Abstract
Recent studies using the Drosophila central nervous system as a model have identified key molecules and mechanisms underlying stem cell self-renewal and differentiation. These studies suggest that proteins like Aurora-A, atypical protein kinase C, Prospero and Brain tumor act as key regulators in a tightly coordinated interplay between mitotic spindle orientation and asymmetric protein localization. These data also provide initial evidence that both processes are coupled to cell cycle progression and growth control, thereby regulating a binary switch between proliferative stem self-renewal and differentiative progenitor cell specification. Considering the evolutionary conservation of some of the mechanisms and molecules involved, these data provide a rationale and genetic model for understanding stem cell self-renewal and differentiation in general. The new data gained in Drosophila may therefore lead to conceptual advancements in understanding the aetiology and treatment of human neurological disorders such as brain tumor formation and neurodegenerative diseases.
Key words: stem cell, progenitor, neuroblast, asymmetric division, self-renewal, differentiation, drosophila, prospero, brain tumor
Introduction
Stem cell self-renewal and differentiation has become a major issue in the aetiology and treatment of various diseases. This is particularly evident in the case of solid tumors such as cancers of the colon, breast, lung and brain. There, surgical interventions allow resection and improve local tumor control. However, the further course of the disease often remains dominated by re-appearance of unscheduled cell proliferation and infiltration of normal tissue. These cells often resist apoptotic stimuli from radiotherapy and virtually all chemotherapeutic agents (reviewed in ref. 1). This therapeutic resistance has been attributed to so-called cancer stem cells due to their unrestrained self-renewal capacity and the ability to maintain tumorigenic potential at the single cell level, thereby evading both resection and radiotherapy (reviewed in ref. 2). These observations have led to the “cancer stem cell” hypothesis suggesting that some cancers arise either from normal stem cells or from progenitor cells in which self-renewal pathways have become aberrantly activated. However, insights into the underlying mechanisms are only starting to emerge and rely on understanding the genetic control of stem cell self-renewal and differentiation (reviewed in refs. 3 and 4).
This is equally true considering the therapeutic potential of stem and progenitor cells in cell replacement and transplantation, especially in neurodegenerative disorders. Prevalent in the aging population, neurodegenerative diseases are characterized by the progressive loss of neurons in the central nervous system (CNS). Due to the lack of understanding of the underlying pathogenic mechanisms, most of the current therapeutic approaches are aimed at alleviating motor and psychiatric symptoms, rather than to prevent or halt the progression of the disease.5–7 The lack of restorative treatments available for neurodegenerative disorders such as Parkinson, Huntington or motor neuron disease have led to rising expectations on the potential of stem cell based therapy that may offer a novel treatment option to slow, halt, or even reverse the progression of these devastating illnesses.8–10 Due to the complexity of human brain structure, it may seem daunting to induce functional recovery, simply by replacing the cells lost by the disease. But, recent studies in animal models have demonstrated that neuronal replacement is possible.11
However, several obstacles need to be resolved before stem cell based therapies can be translated clinically.12 One challenge still is to identify molecular determinants of stem cell proliferation so as to control undesired growth and alterations of genetically engineered stem cells, as well as to manage the over-proliferation of the transplanted neural stem cells. There is also need to know how to pattern stem cells to obtain a more complete repertoire of various types of cells for replacement, especially considering the various cellular sub-types in the CNS. On top of that, a major challenge remains to induce effective functional integration of stem cell-derived neurons into existing neural and synaptic networks with the ultimate goal to restore behavioural deficits caused by progressive neurodegeneration.7,11 It is therefore of major therapeutic interest to understand the genetic control of neural stem cell proliferation and differentiation. Although seemingly unrelated to the human nervous system,13 the CNS of the fruitfly Drosophila has become one of the prime model systems to study the genetic mechanisms regulating neural stem cell self-renewal and differentiation.
Stem Cell Proliferation
In both, invertebrates like the insect Drosophila, and mammals, the major characteristic of stem cells is their ability to self-renew. Using various modes of proliferation, stem cells maintain or expand the available stem cell pool, but they can also generate more specialised progeny that constitute the majority of cells in an adult individual. In multi-cellular organisms, totipotent zygotes generate pluripotent stem cells, which become increasingly restricted in their lineage potential during development, and subsequently give rise to mature tissue-specific, multipotent stem cells.14 Stem cells show either ‘proliferative’ symmetric divisions or ‘differentiative' asymmetric divisions to regulate a balance between the maintenance of stem cell pool and the supply of mature cells (Fig. 1A). It is critical for stem cells to tightly control this balance between the two different modes of division, both during development and adulthood, because, failure in maintaining cellular homeostasis may lead to incomplete tissue or organ development, whereas uncontrolled proliferation can lead to tumorigenesis.3
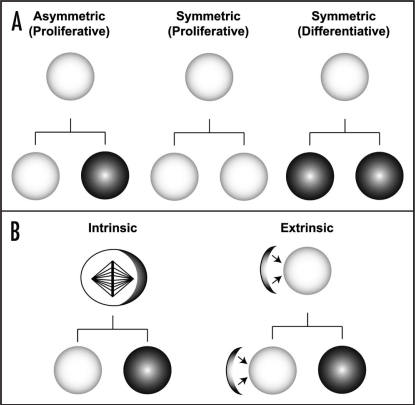
Figure 1.
Modes of stem cell proliferation. (A) Stem cells employ different modes of divisions, which can be either symmetric or asymmetric to regulate a balance between maintaining the number of available stem cells and the supply of differentiating cells. Asymmetric division results in generation of a self-renewing daughter cell (white circles), and a differentiating daughter cell (black circles). Whereas, the symmetric division generates two identical daughter cells, which can be either, the self-renewing stem cells or the differentiating post-mitotic progeny. (B) Asymmetric stem cell division can be regulated by intrinsic or extrinsic mechanisms. Intrinsic mechanisms involve exclusive segregation of the intra-cellular cell fate determinants (black crescent within the stem cell) into the differentiating daughter cell. While, the extrinsic mechanisms rely on the contact with the ‘stem cell niche’ (black crescent adjacent to the stem cell) that provides external cues to maintain self-renewal potential. Thus, the daughter cell that lacks contact to the niche undergoes cell cycle exit and differentiates.
Symmetric cell divisions commonly occur during development of both invertebrates and vertebrates, a phenomena that can also be observed during wound healing and regeneration of tissues.14 This mode of division is defined by the generation of two daughter cells that acquire the same fate, thereby expanding the pool of stem cells required or generating two differentiating daughter cells.15 Asymmetric cell divisions play a key role in generating cellular diversity during development by generating two daughter cells that are committed to different fates in a single division; i.e., simultaneously self-renewing to generate a daughter cell with stem cell properties, as well as to give rise to a more differentiated progeny (Fig. 1A).
Asymmetric stem cell divisions can be controlled by intrinsic mechanisms or the asymmetric exposure to extrinsic cues. Intrinsic mechanisms use apical-basal or planar polarity along the mitotic spindle to asymmetrically segregate cell fate determinants into only one daughter cell (see below). Extrinsic mechanisms rely on contact with the so called ‘stem cell niche’, a cellular microenvironment which provides external cues (reviewed in refs. 16–18). Orientation of its mitotic spindle perpendicular to the niche surface allows the asymmetric segregation of cell fate determinants relative to the external stimuli to maintain self-renewal potential (Fig. 1B). Detailed insights into the genetic mechanisms regulating stem cell proliferation and differentiation are coming from studies using the Drosophila CNS as a model.
Neural Stem Cells in the Drosophila CNS
The Drosophila CNS arises from neural stem cells called neuroblasts (NB), which undergo multiple rounds of stem cell-like divisions. Notably, NBs proliferate during two neurogenic periods.19,20 During the embryonic period of neurogenesis, NBs are specified through lateral inhibition within the mono-layered neuroectoderm, and delaminate as single cells from the epithelium before entering mitosis.21,22 Apical-basal polarity and perpendicularly aligned mitotic spindle allow for asymmetric segregation of neural cell fate determinants into the ganglion mother cell (GMC) upon cytokinesis,22–25 resulting in the generation of two daughter cells with distinct sizes and fate. The larger daughter cell retains Drosophila neural stem cell division and differentiation NB characteristics and continues to divide asymmetrically and self renew, whereas the smaller ganglion mother cell (GMC) daughter usually undergoes a terminal division to produce two post-mitotic neurons/glial cells (Fig. 2). After the completion of embryogenesis, embryonic NBs arrest their cell cycle and remain quiescent until proliferation is restored during larval development.19,20
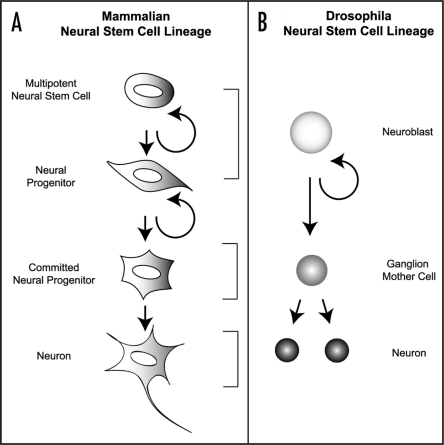
Figure 2.
Neurogenesis in Mammals and Drosophila. (A) In mammalian neurogenesis, a ‘multi-potent’ neural stem cell (NSC) is capable of generating all the lineages in neural specific tissues. A NSC gives rise to a neural progenitor cell which in turn generates a lineage committed progenitor that can directly generate a differentiating neuron. (B) During Drosophila neurogenesis, a neuroblast (NB) divides in a stem cell like fashion to simultaneously give rise to a self-renewing daughter, as well as a smaller differentiating ganglion mother cell (GMC, shown in gray). GMCs are intermediate precursor cells that usually undergo one terminal division to generate two post-mitotic neurons (black).
During the post-embryonic phase of neurogenesis, neurons and glial cells are generated that constitute the majority of the adult Drosophila CNS.26–30 Interestingly, larval NBs do not essentially divide in an apical-basal manner but remain polarized,31,32 and most of them do not shrink with each round of division, as is the case for embryonic NBs.33,34 Rather, many self-renewing larval NBs have the capacity to re-grow back to the size of their parental NB and thereby proliferate for extended periods of time during larval life. As a consequence, postembryonic NBs (pNBs) generate larger lineages of post-mitotic progeny that constitute the majority of the adult CNS through repeated self-renewing, and asymmetric divisions, hence, making them an attractive model system for studying stem cell self-renewal and differentiation.
Several types of larval NBs can be distinguished by their position, size and proliferation pattern (Fig. 3). In the ventral nerve cord (VNC), around 60 pNBs per segment repeatedly divide in an asymmetric manner to form the neurons of the thoracic ganglia, some of which later innervate the wings and legs of the adult animal.27,30 The larval abdominal segments, however, are much smaller which is partially due to the fact that only a small group of embryonic NBs re-enter mitosis and undergo asymmetric cell division.28 Moreover, sexually dimorphic proliferation patterns35 and premature elimination of abdominal pNBs by programmed cell death36–38 also account for the differences in size and function as compared to more anterior segments of the larval and adult CNS.19
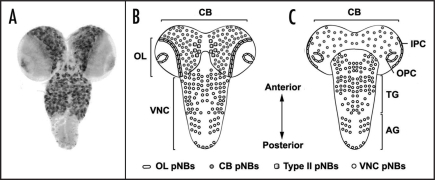
Figure 3.
The Drosophila larval CNS. (A) Overview of post-embryonic neuroblast lineages in the 3rd instar larval CNS, visualised by 1407-Gal4 driven UAS-mCD8::GFP. Schematic representation of dorsal (B) and ventral view (C) of 3rd instar larval CNS. The Drosophila larval CNS is characterised by optic lobes (OL) that consist of the inner (IPC) and outer (OPC) proliferation centres, the central brain (CB) and the ventral nerve cord (VNC) that can be sub-divided into thoracic (TG) and abdominal ganglia (AG). Postembryonic neuroblasts (pNBs) are abundantly localised in the OL, CB and VNC. A recently identified sub-group of type II pNBs are positioned within the dorso-medial region of the CB. Note that abdominal pNBs are not visualized in (A).
pNBs of the larval brain are usually distinguished between two different regions, the optic lobe and the central brain. Optic lobe pNBs arise from two multilayered neuroepithelia called the inner- and outer-proliferation center26 (Fig. 3) and follow a distinct pattern of neurogenesis.39 Thus, optic lobe neuroepithelial cells divide in a proliferative symmetric division mode, thereby expanding the neural stem cell pool at an early phase of larval development. At a later stage, pNBs are generated on the rims of the optic lobe epithelium. These optic lobe pNBs lose their adherens junctions and initiate several rounds of asymmetric cell divisions perpendicular to the epithelial plane,39 thereby generating smaller GMCs which ultimately give rise to differentiating neurons that comprise the visual processing centers of the adult Drosophila brain: lamina, medulla, lobula and lobula plate.40
Of the 100 neuroblasts that can be identified in the embryonic central brain,41 around 85 pNBs re-enter mitosis after quiescence.19,29 These include pNB lineages whose neurons differentiate into specific functional domains of the adult brain such as the central complex involved in courtship behavior and locomotor control, the antennal glomeruli involved in olfactory information processing, and the mushroom bodies which are involved in learning and memory formation.42 Central brain pNBs are heterogeneous in cell cycle length and lineage-specific regulation of self-renewal. This is particularly evident for the four mushroom body pNBs which give rise to 2,500 neurons called Kenyon cells that are part of the memory storage centers in Drosophila.29,43,44 The enormous size of mushroom body pNB lineages is partly due to the fact that these neuroblasts do not enter quiescence at the end of embryogenesis; they rather continue to divide, initially in a symmetric mode of proliferative division, and later switch to asymmetric stem cell divisions, which are maintained until late pupal stages of development.19
Another lineage-specific regulation of self-renewal has been observed for pNBs recently identified in the dorso-medial region of the larval central brain.45–47 Molecular genetic analyses indicate that their mode of division is morphologically symmetrical, but molecularly asymmetrical in that key cell fate determinants are segregated into only one of the two daughter cells.45 These neural stem cells thereby generate secondary, intermediate precursor cells that undergo multiple rounds of self-renewing transit-amplifying divisions.46 Based on these morphological and molecular features, dorso-medial central brain pNB lineages and the resulting intermediate transit amplifying GMCs are described as type II lineages,47 compared to the predominant type I NB lineages where repeated asymmetric cell divisions lead to GMCs that usually undergo one terminal symmetric division to generate two differentiating neurons.48 What are the molecular mechanisms regulating this binary switch between neural stem- and progenitor cell self-renewal and differentiation? One way to regulate this switch is unequal distribution of cell fate determinants by asymmetric cell division.
Mechanisms of Asymmetric Stem Cell Division in the Drosophila CNS
To date, a number of key intrinsic and extrinsic factors that control the asymmetric divisions of NBs have been identified. Among these, the most important trait appears to be the axis of polarity defined by the polarised distribution of two evolutionarily conserved protein complexes that facilitates the orientation of the mitotic spindle in an apical-basal manner to allow the asymmetric segregation of basally localized cell-fate determinants from the NB to the smaller daughter cell, the GMC (Fig. 4). Because there are numerous recent reviews regarding this topic (reviewed in refs. 17, 22, 24, 25, 49–54), only key and novel findings as well as their implications are summarized and reviewed here.
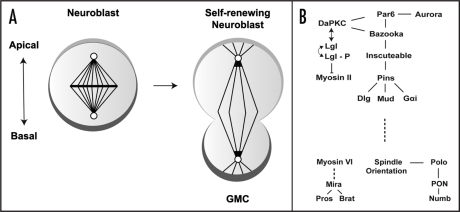
Figure 4.
Molecular mechanisms involved in asymmetric neuroblast division. (A) Apical-basal polarity is used to establish asymmetric localization of cell fate determinants (dark gray crescent) to the basal cortex around late metaphase, before they finally segregate into the smaller daughter, ganglion mother cell (GMC) that undergoes differentiation. Whereas the apically localised protein (light gray crescent) simultaneously ensure stem cell self renewal and maintaining spindle orientation. (B) Summary of key proteins involved in intrinsic asymmetric neuroblast division. See text for details.
Polarity formation.
In the Drosophila embryo, polarisation is established when a neuroblast becomes specified in the polarised epithelium of the neuroectoderm.22 The delaminating neuroblast inherits the protein complex that establishes apical-basal polarity, consisting of the evolutionarily conserved PDZ domain proteins Par-3,55,56 Par-6 (DmPar6),57 and the Drosophila atypical protein kinase C (DaPKC),58–60 from the overlying neuroectoderm, where they are required for maintaining epithelial polarity. Inheritance of this particular set of proteins in a molecular complex to the apical cortex of the embryonic neuroblasts appears to define the orientation of the mitotic spindles as well as the subsequent asymmetric segregation of cell fate determinants to the basal cortex.
Once the neuroblast is delaminated, the mitotic spindle aligns perpendicular to the epithelial plane,61 and an adaptor protein called Inscuteable (Insc)62 binds to the apical protein complex through Bazooka. Inscuteable, in turn, recruits another adaptor protein, Partner of Inscuteable (Pins) that contains three GoLoco domains,63 that bind the heterotrimeric G-protein a-subunit Gai into the complex to form an apical crescent at late interphase/early prophase.64 Binding of Gai to all three of the GoLoco domains enables Pins to recruit an additional protein called Mushroom body defect (Mud),65–67 which is the Drosophila homolog of the microtubule and dynein binding protein NuMA.68,69 Mud is thought to interact with the astral microtubules to ‘fix’ one of the spindle poles on the apical cortex of the neuroblast, thus contributing to the orientation of the mitotic spindle. Pins also binds to a membrane associated guanylyl kinase protein called Discs large (Dlg), that is known to interact with Kinesin heavy chain 73 (Khc-73), localized at the plus ends of astral microtubules. These interactions polarise the complex of proteins localized at the apical cortex of neuroblasts in the direction of the mitotic spindle, which aligns perpendicular to the overlying epithelial plane.64,70,71
Accordingly, mutations in Mud protein cause defects in spindle orientation, leading to over-proliferation of larval central brain neuroblasts due to failure in asymmetric segregation of cell fate determinants.65–67 Moreover, mutation in any component of the apical complex results in mis-localization of the cell fate determinants around metaphase, although, basal crescent formation can occur later on, through a rather enigmatic mechanism called ‘telophase rescue.’72 Molecules of the apical complex therefore direct apical-basal spindle orientation in dividing neuroblasts, and thereby establish an axis of polarity along which cytokinesis takes place. This in turn enables proper asymmetric segregation of cell fate determinants into only one of the resulting daughter cells (Fig. 4).
Asymmetric protein localization.
Once an axis of polarity is established, asymmetric cell division is regulating a binary switch between self-renewal and differentiation. This is mainly achieved by the asymmetric localization and subsequent unequal segregation of fate determinants that promote either stem cell identity or intermediate progenitor cell identity which continues to terminally divide into two differentiating post-mitotic cells. According to the embryonic neuroblast axis of polarity, apically localized proteins are maintained in self-renewing neuroblasts whereas basally localized proteins are segregated into differentiating GMCs.
One of the key substrates that are required for the asymmetric segregation of cell fate determinants include the cortically localized tumour suppressor proteins Dlg and Lethal (2) giant larvae (Lgl).58,73–75 Lgl is a cytoskeletal protein known to specify the basolateral domain and to restrict DaPKC, Bazooka and DmPar6 to the apical cortex.76 Although Lgl does not directly influence spindle orientation and apical localization of the Par complex, phosphorylation of Lgl by DaPKC leads to Lgl inactivation, or exclusion of Lgl from the apical cortex,58 thereby restricting cortical recruitment of basal cell fate determinants. This is in line with Lgl mutants studies, in which the cell fate determinant adaptor protein Miranda (Mira) mis-localizes to the cytoplasm. As a result, Lgl mutant neural lineages lead to multiple pNBs due to occasional ectopic self-renewal,59,77 suggesting that Lgl inhibits uncontrolled neural stem cell self-renewal. Furthermore, overexpression of a membrane-targeted DaPKC, but not a kinase-dead mutant isoform results in increased numbers of larval brain neuroblasts, whereas a decrease in DaPKC expression reduces neuroblast numbers. Genetic interaction experiments showed that Lgl, DaPKC double mutants have normal numbers of neuroblasts and that DaPKC is fully epistatic to Lgl, suggesting that DaPKC directly promotes neuroblast self-renewal.77
Together, these data suggest that DaPKC and Lgl are key players in the establishment and maintenance of apical polarity, thereby providing NBs with the capacity to self-renew. A main question arising from these studies is which mechanisms and molecules are directing DaPKC and Lgl to the apical cortex of a dividing neural stem cell? A partial answer to that comes from recent data suggesting that the mitotic kinase Aurora-A (AurA) is required for the asymmetric localization of DaPKC.78–80 These data suggest that AurA does so via phosphorylation of DmPar6, a member of the apical complex, which in turn prevents an interaction between DmPar6 and DaPKC. Subsequently, phosphorylated DaPKC can act independently of DmPar6 and is able to phosphorylate Lgl, leading to Lgl inactivation/exclusion of Lgl from the apical cortex, a crucial step in restricting cortical recruitment of basal cell fate determinants.58,77 Within the Par complex, this sequence of events leads to the exchange of Lgl for Bazooka, which in turn enables phosphorylation of the cell fate determinant Numb and its subsequent segregation into the differentiating GMCs.80
The important impact of these new data is that they provide a direct link between asymmetric protein localization and mitotic spindle orientation. A linkage between mitotic spindle and apical cortex had already been established with the identification of the Mud/NuMa protein and its role in regulating NB self-renewal via proper spindle-orientation. However, mutant Mud does not alter cortical polarity,65–67 whereas mutant AurA does.78–80 This difference is far from being obvious, as both proteins localize to the centrosomes and mutants of AurA and Mud exhibit similar defects in spindle orientation.65–67,78–83 A possible explanation to this apparent discrepancy comes from genetic interaction data indicating that AurA controls mitotic spindle orientation in dividing neuroblasts by regulating the asymmetric localization of Mud.79 Moreover, AurA seems not only to act on Mud and DmPar6, but also on Notch signalling. Mutational inactivation of AurA leads to ectopic activation of Notch,79 which in its cleaved, intracellular form is able to promote self-renewal and to suppress differentiation of type II pNB lineages in the larval central brain (Diaper and Hirth F, unpublished).
Based on these data, it is conceivable that AurA acts via Mud to orient mitotic spindles required for the establishment of a proper division plane, which is a prerequisite for the unequal segregation of cell fate determinants during NB cytokinesis. Simultaneously, asymmetric protein localization is achieved, at least in part by AurA acting on DmPar6 and in turn via phosphorylation of DaPKC followed by that of Lgl. Such a dual role of AurA linking asymmetric protein localization and mitotic spindle orientation could explain to some extend why in AurA and Mud, but also in DaPKC and Lgl mutants, the net result is the same: supernumerary pNB-like cells at the expense of differentiating neurons.
Basal cell fate determinants.
Self-renewal and differentiation is not only regulated in proliferating NBs. Another stringent control for this binary switch is executed in GMCs that are destined to exit the cell cycle by terminal, symmetric division, thereby generating the majority of neurons that constitute the adult CNS in Drosophila. GMC fate is determined by the exclusive inheritance of key differentiation factors such as the Notch repressor Numb,84 the NHL-domain protein Brain tumour (Brat)85 and the homeodomain transcription factor Prospero85–87 which are collectively known as cell fate determinants. Basal targeting of these cell fate determinants in dividing NBs is achieved via their adaptor proteins, Partner of Numb (Pon)88 and Miranda,89,90 respectively.
Previous experiments showed that Numb is involved in self-renewal and differentiation, as mutant Numb pNB type II lineages over-proliferate at the expense of differentiating neurons.46,78,79 Segregation of Numb into GMCs is regulated by Pon in a cell cycle-dependent manner, and recent data provide evidence that Polo, a key cell cycle regulator itself, is critically required for this event by direct phosphorylation of Pon.91 Accordingly, mutant polo affects the asymmetric localization of Pon, Numb and DaPKC and supernumerary NB-like cells are produced at the expense of neurons. Overexpression of Numb in polo mutant pNB lineages is able to suppress over-proliferation, indicating that Polo inhibits progenitor self-renewal by regulating the localization and function of Numb. As is the case for AurA, polo function therefore provides another link between cell cycle regulation and asymmetric protein localization. However, the mechanism by which Numb directly or indirectly regulates cell cycle activity and proliferation is poorly understood.
In contrast to Numb, insights into this link are coming from studies on Prospero (Pros), another crucial GMC fate determinant. Pros mRNA and protein is already detectable in dividing NBs where it is transported via its adaptor Miranda to the basal side.92–95 Cytokinesis segregates Prospero solely into the GMC where Mira degrades, thereby releasing Prospero from the cortex, which then translocates into the nucleus.96,97 Nuclear activity of Prospero, in turn, inhibits cell cycle progression of the GMC by repressing cell cycle regulators such as cyclin A, cyclin E and the Drosophila cdc25 homologue, string,98 as well as by activating the expression of dacapo,99 a cyclin-dependent kinase inhibitor, ultimately leading to terminal differentiation of the GMC into two post-mitotic neurons/or glial cells. Moreover, genome-wide expression profiling using prospero loss and gain-of function embryos as a template100 indicate that Prospero represses NB-specific apical polarity genes like inscuteable, bazooka and DaPKC, and activates expression of neural differentiation genes such as fushi tarazu and even skipped.101 In addition, mutant analyses provide in vivo evidence that loss of pros results in enlarged pNB lineages essentially devoid of differentiating, post-mitotic neurons.102–104 Instead, the vast majority of cells within these mutant clones show sustained expression of stem cell markers and increased mitotic activity, eventually leading to neoplastic tumor formation.102 These data indicate that loss of pros causes a transformation of GMCs into stem-like cells that are unable to exit the cell cycle and continue to proliferate. Considering the binary role of pros in wildtype GMCs,98–100 these data suggest that in pros mutants pNB lineages, stem cell self-renewal is not repressed and differentiation not initiated. It is, therefore, reasonable to conclude that Prospero is a gate-keeper in regulating self-renewal and differentiation in GMCs.
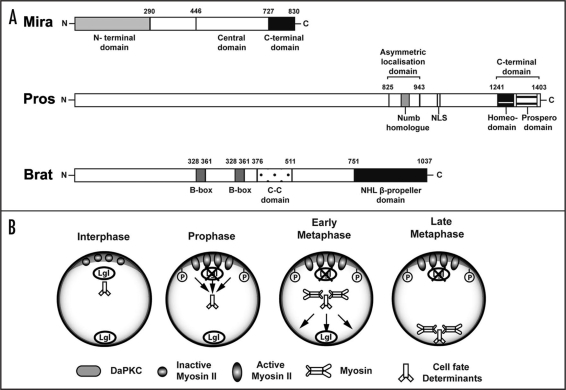
Another recently identified cell fate determinant appears to be Brain Tumor (Brat). brat encodes a member of the conserved NHL family of proteins105–107 and is characterized by the presence of a C-terminal NHL domain, a coiled-coil region and two N-terminal Zinc binding B-boxes (Fig. 5A). Similar to pros, brat mutation results in over-proliferating pNB lineages at the expense of differentiating neurons.46,102–104 Brat mutant pNB clones show cortical mis-localization of Miranda and the loss of nuclear pros,103 suggesting that these proteins may play a role in the same molecular pathway. To bolster this view, ectopic expression of Pros can rescue the tumour formation in Brat mutants in the larval central brain.102 However, Brat localization remains unaffected in Pros mutants, demonstrating that Pros may act downstream of Brat.102 Furthermore, Mira mutants lead to mis-localization of Brat and Pros (Kim D and Hirth F, unpublished).
Figure 5.
Basal cell fate determinants and potential mechanism of asymmetric localization. (A) Cell fate determinant proteins and their functional domains. Miranda (Mira) protein consists of the central domain that provides binding sites for the cell fate determinants carrying them as its cargo proteins. The N-terminal domain of Mira is required for its association to the membrane, whereas, the C-terminal domain plays a role in cortical localization, and release of its cargo proteins upon cytokinesis. The Prospero (Pros) protein contains an asymmetric localization domain required for binding to Miranda. Within this region lies the Numb homolog domain, that may indicate a common asymmetric segregation mechanism shared with Numb. Other well established domains include the nuclear localizing signal (NLS), the homeodomain required for DNA binding, as well as the prospero domain that masks nuclear exclusion. Brain tumor (Brat) protein is characterised by the presence of two N-terminal Zinc binding B-boxes, a coiled-coil region, and the C-terminal NHL β-propeller domain, which can bind the central domain of Miranda. (B) Basal protein targeting of cell fate determinants. When Lgl is phosphorylated and inactivated by DaPKC at the apical cortex, Myosin II becomes activated around early prophase to exclude Mira and its cargo proteins from the cortex into the cytoplasm. Subsequently, Mira may bind to Myosin VI to be transported basally to form a crescent around metaphase. See text for details.
These results indicate that Mira is essential for the asymmetric localization of Brat and Pros, which is in line with the fact that Pros binds to the central Pros-binding domain of Miranda (Fig. 5),108 and Brat binds to the coiled-coil cargo binding domain of Miranda (Fig. 5A) as cargo proteins.104 Moreover, the interaction between the NHL domain of Brat and the C-terminal domain of Mira104 appears to be essential for promoting asymmetric localization of Pros to the GMC, where it is required for cell cycle exit and neuronal fate determination. Thus, it is conceivable that Mira and its cargo proteins Brat and Pros maybe transported across the dividing NB as a complex. But what drives basal protein targeting of adaptor proteins and their respective cell fate determinants?
Mechanisms of Basal Protein Targeting
Previous studies suggested that the localization of Mira and Pros appear to be dependent on actin,109 as well as on motor proteins, Myosins in particular.73,110 These studies indicated an interaction between Lgl with a plus-end directed motor, myosin II.73 Subsequent experiments showed that Spaghetti Squash (Sqh), the regulatory light chain of Myosin II, is required in embryonic neuroblasts both, to organize the actin cytoskeleton, thereby enabling determinants to localize to the cortex, and to confine determinants to the basal side.111 These data suggested that Myosin II is one of the motor proteins involved in basal localization of the cell fate determinants. In line with this, Mira was also found to physically interact with Zipper, the heavy chain of myosin II.73 Thus, non-phosphorylated Lgl can negatively regulate Myosin II in embryonic NBs by directly binding to it. The model proposed by Barros et al.111 therefore suggested that DaPKC-mediated phosphorylation and inactivation of Lgl at the apical cortex, leads to activation, and movement of Myosin II along the cortex towards the cleavage furrow to exclude Mira protein from the cortex into the cytoplasm (Fig. 5B). Hence, Myosin II appears to be responsible for cortical exclusion of Mira rather than direct active transport.111
In Myosin II mutant studies, cell fate determinants failed to form a basal crescent in embryonic NBs,73 notably Mira is mis-localized uniformly around the cortex.112 Similarly, reduced Myosin VI (Jaguar) activity in embryos, leads to a failure in basal crescent formation as well, with Mira mis-localising to the cytoplasm in patches.110 Myosin VI transiently accumulates in the basal cortex, partially co-localizes with Mira during metaphase (Fig. 5B), and in vitro studies using Drosophila embryonic extracts also showed physical interaction with Mira. It is therefore feasible that Myosin VI may be the motor protein responsible for transporting Mira to the basal cortex of NBs.112
The distinct phenotype, mode of action, and sub-cellular localization of Myosin II and Myosin VI suggests that they may act at consecutive steps in a single pathway to localize Mira and its cargo proteins to the basal side of dividing NBs. In addition, Erben et al.112 provide some evidence that Myosin II acts upstream of Myosin VI in a common pathway. Thus, the proposed model for the basal protein targeting (Fig. 5B) involves activation of Myosin II around early prophase by DaPKC phosphorylating Lgl, leading to cortical exclusion of Mira and its cargo proteins into the cytoplasm, where it binds to Myosin VI to be transported basally to form a crescent around metaphase.112
However, the current proposed models for basal targeting of cell fate determinants are not without its controversies. Previously, Mayer et al.113 suggested that actin-myosin based cortical transport is incompatible with photo-bleaching experiments that have determined the dynamics of asymmetric Pon localization. Whilst the experiment failed to detect any directional lateral mobility of some of the segregating determinants, such as Numb and Pon, it cannot rule out that acto-myosin based segregation is required for basal targeting of Mira, Pros and Brat. Moreover, Erben et al.112 proposed that while Pon requires Myosin II, its localization does not depend on Myosin VI, thus maybe utilising a distinct mode of localization. Therefore, further experiments employing Brat-Mira-Pros transport are required to address this question, and the validity of the current model proposed by Erben et al.112 remains to be seen.
Growth Control of Stem Cell Self-Renewal and Differentiation
The data discussed above provide compelling evidence that one strategy to regulate neural stem cell-self-renewal and differentiation is asymmetric segregation of fate determinants in a dividing cell. This is achieved, in part, by asymmetric protein localization and related mitotic spindle orientation, thereby providing a template for unequal distribution of key regulators such as AurA, DaPKC, Numb and Pros. Interestingly, however, such a cascade of events does not explain why mutant pNBs continue to proliferate, thereby self-renewing for an extended period of time without progressive volume decline. It is therefore reasonable to assume that in dividing pNBs, asymmetric protein localization and mitotic spindle orientation is tightly linked to cell cycle progression and growth control.53,114 This is particularly evident in the case of continued proliferation in pros mutant pNB clones, which appears to be accompanied by compensatory cell growth. There, pros mutant cells display sustained symmetric divisions without shrinkage in cell size,102 a phenomenon that is usually accompanied with NB division in the embryonic CNS. Thus, in pros pNB mutant clones, a constant cell size appears to be maintained over many rounds of self-renewing divisions, indicating that Pros may also act as a transcriptional repressor on genes involved in growth control. However, genome-wide expression profiling did not identify growth control genes as potential targets of pros, maybe because embryos had been used as a template.100 A possible link between asymmetric protein localization, cell cycle progression and growth control may be provided by Brat.
Previous studies in Drosophila had shown that brat is a translational repressor115 which also functions in the regulation of cell growth and ribosomal RNA synthesis.116 Moreover, Brat mutant cells display enlarged nucleoli116 and pNB type II-derived brat mutant lineages comprise stem-like cells that show continued proliferation apparently accompanied by compensatory cell growth (Kim D and Hirth F, unpublished). Growth and proliferation of brat mutant cells might be perpetuated by dis-inhibited dMyc activity,104 a transcription factor regulating cell growth and proliferation.117 Interestingly, recent data provide evidence that dMyc interacts with Groucho, a transcriptional repressor,118 in the regulation of several target genes involved in neuronal specification and mitotic control in the embryonic CNS.119 However, a direct interaction of Brat and dMyc has not been shown and it is not clear whether increased activity of dMyc alone is able to orchestrate cell cycle progression and growth control in pNB lineages.
The available data rather suggest that Brat activity regulates a large number of direct and indirect targets involved in cell cycle progression and growth control. This notion is supported by genome-wide expression studies using adult wildtype and brat mutant brain tissue as a template.120 These studies identified several potential target genes of Brat, most prominent among them genes involved in cell cycle regulation and translation control, as well as RNA binding/processing, all being upregulated in brat mutant tissue.120 In addition, brat gain of function can inhibit cell growth and ribosomal RNA accumulation, and slowdown cell division cycles.116 Considering its mutant pNB lineage phenotype, these data suggest that brat may inhibit cell growth by limiting the rate of ribosome biogenesis and protein synthesis. Moreover, Brat appears to co-localize both with Mira and Pros in dividing neuroblasts.102–104 Therefore, it is conceivable that the concerted action of Mira, Pros and Brat provides a direct link between asymmetric protein localization, cell cycle progression and growth control.
Based on the above mentioned data, it is tempting to speculate that, as a result of mitotic spindle orientation and asymmetric protein localization, transcriptional activation/repression (for example, via Pros) and ribosome biogenesis/protein synthesis (for example, via Brat) are key mechanisms executing self-renewal or differentiation of neural stem and progenitor cells. Thus, the presence/absence as well as location and amount of protein could determine whether a cell divides in a symmetric or asymmetric proliferative mode (like a pNB), or in a symmetric differentiative mode (like a GMC; see Fig. 1). A striking example for such a scenario has previously been reported for a cell cycle regulator, where specific mutation of cdc2 affects neural progenitor cell division in the embryonic CNS of Drosophila.121 This study suggests that the level of cdc2 kinase activity determines whether and how a progenitor cell divides, either symmetric or asymmetric. However, it is not known whether the amount of cdc2 activity is regulated at the level of gene transcription or mRNA translation, or even at the post-translational level. Interestingly, genome-wide gene expression studies identified cdc2 as a potential target of Brat activity.120 Considering that cell size and cell cycle length are rate-limiting steps in cell division,122 it is likely that differential control of mRNA translation123 is providing means for regulating the level of proteins involved in cell cycle progression and growth control.53 Coupled to asymmetric protein localization and mitotic spindle orientation, this would enable a tight control system for stem and progenitor cell proliferation, and could explain why dysfunction of any of these modules may lead to cancer formation.124 A physical link between regulators of these modules, as seems to be the case for Mira, Brat and Pros, therefore provides a genetic mechanism regulating self-renewal and differentiation of stem and progenitor cells.
Concluding Remarks
The CNS of the fruitfly Drosophila has become one of the prime model systems to study the genetic mechanisms underlying stem cell self-renewal and differentiation. These studies led to the identification of key molecules involved in asymmetric protein localization and mitotic spindle orientation, coupled to cell cycle regulation and growth control. It is becoming apparent that, at least in part, some of these mechanisms and molecules are evolutionarily conserved, and therefore valid in mammals, including man.3,15,17,24,53,123 For therapeutic applications, it will now be important to determine further details of the machinery involved, in order to be able to manipulate its building blocks in vivo. Several key questions need to be addressed to achieve these goals. What mechanisms and molecules define and maintain stemness? Which other molecules, like Prospero and Brat, couple self-renewal with growth and proliferation? How is differentiation achieved and maintained? Considering previous contributions, it is reasonable to assume that Drosophila research will have a significant impact in addressing and answering these questions in the near future.
Acknowledgements
Work in the Hirth laboratory is supported by grants from the UK Medical Research Council (G070149), the Royal Society (Hirth/2007/R2), the Parkinson's Disease Society (G-0714), the Motor Neurone Disease Association (Hirth/Oct07/6233), and King's College NHS Medical Research Trust (JRC19/2007).
Abbreviations
- AurA
aurora-A
- Brat
brain tumor
- Cdc2
cell division cycle 2
- Cdc25
cell division cycle 25
- CNS
central nervous system
- DaPKC
drosophila atypical protein kinase C
- Dlg
discs large
- DmPar6
Drosophila melanogaster partitioning defective 6
- Gαi
G-protein alpha, subunit i
- GMC
ganglion mother cell
- GoLoco
G-protein 0, locomotion defects domain
- Insc
inscuteable
- Khc-73
kinesin heavy chain 73
- Lgl
lethal (2) giant larvae
- Mira
miranda
- Mud
mushroom body defect
- NB
neuroblast
- NHL
NCL-1, HT2A and LIN-41 domain
- NuMa
nuclear mitotic apparatus
- PDZ
post synaptic density 95, discs large and zonula occludens-1 domain
- Pins
partner of inscuteable
- pNB
post-embryonic neuroblast
- Pon
partner of numb
- Pros
prospero
- RNA
ribonucleic acid
- Sqh
spaghetti squash
- VNC
ventral nerve cord
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/8690
References
- 1.Merlo A. Genes and pathways driving glioblastomas in humans and murine disease models. Neurosurg Rev. 2003;26:145–158. doi: 10.1007/s10143-003-0267-8. [DOI] [PubMed] [Google Scholar]
- 2.Stiles CD, Rowitch DH. Glioma stem cells: A midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 3.Caussinus E, Hirth F. Asymmetric stem cell division in development and cancer. Prog Mol Subcell Biol. 2007;45:205–225. doi: 10.1007/978-3-540-69161-7_9. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Bjorklund A, Lindvall O. Cell replacement therapies for central nervous system. Nat Neurosci. 2000;3:537–544. doi: 10.1038/75705. [DOI] [PubMed] [Google Scholar]
- 6.Lindvall O, Kokaia Z, Martinez-Serrano A. Stem cell therapy for human neurodegenerative disorders—how to make it work. Nat Med. 2004;10:1038–1064. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 7.Lindvall O, Kokaia Z. Stem cells for the treatment of neurological disorders. Nature. 2006;441:1094–1096. doi: 10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 8.Peschanski M, Bachoud-Levi AC, Hantraye P. Integrating fetal neural transplants into a therapeutic strategy: the example of Huntington's disease. Brain. 2004;127:1219–1228. doi: 10.1093/brain/awh145. [DOI] [PubMed] [Google Scholar]
- 9.Rosser AE, Zietlow R, Dunnett SB. Stem cell transplantation for neurodegenerative diseases. Curr Opin Neurol. 2007;20:688–692. doi: 10.1097/WCO.0b013e3282f132fc. [DOI] [PubMed] [Google Scholar]
- 10.Einstein O, Ben-Hur T. The changing face of neural stem cell therapy in neurological diseases. Arch Neurol. 2008;65:452–456. doi: 10.1001/archneur.65.4.452. [DOI] [PubMed] [Google Scholar]
- 11.Ormerod BK, Palmer TD, Caldwell MA. Neurodegeneration and cell replacement. Phil Trans R Soc B. 2008;363:153–170. doi: 10.1098/rstb.2006.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamanaka S. Pluripotency and nuclear reprogramming. Phil Trans R Soc B. 2008;363:2079–2087. doi: 10.1098/rstb.2008.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirth F, Reichert H. Conserved genetic programs in insect and mammalian brain development. BioEssays. 1999;21:677–684. doi: 10.1002/(SICI)1521-1878(199908)21:8<677::AID-BIES7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Eckfeldt CE, Mendenhall EM, Verfaillie CM. The molecular repertoire of the ‘almighty’ stem cell. Nat Rev Mol Cell Biol. 2005;6:726–737. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- 15.Morrison SJ, Kimble J. Asymmetric and symmetric stem cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 17.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 18.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truman JW, Taylor BJ, Awad TA. Formation of the adult nervous system. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. New York, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1245–1275. [Google Scholar]
- 20.Maurange C, Gould AP. Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci. 2005;28:30–36. doi: 10.1016/j.tins.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Campos-Ortega JA, Jan YN. Genetic and molecular bases of neurogenesis in Drosophila melanogaster. Annu Rev Neurosci. 1991;14:399–420. doi: 10.1146/annurev.ne.14.030191.002151. [DOI] [PubMed] [Google Scholar]
- 22.Egger B, Chell JM, Brand AH. Insights into neural stem cell biology from flies. Phil Trans R Soc B. 2008;363:39–56. doi: 10.1098/rstb.2006.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jan YN, Jan LY. Asymmetric cell division in the Drosophila nervous system. Nat Rev Neurosci. 2001;2:772–779. doi: 10.1038/35097516. [DOI] [PubMed] [Google Scholar]
- 24.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization and tumourigenesis. JCB. 2008;2:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White K, Kankel DR. Patterns of cell division and cell movement in the formation of the imaginal nervous system of Drosophila melanogaster. Dev Biol. 1978;65:296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- 27.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 28.Prokop A, Technau GM. The origin of postembryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Development. 1991;111:79–88. doi: 10.1242/dev.111.1.79. [DOI] [PubMed] [Google Scholar]
- 29.Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 30.Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- 31.Ceron J, Gonzalez C, Tejedor FJ. Patterns of cell division and expression of asymmetric cell fate determinants in postembryonic neuroblast lineages of Drosophila. Dev Biol. 2001;230:125–138. doi: 10.1006/dbio.2000.0110. [DOI] [PubMed] [Google Scholar]
- 32.Akong K, McCartney BM, Peifer M. Drosophila APC2 and APC1 have overlapping roles in the larval brain despite their distinct intracellular localizations. Dev Biol. 2002;250:71–90. doi: 10.1006/dbio.2002.0777. [DOI] [PubMed] [Google Scholar]
- 33.Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- 34.Fuse N, Hisata K, Katzen AL, Matsuzaki F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr Biol. 2003;13:947–954. doi: 10.1016/s0960-9822(03)00334-8. [DOI] [PubMed] [Google Scholar]
- 35.Taylor BJ, Truman JW. Commitment of abdominal neuroblasts in Drosophila to a male or female fate is dependent on genes of the sex-determining hierarchy. Development. 1992;114:625–642. doi: 10.1242/dev.114.3.625. [DOI] [PubMed] [Google Scholar]
- 36.Peterson C, Carney GE, Taylor BJ, White K. reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- 37.Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- 38.Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 39.Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischbach KF, Hiesinger PR. Optic lobe development. Adv Exp Med Biol. 2008;628:115–136. doi: 10.1007/978-0-387-78261-4_8. [DOI] [PubMed] [Google Scholar]
- 41.Urbach R, Schnabel R, Technau GM. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–3606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- 42.Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. Adv Exp Med Biol. 2008;628:137–158. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- 43.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 44.Heisenberg M. Mushroom body memoir: From maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 45.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;19:3–5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumour suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neuriobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jan YN, Jan LY. Asymmetric cell division. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- 49.Carmena A. Signaling networks during development: the case of asymmetric cell division in the Drosophila nervous system. Dev Biol. 2008;321:1–17. doi: 10.1016/j.ydbio.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gönzy P. Mechansims of asymmetric cell division: Flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 52.Januschke J, Gonzalez C. Drosophila asymmetric division, polarity and cancer. Oncogene. 2008;27:6994–7002. doi: 10.1038/onc.2008.349. [DOI] [PubMed] [Google Scholar]
- 53.Kohlmaier A, Edgar BA. Proliferative control in Drosophila stem cells. Curr Opin Cell Biol. 2008;20:699–706. doi: 10.1016/j.ceb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 56.Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila nuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- 57.Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblast in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 58.Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 59.Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Izumi Y, Ohta N, Itoh-Furuya A, Fuse N, Matsuzaki F. Differential functions of G protein and Baz-aPKC signalling pathways in Drosophila neuroblast asymmetric division. J Cell Biol. 2004;164:729–738. doi: 10.1083/jcb.200309162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaltschmidt JA, Davidson C, Brown NH, Brand AH. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat Cell Biol. 2000;2:7–10. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- 62.Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of Inscuteable in orienting asymmetric cell division in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- 63.Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci USA. 2007;104:14306–14311. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu F, Wang H, Qian H, Kaushik R, Bownes M, Yang X, Chia W. Locomotion defects, together with Pins, regulates heterotrimeric G-protein signalling during Drosophila neuroblast asymmetric divisions. Genes Dev. 2005;19:1341–1353. doi: 10.1101/gad.1295505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- 66.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 67.Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMa homolog Mud regulates spindle orientation in asymmetri cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Zheng C. NuMA: a nuclear protein involved in mitotic centrosome function. Microsc Res Tech. 2000;49:467–477. doi: 10.1002/(SICI)1097-0029(20000601)49:5<467::AID-JEMT9>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 69.Sun QY, Schatten H. Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein. Front Biosci. 2006;11:1137–1146. doi: 10.2741/1868. [DOI] [PubMed] [Google Scholar]
- 70.Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 71.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 72.Wang H, Cai Y, Chia W, Yang X. Drosophila homologs of mammalian TNF/TNFR-related molecules regulate segregation of Miranda/Prospero in neuroblasts. EMBO J. 2006;25:5783–5793. doi: 10.1038/sj.emboj.7601461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohshiro T, Yagami T, Zhang C, Matsuzaki F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:596–600. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- 74.Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- 75.Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 76.Wirtz-Peitz F, Knoblich JA. Lethal giant larvae take on a life of their own. Trends Cell Biol. 2006;16:234–241. doi: 10.1016/j.tcb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 77.Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 78.Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Aurora-A acts as a tumour suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric divsion: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 82.Berdnik D, Knoblich JA. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr Biol. 2002;12:640–647. doi: 10.1016/s0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- 83.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 85.Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- 86.Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 87.Matsuzaki F, Koizumi K, Hama C, Yoshioka T, Nabeshima Y. Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem Biophys Res Comm. 1992;182:1326–1332. doi: 10.1016/0006-291x(92)91878-t. [DOI] [PubMed] [Google Scholar]
- 87.Lu B, Rothenberg M, Jan LY, Jan YN. Partner of Numb co-localizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- 89.Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- 90.Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 91.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen CP, Knoblich JA, Chan YM, Jiang MM, Jan LY, Jan YN. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 95.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. Miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 96.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 97.Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- 98.Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- 99.Liu TH, Li L, Vaessin H. Transcription of the Drosophila CKI gene decapo is regulated by a modular array of cis-regulatory sequences. Mech Dev. 2002;112:25–36. doi: 10.1016/s0925-4773(01)00626-8. [DOI] [PubMed] [Google Scholar]
- 100.Choksi SP, Southall TD, Bossing T, Edoff K, De Wit E, Fischer BE, et al. Prospero acts as a binary switch between self renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 101.Doe CQ, Smouse D, Goodman CS. Control of neuronal fate by the Drosophila segmentation gene even-skipped. Nature. 1988;333:376–378. doi: 10.1038/333376a0. [DOI] [PubMed] [Google Scholar]
- 102.Bello BC, Reichert H, Hirth F. The brain tumour gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 103.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 104.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumour suppressor Brat regulates self-renewal in drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 105.Arama E, Dickman D, Kinchie Z, Shearn A, Lev Z. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene. 2000;19:3706–3716. doi: 10.1038/sj.onc.1203706. [DOI] [PubMed] [Google Scholar]
- 106.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding and cortical release. Mol Cell Neurosci. 1998;12:325–339. doi: 10.1006/mcne.1998.0724. [DOI] [PubMed] [Google Scholar]
- 109.Broadus J, Doe CQ. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr Biol. 1997;7:827–835. doi: 10.1016/s0960-9822(06)00370-8. [DOI] [PubMed] [Google Scholar]
- 110.Petritsch C, Tavosanis G, Turck CW, Jan LY, Jan YN. The Drosophila myosin VI jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Dev Cell. 2003;4:273–281. doi: 10.1016/s1534-5807(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 111.Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle Myosin II promotes the asymmetric segregation of cell fate determinants by cotical exclusion rather than active transport. Dev Cell. 2003;5:829–840. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 112.Erben V, Waldhuber M, Langer D, Fetka I, Jansen RP, Petritsch C. Asymmetric localization of the adaptor protein Miranda in neuroblasts is achievd by the diffusion and sequential interaction of myosin II and VI. J Cell Sci. 2008;121:1403–1414. doi: 10.1242/jcs.020024. [DOI] [PubMed] [Google Scholar]
- 113.Mayer B, Emery G, Berdnik D, Wirtz-Peitz F, Knoblich JA. Quantitive analysis of protein dynamics during asymmetric cell division. Curr Biol. 2005;15:1847–1854. doi: 10.1016/j.cub.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 114.Prokopenko SN, Chia W. When timing is everything: role of cell cycle regulation in asymmetric division. Semin Cell Dev Biol. 2005;16:423–437. doi: 10.1016/j.semcdb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 115.Sonoda J, Wharton RP. Drosophila Brain Tumour is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development. 2002;129:399–407. doi: 10.1242/dev.129.2.399. [DOI] [PubMed] [Google Scholar]
- 117.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18:435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 119.Orian A, Delrow JJ, Rosales Nieves AE, Abed M, Metzger D, Paroush Z, et al. A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc Natl Acad Sci USA. 2007;104:15771–15776. doi: 10.1073/pnas.0707418104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loop T, Leemans R, Stiefel U, Hermida L, Egger B, Xie F, et al. Transcriptional signature of an adult brain tumour in Drosophila. BMC Genomics. 2004;5:24. doi: 10.1186/1471-2164-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tio M, Udolph G, Yang X, Chia W. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature. 2001;409:1063–1067. doi: 10.1038/35059124. [DOI] [PubMed] [Google Scholar]
- 122.Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 123.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boulay JL, Stiefel U, Taylor E, Dolder B, Merlo A, Hirth F. Loss of heterozygosity of TRIM3 in malignant gliomas. BMC Cancer. 2009;9:71. doi: 10.1186/1471-2407-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]