protein. (Right) Heteromeric receptor complex held together by the E5 dimer. (X) represents the mutation in K634R that destroys kinase activity, and (P) represents phosphate on the kinase-negative receptor trans-phosphorylated in response to the E5 protein.
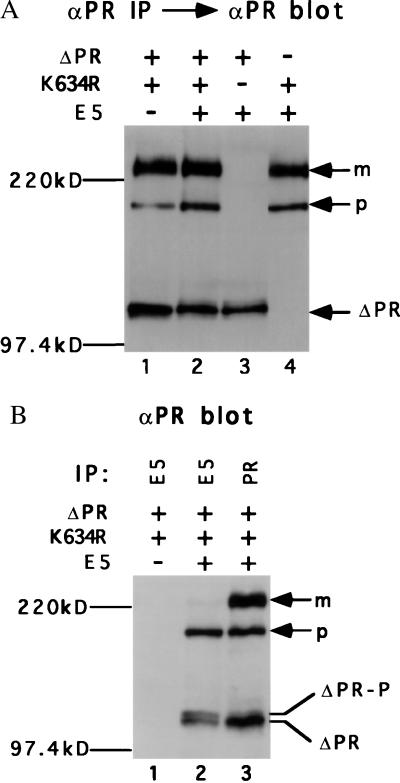
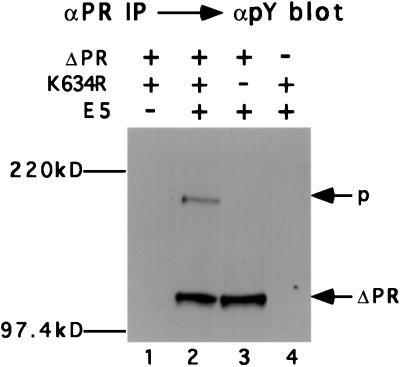
To assess receptor expression, cell extracts were immunoprecipitated and immunoblotted with the αPR antiserum. Cells expressing the full-length kinase-negative receptor (K634R) expressed both mature and immature receptor forms, whereas cells expressing the truncated receptor (ΔPR) expressed a major species that migrates with the mobility of an ≈110-kDa protein (Fig. 3A). To determine the receptor forms able to complex with the E5 protein, cell extracts were immunoprecipitated with the E5 antiserum and immunoprecipitated receptors were identified by immunoblotting with αPR. As shown in Fig. 3B (lane 2), for cells expressing all three proteins, the E5 antiserum coimmunoprecipitated the precursor form of the full-length receptor and two closely migrating species of the truncated receptor, which are described below. The E5 antiserum did not immunoprecipitate the PDGF β receptor from extracts that did not contain the E5 protein (Fig. 3B, lane 1), confirming that the E5 antiserum did not cross-react with the PDGF β receptor. These results demonstrated that the E5 protein formed a stable complex with both full-length and truncated receptors and implied that complexes containing the E5 protein were localized intracellularly. This is consistent with the known localization of the E5 protein to membranes of the Golgi apparatus and endoplasmic reticulum (26).
Figure 3.
Characterization of Ba/F3 cells. Extracts from Ba/F3 cells expressing the indicated proteins were immunoprecipitated with αPR (A) or the indicated antiserum (B). After gel electrophoresis, PDGF β receptor was detected by immunoblotting with αPR. The position of the mature (m) and precursor (p) forms of the full-length PDGF β receptor is shown, as is the position of the truncated receptor (ΔPR) and phosphorylated truncated receptor (ΔPR-P).
To assess the ability of the E5 protein to induce oligomerization between the full-length and truncated PDGF β receptor species, lysates of cells coexpressing both receptors were immunoprecipitated with αPRex antiserum that specifically recognized the ligand-binding domain of the full-length receptor but did not recognize the truncated receptor. Immunoprecipitated receptor species were detected by immunoblotting with αPR that recognized both full-length and truncated receptors, which were readily distinguished by their different electrophoretic mobility. If the two receptor mutants exist independently in cells (Fig. 2, Left), the αPRex antibody should immunoprecipitate only the full-length receptor, whereas αPRex should coimmunoprecipitate truncated receptor associated with the full-length version if these two receptor forms coexist in a heteromeric receptor complex (Fig. 2, Right). As shown in Fig. 4, the αPRex antibody coimmunoprecipitated the truncated receptor from extracts of cells expressing both receptor species and the E5 protein (lane 2) but not from extracts of cells lacking the E5 protein (lane 1). No PDGF β receptor was precipitated from cells coexpressing the E5 protein and the truncated receptor without the full-length receptor (Fig. 4, lane 4), confirming the specificity of αPRex for the full-length form. These results were obtained in multiple, independently derived cell lines and established that expression of the E5 protein induced the formation of complexes containing both full-length and truncated PDGF β receptor molecules. The relatively small amount of truncated receptor in complex with the full-length receptor was consistent with the results of the chemical cross-linking experiments, which demonstrated that only a small fraction of the PDGF β receptor was activated by the E5 protein in transformed cells.
Figure 4.
Oligomerization of the PDGF β receptor in response to the E5 protein. Extracts of Ba/F3 cells expressing the indicated proteins were immunoprecipitated with αPR, which recognizes total PDGF β receptor or with αPRex, which recognizes only the full-length receptor, as indicated. Receptor species immunoprecipitated by these antisera were detected by immunoblotting with αPR. Position of mature (m) and precursor (p) forms of full-length receptor is shown, as is position of truncated receptor (ΔPR).
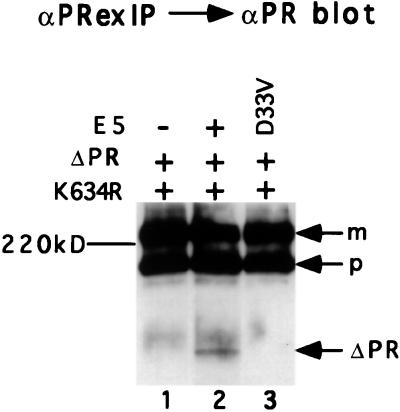
We next determined whether receptor oligomerization required an E5 protein able to complex with the PDGF β receptor. Cells coexpressing the full-length and truncated receptors were infected with retroviruses encoding either the wild-type E5 protein or a transformation-defective E5 protein with an Asp-to-Val mutation at position 33 (D33V). This mutation impaired binding to the full-length and truncated PDGF β receptor and prevented receptor activation and cell transformation (ref. 13; data not shown). The wild-type and mutant E5 proteins were expressed at similar levels in the cells (data not shown). When extracts of these cells were immunoprecipitated with αPRex, the truncated receptor was coimmunoprecipitated from the cells expressing the wild-type E5 protein (Fig. 5, lane 2) but not from cells expressing the E5 mutant (lane 3). Similar results were obtained in extracts from cells expressing a transformation-defective E5 mutant containing a Glu-to-Leu substitution at amino acid 17 that impaired complex formation (ref. 13; data not shown). Therefore, mutant E5 proteins unable to bind the PDGF β receptor were unable to induce receptor oligomerization.
Figure 5.
Receptor oligomerization by wild-type and mutant E5 proteins. Ba/F3 cells expressing the full-length and truncated forms of the PDGF β receptor were engineered to express the wild-type E5 protein or a mutant protein containing the Asp-to-Val substitution at position 33 (D33V). Detergent extracts were immunoprecipitated with αPRex and immunoblotted with αPR to detect truncated receptor associated with the full-length receptor. Position of receptor species is indicated as described in the legend to Fig. 4.
Trans-Phosphorylation of the PDGF β Receptor in Response to the E5 Protein.
We also tested the ability of the E5-activated, truncated PDGF β receptor to catalyze trans-phosphorylation of the kinase-negative receptor. Total PDGF β receptor was immunoprecipitated from cells expressing various combinations of exogenous proteins, and immunoprecipitated proteins were subjected to electrophoresis and immunoblotting with the anti-phosphotyrosine antibody (Fig. 6). Neither receptor species was tyrosine-phosphorylated in the absence of E5 protein expression (Fig. 6, lane 1). In cells expressing the truncated receptor alone, the E5 protein induced abundant receptor tyrosine phosphorylation (Fig. 6, lane 3), confirming that the E5 protein could activate this molecule. As expected, the E5 protein did not induce detectable tyrosine phosphorylation of the full-length kinase-negative receptor when this was the only receptor species expressed in the cells (Fig. 6, lane 4). Strikingly, simultaneous expression of the E5 protein and the two species of receptor resulted in the tyrosine phosphorylation of not only the truncated, kinase-active receptor, but also of the precursor form of the full-length kinase-negative receptor (Fig. 6, lane 2). Thus, the kinase-active receptor was able to trans-phosphorylate only the precursor form of the kinase-negative receptor, the form that associated with the E5 protein. In contrast, the E5 mutants unable to induce receptor oligomerization were also defective for inducing phosphorylation of the kinase-negative receptor (data not shown).
Figure 6.
Trans-phosphorylation of kinase-negative PDGF β receptor. Extracts of Ba/F3 cells expressing the indicated proteins were immunoprecipitated with αPR, and tyrosine-phosphorylated species were detected by immunoblotting with anti-phosphotyrosine antibody. Position of receptor species is indicated as described in the legend to Fig. 4.
To examine tyrosine phosphorylation of the PDGF β receptor in more detail, extracts from cells coexpressing the E5 protein and both species of the PDGF β receptor were analyzed by using immunoprecipitation with various antibodies followed by immunoblotting. The same filter was probed sequentially with αPR and anti-phosphotyrosine antibody, and the bands were identified by superimposing the resulting films. In confirmation of the results shown in Fig. 6, the precursor form of kinase-negative full-length receptor was trans-phosphorylated (Fig. 7A, lane 4). When E5-associated receptor was examined by immunoprecipitation with αE5, both the precursor form of the full-length kinase-negative receptor and the truncated, kinase-active receptor contained phosphotyrosine (Fig. 7A, lane 1). In contrast, as revealed by using αPRex to immunoprecipitate the full-length receptor and any associated truncated receptor, the truncated, kinase-active receptor in a complex with the full-length receptor was not detectably tyrosine-phosphorylated (Fig. 7A, lane 3). This finding implies that when the kinase-active receptor was in the heteromeric receptor complex, it was able to trans-phosphorylate the kinase-negative receptor, but it was not able to phosphorylate itself. Evidently, the tyrosine-phosphorylated truncated receptor isolated from these cells existed exclusively in a homodimeric complex of two truncated receptor molecules.
Figure 7.
Characterization of PDGF β receptor phosphorylation. Extracts of Ba/F3 cells coexpressing the E5 protein, the full-length kinase-inactive receptor, and the truncated kinase-active receptor were immunoprecipitated with αPR, αPRex, or αE5, as indicated. The indicated samples also were treated with protein tyrosine phosphatase (PTPase). After gel electrophoresis, the immunoprecipitated receptor species were detected by probing the filter with anti-phosphotyrosine antibody (A) and αPR (B). The position of receptor species is indicated as described in the legend to Fig. 3.
We confirmed the phosphorylation status of the truncated receptor present in the heteromeric receptor complex by examining its mobility. The truncated receptor consisted of two closely migrating forms (Fig. 7B, lane 4). The less abundant, more slowly migrating form reacted with phosphotyrosine antibody (Fig. 7A, lane 4) and was converted to the more rapidly migrating species by phosphatase treatment (Fig. 7B, lane 5). The E5 antiserum immunoprecipitated similar amounts of both forms of the truncated PDGF β receptor (Fig. 7B, lane 1). The more slowly migrating form of the E5-associated truncated receptor also reacted with the anti-phosphotyrosine antibody (Fig. 7A, lane 1) and was converted to the more rapidly migrating form by phosphatase treatment (Fig. 7B, lane 2). The αPRex antibody coimmunoprecipitated only the more rapidly migrating, unphosphorylated form of the truncated PDGF β receptor (Fig. 7B, lane 3). We conclude that the truncated receptor present in the heteromeric complex with the kinase-negative receptor was not tyrosine-phosphorylated.
DISCUSSION
The PDGF β receptor can be activated by two strikingly dissimilar proteins, PDGF and the E5 oncoprotein of bovine papillomavirus. Studies of the interaction between the E5 protein and the PDGF β receptor will help determine the mechanism of viral transformation and establish how receptors are activated by proteins that do not resemble their normal ligands. The results presented here provide biochemical evidence that the PDGF β receptor in the activated complex is dimeric and strongly suggest that E5-induced dimerization of the PDGF β receptor is the crucial trigger that drives receptor activation and cell transformation.
Expression of the E5 protein resulted in the appearance of a form of PDGF β receptor that, when cross-linked, comigrated with dimeric receptor induced by PDGF treatment, suggesting that the E5 protein induced dimerization of the PDGF β receptor. Most of the tyrosine-phosphorylated PDGF β receptor in E5-transformed cells was in this oligomeric form, indicating that the great majority of E5-activated receptor was oligomeric. However, E5-induced dimeric receptor was apparent only by phosphotyrosine blotting, whereas immunoblotting for total PDGF receptor detected primarily monomeric receptor. Thus, the E5 protein activated only a small fraction of the PDGF β receptor in cells, probably because the E5 protein is located largely in the endoplasmic reticulum and Golgi apparatus, whereas most of the PDGF β receptor is located at the cell surface.
In our coimmunoprecipitation analysis of cells coexpressing full-length and truncated receptors, there was no detectable association between the truncated and the full-length receptor in the absence of the E5 protein. However, expression of the E5 protein resulted in the coimmunoprecipitation of the truncated receptor by the antiserum specific for the full-length receptor, confirming that the E5 protein induced oligomerization of a small fraction of the receptor. Tyrosine phosphorylation of the precursor form of full-length, kinase-negative receptor occurred when it was expressed together with the kinase-active receptor and the E5 protein, providing additional, independent evidence that the E5 protein induced at least transient association between kinase-active and kinase-negative receptors. In contrast, the truncated, kinase-active PDGF β receptor that was stably associated with the kinase-negative receptor was not phosphorylated. This implies that there was only a single molecule of kinase-active receptor in the heteromeric receptor complex, because if there was more than one, it too, presumably, could be trans-phosphorylated. Thus, the heteromeric complex is likely to contain two molecules of the PDGF β receptor and not higher order receptor oligomers. Similarly, the absence of phosphorylated, truncated receptor in the heteromeric receptor complex suggested that trans-phosphorylation is catalyzed by the active subunit within each E5-induced dimer and not by a separate, active dimer. In contrast, structural information suggests that trans-phosphorylation occurs between dimers of the ligand-activated fibroblast growth factor receptor 1 (27).
Several lines of evidence indicate that complex formation between the E5 protein and the PDGF β receptor is required for receptor activation. First, an E5 protein containing an epitope tag preferentially bound the precursor form of the receptor in C127 cells and activated only this form (17). Second, mutations in the E5 protein or in the receptor that impaired complex formation also impaired receptor activation (refs. 13, 16, and 21; unpublished data). Third, in the Ba/F3 cells examined here, the precursor was the only form of the full-length receptor that formed a complex with the E5 protein and was the only form trans-phosphorylated by the kinase-active receptor. Finally, transformation-defective E5 point mutants unable to bind the receptor did not induce receptor oligomerization or trans-phosphorylation. We conclude that the E5 protein must bind the PDGF β receptor to induce activation. Thus, our results suggest a simple model by which the E5 protein causes ligand-independent receptor activation. When the E5 protein and the PDGF β receptor are coexpressed in the membranes of the secretory pathway, interactions between the transmembrane and juxtamembrane domains of the two proteins occur and mediate complex formation between the dimeric E5 protein and two molecules of the PDGF receptor. In the resulting dimeric receptor complex, each receptor molecule is able to catalyze trans-phosphorylation of the other, resulting in full catalytic activity. It is not known whether E5-activated intracellular receptor is competent to deliver a proliferative signal or whether some activated receptor must be present at the cell surface for proliferative signaling. Binding of dimeric PDGF to the extracellular domain of the receptor causes receptor dimerization at the cell surface, which allows trans-phosphorylation of the receptor at a specific tyrosine in the activation loop (3, 5, 24, 25, 28–36). Trans-phosphorylation is thought to result in a conformational change in the loop that releases inhibitory constraints on substrate binding and increases the catalytic activity of the enzyme (33, 36). E5 (or PDGF) binding or consequent receptor dimerization may induce a conformational change in the catalytic domain of the receptor that facilitates trans-phosphorylation; alternatively, the mere close juxtaposition of two receptor molecules in the complex may be sufficient to allow trans-phosphorylation and activation.
Because the phosphorylated receptor in the heteromeric receptor complex was kinase-negative, it could not carry out additional phosphorylation reactions, and the activation process appeared to be frozen after an initial phosphorylation step. In this complex, the truncated receptor was unable to catalyze intramolecular autophosphorylation, indicating that the first phosphorylation events after E5-induced receptor dimerization occurred obligatorily in trans. It has not been determined whether intramolecular autophosphorylation of the PDGF receptor occurs in response to ligand addition. Further characterization of the interaction between the PDGF β receptor and the E5 protein may provide general insights into the mechanism of receptor tyrosine kinase activation.
Acknowledgments
We thank A. Kazlauskas for DNA encoding ΔPR and K634R. We also thank O. Klein and P. Irusta for critical comments on this manuscript and J. Zulkeski for assistance in preparing this manuscript. C.H. was supported in part by a Medical Scientist Training Program training grant from the National Institutes of Health. This work was supported by grants from the National Cancer Institute (CA37157 and CA16038).
ABBREVIATION
- PDGF
platelet-derived growth factor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Yarden Y, Schlessinger J. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Trends Biochem Sci. 1988;13:443–447. doi: 10.1016/0968-0004(88)90219-8. [DOI] [PubMed] [Google Scholar]
- 3.Heldin C-H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 5.Claesson-Welsh L. J Biol Chem. 1994;269:32023–32026. [PubMed] [Google Scholar]
- 6.Surti, T., Klein, O., Ascheim, K., DiMaio, D. & Smith, S. O. (1998) Proteins Struct. Funct. Genet., in press. [PubMed]
- 7.Petti L, Nilson L A, DiMaio D. EMBO J. 1991;10:845–855. doi: 10.1002/j.1460-2075.1991.tb08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMaio D, Petti L, Hwang E-S. Semin Virol. 1994;5:369–379. [Google Scholar]
- 9.Drummond-Barbosa D, Vaillancourt R R, Kazlauskas A, DiMaio D. Mol Cell Biol. 1995;5:2570–2581. doi: 10.1128/mcb.15.5.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein D J, Li W, Wang L-M, Heidaran M A, Aaronson S A, Shinn R, Schlegel R, Pierce J H. J Virol. 1994;68:4432–4441. doi: 10.1128/jvi.68.7.4432-4441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen B D, Goldstein D J, Rutledge L, Vass W C, Lowy D R, Schlegel R, Schiller J. J Virol. 1993;67:5303–5311. doi: 10.1128/jvi.67.9.5303-5311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilson L A, DiMaio D. Mol Cell Biol. 1993;13:4137–4145. doi: 10.1128/mcb.13.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilson L A, Gottlieb R, Polack G W, DiMaio D. J Virol. 1995;69:5869–5874. doi: 10.1128/jvi.69.9.5869-5874.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riese D J, DiMaio D. Oncogene. 1995;10:1431–1439. [PubMed] [Google Scholar]
- 15.Staebler A, Pierce J H, Brazinski S, Heidaran M A, Li W, Schlegel R, Goldstein D J. J Virol. 1995;69:6507–6517. doi: 10.1128/jvi.69.10.6507-6517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein O K, Polack G W, Surti T, Kegler-Ebo D, Smith S O, DiMaio D. J Virol. 1998;72:8921–8932. doi: 10.1128/jvi.72.11.8921-8932.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petti L, DiMaio D. Proc Natl Acad Sci USA. 1992;89:6736–6740. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petti L, DiMaio D. J Virol. 1994;68:3582–3592. doi: 10.1128/jvi.68.6.3582-3592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein D J, Andresson T, Sparkowski J J, Schlegel R. EMBO J. 1992;11:4851–4859. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sparkowski J, Mense M, Anders J, Schlegel R. J Virol. 1996;70:2420–2430. doi: 10.1128/jvi.70.4.2420-2430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petti L M, Reddy V, Smith S O, DiMaio D. J Virol. 1997;71:7318–7327. doi: 10.1128/jvi.71.10.7318-7327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazlauskas A, Durden D L, Cooper J A. Cell Regul. 1991;2:413–425. doi: 10.1091/mbc.2.6.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishayee S, Majumdar S, Khire J, Manjusri D. J Biol Chem. 1989;264:11699–11705. [PubMed] [Google Scholar]
- 25.Seifert R A, Hart C E, Phillips P E, Forstrom J W, Ross R, Murray M J, Bowen-Pope D F. J Biol Chem. 1989;264:8771–8778. [PubMed] [Google Scholar]
- 26.Burkhardt A, Willingham M, Gay C, Jeang K-T, Schlegel R. Virology. 1989;170:334–339. doi: 10.1016/0042-6822(89)90391-7. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadi M, Schlessinger J, Hubbard S R. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 28.Eriksson A, Rorsman C, Ernlund A, Claesson-Welsh L, Heldin C-H. Growth Factors. 1992;6:1–14. doi: 10.3109/08977199209008867. [DOI] [PubMed] [Google Scholar]
- 29.Hammacher A, Mellström K, Heldin C-H, Westermark B. EMBO J. 1989;8:2489–2495. doi: 10.1002/j.1460-2075.1989.tb08385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heldin C-H, Ernlund A, Rorsman C, Rönnstrand L. J Biol Chem. 1989;264:8905–8912. [PubMed] [Google Scholar]
- 31.Kanakaraj P, Raj S, Khan S A, Bishayee S. Biochemistry. 1991;30:1761–1767. doi: 10.1021/bi00221a005. [DOI] [PubMed] [Google Scholar]
- 32.Kelly J D, Haldeman B A, Grant F J, Murray M J, Seifert R A, Bowen-Pope D, Cooper J A, Kazlauskas A. J Biol Chem. 1991;266:8987–8992. [PubMed] [Google Scholar]
- 33.Baxter R M, Secrist J P, Vaillancourt R R, Kazlauskas A. J Biol Chem. 1998;273:17050–17055. doi: 10.1074/jbc.273.27.17050. [DOI] [PubMed] [Google Scholar]
- 34.Fantl W J, Escobedo J A, Williams L T. Mol Cell Biol. 1989;9:4473–4478. doi: 10.1128/mcb.9.10.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fantl W J, Escobedo J A, Martin G A, Turck C W, Rosario M del, McCormick F, Williams L T. Cell. 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- 36.Hubbard S R, Mohammadi M, Schlessinger J. J Biol Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]