Abstract
The central nervous system (CNS) is a large network of interconnecting and intercommunicating cells that form functional circuits. Disease and injury of the CNS are prominent features of the healthcare landscape. There is an urgent unmet need to generate therapeutic solutions for CNS disease/injury. To increase our understanding of the CNS we need to generate cellular models that are experimentally tractable. Neural stem cells (NSCs), cells that generate the CNS during embryonic development, have been identified and propagated in vitro. To develop NSCs as a cellular model for the CNS we need to understand more about their genetics and cell biology. In particular, we need to define the mechanisms of self-renewal, proliferation and differentiation—i.e. NSC behavior. The analysis of pluripotency of embryonic stem cells through mapping regulatory networks of transcription factors has proven to be a powerful approach to understanding embryonic development. Here, we discuss the role of transcription factors in NSC behavior.
Key words: neural stem cells, transcription factors, self-renewal, proliferation, differentiation
Introduction
The most complex organ of the body, the central nervous system (CNS), comprising the brain and the spinal cord, is composed of three distinct cell types; astrocytes, oligodendrocytes and neurons. These cells help form a complex network of connections that facilitate electro-chemical signaling with the neuron taking center stage. There are approximately 100 billion neurons in the brain and a typical neuron has about 1,000 to 10,000 synapses (that is, it communicates with 1,000–10,000 other neurons, muscle cells, glands, etc). Thus the brain is made-up of trillions of connections. This complexity is further enhanced by the fact that there are many different neuronal cell-types specialized to have particular morphology, connectivity and work with distinct neuromodulators and neurotransmitters.
Diseases of the brain affect millions of people worldwide and are becoming increasingly prominent as the population ages. CNS diseases where neural stem cells (NSCs) could be useful as cellular models or to provide therapeutic solutions include; Alzheimer, Parkinson, stroke, Huntington, Lou Gehrig (ALS) and the devastating disease of childhood—Batten disease. In fact, NSCs are already in clinical trials for the treatment of stroke and of Batten's disease.1 [Neurospheres contain both NSCs and Neural Progenitors (NPs). Since there are no definitive markers for NSCs or NPs the two populations cannot be separated. The difference between NSCs and NPs is that the latter has limited replication abilities will not passage and is likely to be uni or bipotent].
If we are to tackle the complexity of the CNS and generate solutions for CNS disease states it is essential that we generate therapeutic cellular models. The discovery of NSCs of embryonic and adult CNS2,3 has opened up the possibility to develop cellular models of the CNS. NSCs are multipotent cells that can be defined simply as cells that have the ability to self-renew and generate the major cell types of the CNS. The NSC characteristic of self-renewal has been linked to cancer and it is important to investigate this link further. The role of NSC in cancer is supported by the work of Singh et al.4 where they isolated CD133 positive cells from human tumors. The CD133 positive cells gave rise to tumors in vivo in NOD-SCID mice, were serially transplantable and phenocopied the patients' original tumor.4 Thus, the NSC property of self-renewal is important to understand from a brain cancer point of view and may provide targets for anti-cancer drugs. NSCs can be cultured for extended periods of times and this allows the generation of large numbers of specific cell-types such as dopaminergic neurons. Transcription factors (TFs) play prominent roles in developmental processes and have provided excellent tools to understand stem cell-lineage specification. The generation of specific cell-types from NSCs will be important to model neural development and disease states.
TFs and Pluripotency
Several independent studies have sought to define ‘stemness’ by attempting to identify a set of conserved genes that govern key regulatory pathways of stem cells.5,6 In an effort to elucidate a common transcriptional profile attributable to ‘stemness’, two independent studies used hematopoietic stem cells (HSCs), embryonic stem cells (ESCs) and NSCs to perform a genome wide gene expression microarray analysis. Consistent in both reports is that there is a subset of six genes that are common to ESC, HSC and NSC. However, when a third dataset was analyzed only a single gene remained in the common pool.7 Thus it seems that the concept of ‘stemness’ via gene profiling is rather vague. A more successful approach to understanding ‘stemness’ has been the identification of proteins, and in particular TFs, that play crucial roles in pluripotency. Work over the last ten years has revealed that the TFs, OCT4, SOX2 and NANOG, play dominant roles in the maintenance of pluripotency. In mouse embryos lacking Oct4, the pluripotent inner cell mass fail to develop and thus cannot survive past the blastocyst stage.8 Sox2-null embryos show defective epiblasts and die immediately after implantation.9 Similarly, the lack of Nanog resulted in embryos failing to develop an epiblast.10 Thus, these three factors appear to be critical for embryonic development. Indeed, genome-wide studies in both mouse11,12 and human ESCs13 revealed that these factors co-occupy and share a substantial portion of target genes that form a characteristic network that maintains cellular pluripotency. Furthermore, the ability of these three TFs to reprogram somatic cells14,15 is compelling evidence of their role as key regulators of pluripotency.
Central to the maintenance of the pluripotent network is a tight balance of the levels of the TFs. While self-renewal and an ESC state are preserved by overexpressing NANOG, ESCs that are depleted of NANOG are driven towards endodermal lineages.10,12 Similarly the role of OCT4 as a gatekeeper in the decision between pluripotency and lineage specification8,16 can be predicted by its concentration. Its expression is high in undifferentiated ESCs, and decreases during differentiation.7 Precise levels of OCT4 are required for the maintenance of pluripotent ESCs as reduction of OCT4 expression to 50% or less induces trophectodermal differentiation, while overexpression causes differentiation to primitive endoderm and mesoderm.18–20 As OCT4 is able to cooperate with SOX2,9 and is involved in the reciprocal regulation of each other's expression21 to mediate for instance NANOG activity,22 tweaking the levels of SOX2 also skews the transcriptional network inadvertently. Essentially, elevating the levels of SOX2 decreases expression of its own gene and inhibits SOX2:OCT4 targets like Oct4 and Nanog.23 In addition, eliciting small increases in SOX2 protein via an inducible system triggers the differentiation of ESCs that gives rise to cell types that exhibits neuroectoderm, mesoderm and trophectoderm markers.24 Reducing the level of SOX2 in contrast, promote the differentiation of ESCs into trophectoderm-like cells.21 Thus, these data suggest that a precise level of SOX2 and OCT4 is important to maintain the pluripotent state.
The success of the regulatory network-TFs-pluripotency approach to understanding ESCs suggests that a similar approach applied to NSCs may give insight to the biology of these cells. Important questions that could be addressed include; (1) which TFs are essential for NSC self-renewal, proliferation and differentiation? (2) can this information be used to define markers for NSCs? and (3) are TFs that control the NSC self-renewal potential anti-cancer targets? Although TFs clearly play important roles in stem cell behavior it is important to realize that TFs are part of an intricate network of cell signaling pathways that respond to cell-cell contact, growth factors and cytokines released in autocrine and paracrine fashion. Before discussing specific TFs and their role in NSC behavior we start the review by highlighting three important cell signaling pathways of NSCs; Wnt, Notch and Sonic hedgehog (Shh).
Three Major Cell Signaling Pathways of NSCs
The presence of Wnt/β-catenin pathway within the sub-ventricular zone (SVZ) suggests a role for β-catenin in neural development.25 β-Catenin is a central and essential component of the canonical Wnt signaling pathway that functions by activating TCF/LEF TFs.26–28 Conditional mutation of β-catenin results in elimination of the cells at the midhindbrain boundary,29 decreases in the overall size of the nervous system and the neuronal precursor population.30 On the other hand, continuous expression of β-catenin resulted in marked generalized hypercellularity of the brain.31 In NSCs cultures, the addition of Wnt protein caused an increase in survival of NSCs and more efficient colony initiation.25 However, depending on the stage of development Wnt/β-catenin pathway switches its role into triggering neuronal differentiation.32 β-catenin and its downstream partners (TCF/LEF) control the balance between progenitor expansion and differentiation.27,30 It has been proposed that β-catenin alone stimulates neuronal differentiation, whereas β-catenin along with Fgf2 inhibits neuronal differentiation.33
Notch signaling has also been implicated in regulating the balance between neuronal differentiation and progenitor expansion.34,35 The bHLH genes Hes1 and Hes5 which are essential effectors of Notch signaling encode transcriptional repressors and regulate the maintenance of cells in the undifferentiated state and repress neuronal differentiation.36,37 Embryonic NSCs change their characters over time from Hes-independent neuroepithelial cells, transitory Hes-dependent neuroepithelial cells to Hes-dependent radial glial cells.36 Hes-related bHLH genes, Hesr1 and Hesr2 are also expressed by NSCs and NPs in the embryonic brains and act as Notch signaling effectors. Hesr1/2 regulates NSC maintenance, possibly in conjunction with HES proteins.38 Notch signaling seems to be an important signaling pathway in distinguishing stem cells from more limited progenitors in a variety of tissues. Knockdown of the canonical Notch effector C-promoter binding factor 1 (CBF1/RBP-J) promotes the conversion of NSCs to NPs, whereas activation of CBF1 is insufficient to convert NPs back to NSCs.39 The results from conditionally ablated transcription factor RBP-J indicated that the RBP-J-mediated signaling might inhibit the differentiation of NSCs into NPs.40 Mammalian Musashi-1 augments Notch signaling through the translational repression of its target mRNA, mNumb, thereby contributing to the maintenance of NSCs/NPs.41
Shh-Gli signaling is another key pathway that is involved in nervous system development by modulating precursor proliferation in different regions of the brain like neocortex, cerebellum and tectum. Shh has also been implicated in cell proliferation and growth of the late embryonic and postnatal dorsal brain.42–44 Gli-1 expression in Nestin positive NSCs/NPs increases precursor and clonogenic stem cell number in vivo and in vitro.45 E18.5 cortical tissue deficient in Gli-2 or Gli-3 the downstream mediators of Shh showed reduced primary and secondary neurosphere formation.46 Gli-2-specific shRNA in NSCs in vivo and in vitro inhibited cell proliferation and the expression of Sox2 and other NSC markers, including Hes1, Hes5, Notch1, CD133 and Bmi-1.47 Taken together, it appears that Wnt, Notch and Shh signaling pathways play essential roles in the maintenance of NSCs.
NSC Self-Renewal
One of the defining features ascribed to NSCs, and stem cells in general, is the ability to self-renew; to generate duplicate multipotent copies of themselves. At the surface the concept of self-renewal seems straightforward. However, to assay self-renewal is more complicated. The most widely used assay for NSC self-renewal is carried out in vitro by measuring neurosphere formation through passaging of cultures. The number of multipotent neurospheres generated during passaging is taken as a reflection of the self-renewal activity. Table 1 lists the main transcription factors involved in self-renewal.
Table 1.
TFs and NSC self-renewal
| Transcription factors | Comments |
| HES1 and HES5 (bHLH) | Decreased self-renewal by secondary sphere formation.37,147Hes1-/- and Hes5-/- mice exhibit premature exit for differentiation.36,48,147 |
| CBF-1 (CSL) | CBFE-EGFPhi cells generate large primary and secondary neurospheres in comparison to CBFRE-EGFPlo/neg cells.39 |
| SOX2 (HMG) | Sox2 positive cells contain self renewing properties in vitro.54,148 Enlarged ventricles in the deficiency and conditional inactivation of Sox2 in mice.52,54 |
| HMGA2 (HMGA) | Hmga2 promotes the self renewal of fetal and young-adult stem cells.64Hmga2-/- mice shows defects in neural stem cell frequency. Hmga2 is regulated by the expression of let-7 microRNA in old adult mice.64 |
| BMI-1 (PC) | Bmi1-/- neurospheres are smaller than wild-type.66,83,149 Smaller cerebellum and thinner molecular and cellular layers (reduced expansion of cells).66,83,149 |
| GLI-2, GLI-3 (ZFP) | Gli-2-/-, Gli-3-/- cortical cells are unable to passage more than four times and do not retain multipotency.46 The mutant mice display reduced cortical architecture. |
The table lists TFs followed by the protein family they belong to. Comments in the right hand column highlight information that supports the role of these TFs in NSC self-renewal. bHLH, basic helix-loop-helix; CSL-, HMG, high mobility group; PC, Polycomb; ZFP, zinc finger proteins.
HES1/5.
Mice that lack both Hes1-/-/Hes5-/- or mis-expression studies suggest a role for HES1/5 in self-renewal of NSCs.36,48 The secondary sphere forming capability is reduced in telencephalic cells that lack HES1 and HES5.
CBF-1.
Notch signaling has been linked to the stem cell state as mentioned above. The NICD-CBF1 complex generated upon Notch activation targets Hes1/5 genes. Mizutani et al. 2007,39 have analyzed the role of CBF-1 further. Knockdown of CBF-1 induced neurogenesis. Mizutani et al. 2007,39 then generated CBF-1 promoter fusions linked to EGFP and identified two discreet populations of cells in vivo, EGFPhi and EGFPlo. On isolating these two populations by using CD133 selection they found that the EGFPhi cells formed greater numbers of multipotent neurospheres and had 3.5–3.8-fold greater expression of Hes1/5. In contrast, EGFPlo cells had higher expression of Mash1. They propose that Notch signaling may allow NSCs to be distinguished from NPs.
SOX2.
SOX TFs with a high-mobility-group (HMG) DNA binding domain have been shown to have homologous roles in specification and maintenance of NP identity in the CNS and the peripheral nervous system. The SOXB1 factors (Sox1, Sox2 and Sox3) which are transcriptional activators are co-expressed in the proliferating NSCs/NPs of embryonic and adult CNS.49–51 A reduction in SoxB1 levels leads to precocious neural differentiation and to the depletion of the progenitor pool, whereas misexpression of SoxB1 family members can block neuronal differentiation and maintain the progenitor population.49,52–54 SOXB1 transcriptional factors antagonize the neuronal differentiation that is induced by the bHLH proneural proteins MASH1 and the NGNs49,55 and proneural proteins can directly bind and inhibit SOXB1 protein function. Proneural factors also upregulate Sox21 (SoxB2 group) expression which represses Sox1-3 activity inducing downregulation of progenitor markers, cell cycle exit and neuronal differentiation.56 Thus, the balance of SOXB1 and proneural activity determines the activation of neurogenesis.
The Sox2 enhancer, termed Sox2 regulatory region 2 (SRR2), that is specific to ESCs also functions in NSCs/NPs and drives strong expression in these cells. Chromatin immunoprecipitation assays reveal interactions of class III POU proteins, such as BRN1 and BRN2 with SOX2 at SRR2 in NSCs/NPs.57,58 POUIII transcriptional factors BRN1, BRN2, BRN4 and OCT6 are widely expressed in the developing CNS with extensive regional overlap.59–61 In the ventricular zone of the embryonic spinal cord nestin expression is seen in the regions co-expressing SOXB1 and BRN2 proteins. Group B1 and group C SOX proteins interact with POUIII TFs and activate the nestin neural enhancer.62 However, a switch in POU TFs from BRN1/2 to BRN3a occurs in post-mitotic cells.63
HMGA2.
High mobility group A2 (HMGA2) is a chromatin associated protein that potentiates the activity of TFs. In a recent analysis HMGA2 was found to be expressed at high levels in fetal cells and declined with age.64 Hmga2 KO mice show reduced stem cell numbers throughout the CNS. Nishino et al. 2008,64 derived a self-renewal index (secondary neurosphere numbers/primary neurosphere numbers) for neurosphere formation and used this to compare KO mice with wild-type controls. HMGA2 KO reduced self-renewal of NSCs by 70% and this could be reversed by expression of Hmga2 in the KO cells. The HMGA2 KO neurospheres were multipotent but much smaller than wild-type controls.
BMI-1.
A polycomb family transcriptional repressor, BMI-1 has also been shown to be required for the maintenance of NSCs/NPs.65,66 Bmi-1 knockout studies have shown progressive postnatal growth retardation and neurological defects.66 shRNA mediated Bmi-1 reduction causes defects in embryonic and adult NCSs cell maintenance.65 BMI-1 maintains NSCs by repressing the cyclin-dependent kinase inhibitors, p16Ink4a and p19Arf as well as p21-Rb pathway.65
Gli-2/3.
Cortical mutant cells from Gli-2, Gli-3 KO mice fail to form both primary and secondary neurospheres.46 The GLI pathway regulates expression of several NSCs/NPs markers such as Sox2, Hes1, Hes5, Notch1, Bmi-1 and CD-133.47 This novel circuit of TFs is important for self-renewal of the NSCs cells from embryonic CNS.
TLX.
The orphan nuclear receptor TLX has been shown to maintain adult NSCs in an undifferentiated proliferative state. In vivo, TLX mutant mice show a loss of cell proliferation and reduced nestin labeling in the neurogenic areas of the adult brain and in vitro, TLX null cells fail to proliferate.67 One mechanism by which TLX regulates maintenance of NSCs is by recruiting histone deacetylases to its downstream targets to repress their expression.68
NSC Growth
NSC/NP growth is regulated at two levels by TFs. The first is at the level of the cell cycle. The second is at the level of early differentiation. The main methods used to measure NSC growth are, (1) neurosphere size, (2) rates of BrdU incorporation and (3) number of cells in vivo in particular CNS locations.
SoxB1.
The SoxB1 genes are thought to be critical in maintaining the NSC state. The main mechanism seems to be through inhibition of differentiation. Some evidence exists to support a role for these genes in proliferation. All SoxB1 null mutants have defects in brain development.52,69,70 SOX1 overexpression induces expansion of the NP pool in vivo followed by neuronal differentiation.71 But in vitro overexpression of SOX1 promotes neural differentiation.72 Sox2 expression correlates with proliferating NSCs/NPs in vivo and in vitro.53 Sox2 conditional KO mutants have less proliferating cells in vivo and form less primary neurospheres in culture.54 However, subsequent passaging and differentiation of mutant cells were unaffected. Sox3 has also been shown to express in proliferating cells in vivo and in vitro.73 Whether Sox2 and 3 directly control proliferation is not clear. Table 2 lists the main TFs involved in proliferation.
Table 2.
TFs and NSCs proliferation
| Transcription factors | Comments |
| SOXB1 (SOX1, 2 and 3) (HMG) | All Sox null mutants showed defect in brain development.52,69,70 SOX1 overexpression expands progenitor pool moderately in vivo.71 Sox2 and 3 expression correlate with proliferating cells in vivo and in vitro53,73 Conditional Sox2 mutants have less proliferating cells in vivo.54 |
| GLI-1 (ZFP) | shRNA knockdown inhibits proliferation in vitro.78 Overexpression in vivo resulted in enlarged brain and expanded precursor pool.45 |
| GLI-2 and GLI-3 (ZFP) | Null mutants die at birth and show reduced SVZ/VZ volume.75,76 Mutant NSCs showed reduced proliferation in culture.46 Truncated/sh-Gli2 inhibit cell proliferation in vivo and in vitro.47 |
| HES1 and HES5 (bHLH) | Neurospheres from Hes1-/-–Hes5-/- telenchephalon were significantly smaller than the wild type.37 Anti-sense knockdown of Hes1 resulted in less BrdU incorporation in neurosphere culture.81 In vitro overexpression of Hes1 induces proliferation.82 However, overexpression of Hes1 and 5 in vivo inhibits neurogenesis without expanding the precursor pool.37 |
| ID2 (bHLH) | ID2-/- NSCs proliferate slower and form smaller neurospheres in culture.143 |
| ID4 (bHLH) | Absence of ID4 compromises the proliferation of NSCs in the ventricular zone.144 |
| OLIG2 (bHLH) | Expressed in proliferating NSCs in culture. Null mutant cells showed reduced proliferation in culture.145 |
| BMI-1 (PC) | Null mutant haves less proliferating NSCs/NPs in vivo.66 Mutant cells form smaller neurospheres in culture.146 |
The table lists TFs followed by the protein family they belong to. Comments in the right hand column highlight information that supports the role of these TFs in NSC proliferation. bHLH, basic helix-loop-helix; HMG, high mobility group; PC, Polycomb; ZFP, zinc finger proteins.
GLI family.
The Gli family of TFs, consisting of Gli-1, 2 and 3 are the main mediators of the Hedgehog signaling pathway which is well known to regulate NSC proliferation and self-renewal.74 Both Gli-2 and Gli-3 null mutants die at birth.75,76 They displayed a much reduced SVZ/VZ as well as cortex. In vitro culture of NSCs from these mutants showed greatly reduced cell proliferation and neurosphere formation.46 Gli-1 null mutant on the other hand appeared normal,77 and NSCs derived from these mice form multipotent neurospheres that can be maintained over multiple passages.78 However, knockdown of Gli-1 with shRNA impaired proliferation and neurosphere formation. In addition, overexpression of Gli-1 in vivo resulted in enlarged brains and expanded precursor pools.45 The authors also showed using an inducible Gli-1 expression system that the number of neurospheres formed correlated with the level of Gli-1 expression with higher levels giving rise to more neurospheres.
Gli-2 has been reported as a novel regulator of Sox2 expression, which is essential for the maintenance of NSCs. Besides, neocortical cells from Gli-2 mutant mice showed compromised neurosphere forming abilities.46 Primary cultures of E18.5 Gli3 mutant neocortices in full NSC media yielded transiently forming clumps that rapidly degenerated, whereas NSC cultures from wild-type siblings formed stable neurospheres.46
HES1 and HES5.
TFs of the basic-helix-loop-helix (bHLH)1 family play important roles in regulation of neurogenesis in the CNS. Hes1 is expressed at high levels in the VZ of the developing CNS. Persistent expression of Hes1 severely perturbs differentiation of NPs in the CNS. NPs infected with Hesl-transducing retrovirus stayed in the VZ/SVZ or the ependymal layer and did not differentiate into neurons or glial cells.79 Hes5, a neural-specific factor, shows a similar expression pattern in the developing CNS to that of Hes1. Hes5 is expressed at high levels throughout the VZ of the developing CNS, but the level decreases as neural differentiation proceeds.80 Thus, HES1 and HES5 encode transcriptional repressors and maintain the number and status of undifferentiated NSCs and NPs cells in the developing CNS.36
When the NSCs exit the cell cycle in the VZ of the neural tube, these cells express Notch ligands (Delta and Jagged) on the cell surface and activate the Notch of neighboring progenitor cells. In the Notch-activated cells, intracellular domain (ICD) is released and forms a complex with the DNA binding protein RBP-J in the nucleus. This complex induces Hes1 and Hes5 expression. Hes1 and Hes5 repress both the expression and activity of Mash1, Math3 and Ngn2 by binding to their promoter and recruiting the corepressor TLE/Grg, and then neuronal differentiation is inhibited.35
There are also some evidence for involvement of Hes1 and 5 in NSC proliferation. In vitro, NSCs from Hes1-/- and Hes5-/- mutants form fewer and smaller neurospheres compared to wild type.37 In addition, anti-sense knockdown of Hes1 resulted in less BrdU incorporation in human neurosphere cultures.81 In vitro overexpression of Hes1 in granule neuron precursor induced proliferation.82 However, in vivo overexpression of both HES1 and 5 inhibited neurogenesis without expanding the NP pool.37 Thus, the role of Hes genes may be to maintain precursor cells in a proliferation competent state rather than regulating their cell cycle directly.
BMI-1.
NSCs depend increasingly on BMI-1 for proliferation as development proceeds from embryonic through adult stages.65 BMI-1 promotes NSC self-renewal, maintenance and development in the nervous system by repressing the p16Ink4a and p19Arf senescence pathways. Deletion of Ink4a and Arf from BMI-1 knockout mice partially rescued NSC self-renewal and NSC frequency.83 However, using lentiviral-delivered shRNAs in vitro and in vivo, Fasano et al. 2007,65 found no evidence of an increase in either p16Ink4a or p19Arf at any developmental stage 48 hours after reduction of Bmi-1. Instead, the cell cycle inhibitor p21/Cip1 was rapidly upregulated. In support of their role in proliferation, stable expression of Bmi-1 in human HSCs promotes long term in vitro expansion of these cells.84 Expression of BMI-1 in astrocytes has been shown to convert these terminally differentiated cells to NSC-like cells that were able to proliferate and form multipotent neurospheres that self-renewal.85
HESR1 and HESR2.
Hes-related bHLH genes, termed Hesr genes (also known as Hey, HERP, HRT, CHF and gridlock) have been identified as immediate transcriptional targets in Notch signaling.86 HES and HESR proteins differ primarily in that Hes proteins contain a conserved proline residue in the basic region, while Hesr proteins do not. Hers1 and Hers2 are expressed by NSCs in developing brain. It has recently been reported that HESR1 and HESR2 negatively regulate neuronal bHLH genes and promote maintenance of NSCs in the developing brain.38 Iso T, et al.87 revealed that HESR and HES proteins exert synergistic effects by forming heterodimers. Thus, it is possible that HESR and HES cooperatively regulate maintenance of NSCs. Table 3 lists the main TFs involved in repression of differentiation.
Table 3.
TFs and repression of NSC differentiation
| Transcription factors | Comments |
| HES1/5 (bHLH) | Hes-/- embryos prematurely differentiate into neurons.36 |
| HESR1/2 (bHLH) | Misexpression of Hesr1 and Hesr2 by electroporation in mouse brain at embryonic day 13.5 transiently maintains neural precursor cells.38 |
| ID4 (HLH) | Premature differentiation of ID4-/- cortical stem cells.144 |
| SOX1/2/3 (SOXB1) (HMG) | A reduction in SoxB1 levels leads to (HMG) precocious neural differentiation whereas misexpression of SoxB1 family members can block neural differentiation.49,52–54 |
| REST (ZFP) | Inhibition of Rest in hippocampal NSCs leads to activation of neuronal specific genes.150 |
| POU3F2/F3 (ZFP) | Interact with SOX2 and control expression of NSC/NPs genes like nestin, BFAB.57,151 |
The table lists TFs followed by the protein family they belong to. Comments in the right hand column highlight information that supports the role of these TFs in repression of NSC differentiation. bHLH, basic helix-loop-helix; HMG, high mobility group; ZFP, zinc finger proteins.
REST.
REI silencing transcription factor (REST) or neuron restrictive silencer factor (NRSF) is expressed throughout early development where it regulates a large network of neuronal genes.88 REST had been implicated in the transcriptional networks that regulate ESC pluripotency, as the Rest gene is a target of Oct4, Sox2 and Nanog binding.12 However, REST appears to have quite distinct transcription networks in NSCs compared to ESC.89 REST is able to both silence and repress neuronal genes in embryonic hippocampal NSCs by creating a chromatin environment that contains both repressive local epigenetic signature (characterized by low levels of histones H4 and H3K9 acetylation and elevated dimethylation of H3K9) and H3K4 methylation.88
NSC Differentiation
NSC differentiation can be seen as a two step process, where committed progenitors are first formed (early differentiation) followed by the generation of neurons, astrocytes and oligodendrocytes (terminal differentiation). Here we focus on TFs involved in early differentiation (Fig. 1). A large number of factors have been identified that promote terminal differentiation of committed progenitors and these are mentioned in Table 4 but not discussed further here. The generation of committed progenitors depends on the interplay of factors contributing to lineage initiation and specification, lineage commitment, cell cycle exit and feedback loops inhibiting expression of neural stem/progenitor-related TFs.
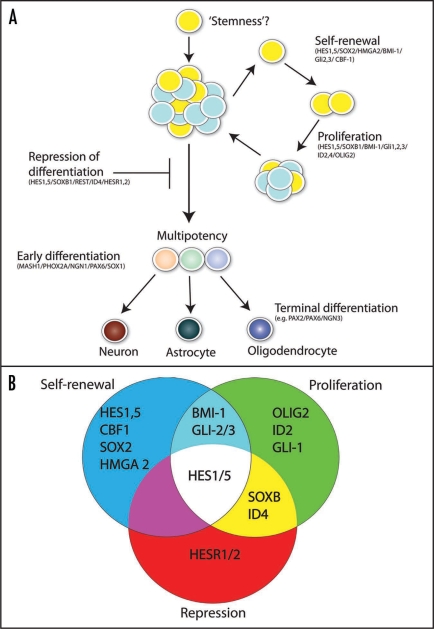
Figure 1.
(A) TFs and NSC behavior. The figure outlines NSC behavior and the TFs involved in each step. The starting cell (yellow) that gives rise to neurospheres is likely to be a NSC. What defines ‘stemness’ in the context of the NSC is currently unknown. The NSC undergoing ‘self-renewal’ generates a copy of itself. The next phase is proliferation where NSCs generate NPs (blue) and a neurosphere forms. A number of TFs are implicated in repression of differentiation of these NPs keeping the neurosphere growing. If differentiation signals are imposed the NSCs/NPs leave the cell cycle and form committed progenitors (light brown, green and blue) which then go on to form terminally differentiated cells; neurons, astrocytes and oligodendendrocytes (dark brown, green and blue). The TFs implicated in each of the steps are placed in brackets. For further information on TFs see Tables 1–4 and text. (B) Overlapping functions of TFs. The three processes that are involved in neurosphere formation are presented as circles in a Venn diagram. Self-renewal—blue, proliferation—green and repression of differentiation—red. Overlap in circles shows TFs that have more than one function.
Table 4.
TFs and NSCs differentiation
| (A) TFs involved in neurogenesis | |
| Transcription factors | Comments |
| PITX3 (HOM) DA neurons | Potentiates Nurr1 in specifying for the phenotype.152 |
| FOXA1/A2 (WH) DA neurons | Specification of phenotype by regulating Ngn2 expression; regulates Nurr1, engrailed 1, aromatic l-amino acid decarboxylase and tyrosine hydroxylase during development.142 |
| NGN2 (bHLH) DA neurons | Differentiation of Nurr1-positive DA neurons from NSCs.153 |
| L3/LHX8 (HOM) Cholinergic neurons | Development or maintenance of cholinergic neurons in the basal forebrain.154 |
| GATA2 (ZFP) GABAergic neurons | Activate genes specific to the GABAergic neurons subtype in the midbrain. Absence leads to differentiation into glutaminergic neurons.155 |
| PAX2 (PB) GABAergic neurons | Specification of interneurons in the dorsal horn.156 |
| NGN2 (bHLH) GABAergic neurons | Region-specific functions: Increased GABA neurons in the hindbrain; decreased GABA neurons in the forebrain.157 |
| MASH1 (bHLH) GABAergic neurons | Region-specific functions: Promotes GABA neurons in the forebrain; decreased GABA neurons in the hindbrain.157 |
| PTF1a (bHLH) GABAergic neurons | Favors GABAergic over glutaminergic neurons.168 |
| GATA2 (ZFP) Serotonergic neurons | Development of serotonergic neurons in the rhombomere region155 and organization of serotonergic dorsal raphe cells.159 |
| EAGLE (Eg) (ZFP) Serotonergic neurons | Regulates serotonergic neuron development by regulating the expression of serotonin transporter.160 |
| PET-1 (ETS) Serotonergic neurons | Differentiation and maintenance of cells in hindbrain raphe nuclei.161 |
| LMX1b (HOM) Serotonergic neurons | Serotonergic neuron specification162 and maintenance.163 |
| (B) TFs involved in astrogliagenesis | |
| Transcription factors | Comments |
| OLIG2 (bHLH) | Expressed in immature astrocytes; deletion leads to loss of astrocytes in the cerebral white matter164 but its nuclear export is required for astrocytes differentiation.165,166 |
| Stem cell leukaemia (SCL) (bHLH) | Astrocyte specification in ventral neural tube.167 |
| PAX6 (HOM) | Promotes maturation.168 |
| MCP-1-induced protein (MCPIP) (Novel) | Overexpression increases GFAP expression and astrocytic morphology.169 |
| NF1A (NF) | Differentiation of astrocytes precursors.170,171 |
| STAT3 (ZFP) | Astrocyte differentiation.106 |
| CSL (ZFP) | Activates GFAP promoter.106 |
| (C) TFs involved in oligodendrogliagenesis | |
| Transcription factors | Comments |
| SOX4 (HMG) | Present in OPCs, inhibit myelination.173,174 |
| SOX5/6 (HMG) | Present in OPCs, inhibit Sox9 and Sox10.175 |
| SOX8 (HMG) | Oligodendrocyte specification, together with Sox9.176 |
| SOX9 (HMG) | Contribute to glial177 and oligodendrocyte specification, together with Sox8.176 |
| SOX10 (HMG) | Terminal differentiation, activating myelinating genes.178,179 |
| SOX11 (HMG) | Present in OPCs.173 |
| SOX17 (HMG) | Oligodendrocyte differentiation.180 |
| MASH1 (bHLH) | OPC specification.107,181 |
| NGN3 (bHLH) | Oligodendrocyte development, possibly maturation and myelination.107 |
| OLIG1 (bHLH) | Oligodendrocyte specification188 with controversial role in maturation.184,185 |
| OLIG2 (bHLH) | Oligodendrocyte maturation186 cooperates with Nkx2.2.192 |
| HES5 (bHLH) | Inhibits OPC differentiation.188,189 |
| ID2/4 (HLH) | Inhibits OPC differentiation.188–190 |
| KROX24 (ZFP) | Maintenance of OPCs, and during immediate stage of differentiation.191 |
| MyT1 (ZFP) | OPC proliferation and differentiation.192 |
| ZFP488 (ZFP) | Oligodendrocyte differentiation, cooperating with OLIG2.73 |
| TST1/OCT6/SCIP/BRN1/2 (ZFP) | Downregulated in early phases of oligodendrocyte development.193 |
| YINYANG 1 (YY1) (ZFP) | Oligodendrocyte differentiation194 by repressing Tcf4 & ID4.111 |
| ATF5 (ZFP) | Expressed in OPCs, inhibit differentiation.196 |
| NKX2.2 (HOM) | Oligodendrocyte maturation.197 |
| NKX6.2 (HOM) | Expressed in myelinating oligodendrocytes.198 |
| HOXB4/A2 (HOM) | Expressed throughout oligodendrocyte development.199 |
| MSX1 (HOM) | Overexpression in mouse neurospheres promoted oligodendrogenesis.200 |
The table lists TFs followed by the protein family they belong in. Comments in the right hand column highlight information that supports the role of these TFs in NSC differentiation. HOM, homedomain; WH, winged helix; bHLH, basic helix-loop-helix; HMG, high mobility group; ZFP, zinc finger proteins. PB-, ETS, ETS domain. In (A) neuron type is given in brackets.
MASH1.
NSCs/NPs express high levels of bHLH factors that besides forming a network to maintain cells in their undifferentiated state, can repress proneural genes. The inhibitory bHLH HES1 protein inhibits the transcriptional activity of proneural gene mammalian achetescute homologue (MASH1); indirectly by binding to promoter sequences recognized by MASH1, and directly by heterodimerizing with MASH1 such that it cannot heterodimerize with E47 transcription factor to activate other proneural genes.90 In fact, mouse embryonic NSCs overexpressing HES1 failed to differentiate into neuron and glial cells79 while loss of HES1 in the olfactory epithelium increased both the level of MASH1 and MASH1-positive NSCs in the olfactory placode.91
While HES1 negatively regulates the function of MASH1 in neuronal differentiation, NUMB2 and NUMB4 increase MASH1 expression, with concurrent expression of Delta1 and Tuj1.92 This induction occurs only when the levels of proneural NUMB2 and 4 was in a 2-molar excess of NUMB1. In addition, the binding of myocyte enhancer factor 2C (MEF2C) and Ca2+/calmodulin-dependent kinase II, induced by apoptosis signal-regulating kinase 1 (ASK1), on the MASH1 promoter can also upregulate the expression of MASH1.93 Overexpression of MASH1 in neural crest stem cells induces morphological differentiation and expression of neuronal markers.94 This effect was in part exerted by its ability to upregulate the expression of the paired homeodomain transcription factor PHOX2a, which in turn induces the expression of cyclin-dependent kinase inhibitor p27 (Kip1) to coordinate cell cycle exit.95 Besides activating PHOX2a, MASH1 was shown to regulate the expression of Neurogenin-1 (NGN1) and subsequently NeuroD, in the olfactory neuron progenitors.96 Brief overexpression of NGN1 and NeuroD in Xenopus laevis ectodermal explants revealed a spectrum of neural genes regulated by these proneural bHLH TFs.97 Such genes include Math3, HEN1, Dll1, Elavl3, Gadd45g, MyT1 and Hes-6.
SOX1.
SOX1 affects neurogenesis. Kan et al. reported that SOX1 binds to the promoter of HES1 and suppresses its expression, disrupts cell cycle by preventing cells from entering the G2 phase, and directly drives the promoter activity of NGN1.72 SOX1 also suppresses β-catenin-mediated TCF/LEF signaling by binding to β-catenin itself. Thus, SOX1 promotes neurogenesis through multiple independent pathways.
PAX6.
GATA-2,98 and PAX6,99 promote differentiation by inducing the transcription of negative regulators of cell cycle. These proneural factors can inhibit the expression of TFs that maintain the NSC state. PAX6 can induce the expression of NGN2,99 which then downregulates the expression of SOX1-3.49 HES6.2 is upregulated by NGN1/2 and synergizes with these proneural factors to promote neuronal differentiation by repressing HES5 and inhibiting downstream Notch effectors.100 Proneural TFs can also inhibit gliogenesis. Examples of such TFs include NGN1, functioning by sequestering the CREB binding protein (CBP)—mothers against decapentaplegic homolog 1 (SMAD1) transcription complex from promoters of astrocyte differentiation genes and by inhibiting signal transducer and activator of transcription (STAT)-3 which would otherwise induce astroglial lineage.101
Despite a variety of TFs known to play a role in neuronal lineage initiation and specification, much less is known about astroglial differentiation. Various signaling molecules can induce the differentiation of NSCs towards the astroglial lineage. Such factors include cytokines belonging to the interleukin (IL)-6 family, those of the activin/Bone Morphogenetic Protein (BMP) family, ciliary neurotrophic factor, and recently Nogo-66. These ligands signal through various pathways which converge in the nucleus to activate STAT3 and SMAD1,102–104 to induce the transcriptional expression of glial proteins such as glial fibrillary acidic protein.105 In addition, Notch signaling activates the GFAP promoter via the CSL DNA-binding protein.106
Interestingly, MASH1 is expressed in oligodendrocyte progenitors and might have a role in specifying the differentiation of oligodendrocytes from immature glial cells.107 Other oligodendrocyte-specifying TFs include bHLH proteins OLIG1 and OLIG2 that are activated by Shh108 and bFGF signals.109 OLIG2 regulates the development of oligodendrocytes by enabling the differentiation of NG2-positive synantocytes into the oligodendroglial lineage.110
MicroRNA
Post-transcriptional gene regulators, such as microRNAs, are likely to be important for controlling the balance between self-renewal and differentiation in NSCs. MicroRNAs, a family of small (∼22 nucleotides long), non-coding RNAs similar to the siRNAs involved in RNA silencing, have been shown to play important roles in diverse processes including apoptosis, fat metabolism, cancer, major signaling pathways, tissue morphogenesis and development.
MicroRNAs originate from stem-loop precursors in the genome. Transcription produces primary microRNA transcripts (pri-microRNAs), which are then cleaved by the nuclear RNase III enzyme Drosha to release precursor microRNAs.111 After Drosha processing, pre-microRNAs are exported out of the nucleus by the nuclear transport receptor Exportin-5, in a process requiring the hydrolysis of GTP to GDP.112,113 Pre-microRNAs are next cleaved by the cytoplasmic RNase III enzyme Dicer to produce ∼22 nucleotide microRNA duplexes.114,115 After Dicer processing, one strand of the microRNA duplex is usually degraded while the other persists as a mature microRNA.116 The strand that has a less thermodynamically stable 5′ end is thought to be incorporated into effector complexes called microRNA-containing RNA-induced silencing complexes (miRISCs).117,118 These miRISCs recognize and bind to target mRNAs to modulate their expression.
MicroRNAs modulate target expression in two different ways: by directing transcript degradation or inhibiting translation.119 In plants and very rarely in animals, microRNAs bind to highly complementary microRNA binding sites in target mRNAs to guide sequence-specific cleavage. This process is similar to RNA interference.120 In animals, microRNAs bind to partially complementary microRNA binding sites and repress translation. This repression is achieved by interfering with translation or by guiding degradation processes that are initiated by mRNA deadenylation and decapping.121
A number of microRNAs exhibit distinct spatial and temporal expression patterns during development.122–124 Additionally, some microRNA expression patterns show species conservation, e.g., miR-1 in muscles, miR-124 in the CNS and miR-10 in anterior-posterior patterning.123 These observations indicate that microRNAs may be involved in the specification and maintenance of tissue identity and other facets of development.
About 70% of the microRNAs identified by 2005 were expressed in mammalian brains, suggesting possible roles of these microRNAs in neural function.124–126 Studies in invertebrate model systems have identified lsy-6, the first microRNA found to play a role in neuronal patterning,127 and miR-9a, which ensures the generation of the precise number of neuronal precursor cells during development.128 In vertebrate models, the restoration of a single microRNA (miR-430) in zebrafish modified to prevent production of endogenous microRNAs ameliorated deficits in neuroectodermal development and neuronal differentiation.129
In addition to being important regulators of vertebrate CNS development,125,126,129 microRNAs also play key roles during neural differentiation in vitro.125,130 During neural differentiation, Smirnova et al.130 demonstrated that the most highly expressed microRNAs in adult brain, miR-124 and miR-128, were preferentially expressed in neurons; miR-23 was restricted to astrocytes; miR-26 and miR-29 had stronger expression in astrocytes than neurons; and miR-9 and miR-125 were fairly evenly distributed.130 Overexpression of miR-124, miR-128 and miR-9 in NPs decreased astrocyte differentiation, whereas inhibition of miR-9 alone or in combination with miR-124 led to reduced neurogenesis.131
Studies which show that Dicer-deficient mice lacking mature microRNAs die at embryonic day 7.5 and lack multipotent stem cells support a role for microRNAs in stem cell self-renewal.132,133 Indeed, Rybak et al. demonstrate that microRNAs let-7 and mir-125 and the pluripotency factor Lin-28 participate in an autoregulatory circuit that controls microRNA processing during neural stem cell commitment.134 Changes in expression of let-7 and one of its known targets, the transcriptional regulator HMGA2, during aging may contribute to the decline in NSC function.64 In human glioma neurosphere cultures, miR-128 has been shown to specifically block glioma self-renewal via post-transcriptional regulation of the NSC self-renewal factor Bmi-1.135
Laminin γ1 and integrin β1, which are highly expressed in NSCs/NPs cells and repressed upon neuronal differentiation, were recently identified as targets of miR-124.136 MiR-124 also regulates the small C-terminal domain phosphatase 1 (SCP1), a phosphatase implicated in neural development, further supporting its role in neurogenesis.137 Furthermore, let-7 has been shown to target Hunchback, a gene which regulates the temporal identity of neuroblasts.138–140 The identification of these and other microRNA targets in the NSCs will help us to better understand the role of microRNAs in regulating of neural stem cell self-renewal and differentiation.141
Conclusion
Taking the success of linking TFs to pluripotency we have attempted in this review to examine the data that links TFs to NSC behavior (Fig. 1). TFs have been found to affect NSC self-renewal, proliferation, repression of differentiation and differentiation (early and late). Some of the TFs involved in these behaviors have overlapping functions (Fig. 1B). For example, BMI-1 and GLI-2/3 have roles in self-renewal and proliferation. Interestingly, HES1 and 5 are the only factors that play a role in all three pre-differentiation steps (Fig. 1B). Further identification of TFs and their roles will help elucidate the regulatory networks that control NSC behavior and this information will be crucial for the use of NSCs as cellular models of development and disease.
Acknowledgements
We are indebted to Hanaa Goolbar for her help in preparing this manuscript. This work was supported by A-STAR.
Abbreviations
- CBF-1
notch effector C-promoter binding factor-1
- CNS
central nervous system
- EGFP
enhanced green fluorescent protein
- ESC
embryonic stem cell
- HSC
hematopoietic stem cell
- ID
inhibitor of DNA binding
- KO
knock-out
- NICD
notch intracellular domain
- NSC
neural stem cell
- NP
neural progenitor
- OPC
oligodendrocyte precursor cell
- Sh
short hairpin
- Shh
sonic hedgehog
- SVZ
subventricular zone
- TFs
transcription factors
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- TLX
human homologue of the drosophilia tailess gene
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/8803
References
- 1.Choi CQ. A stroke for stem cells. Sci Am. 2007;296:8–9. [PubMed] [Google Scholar]
- 2.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 6.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 7.Fortunel NO, Otu HH, Ng HH, Chen J, Mu X, Chevassut T, et al. Comment on “Stemness”: “transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 8.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 9.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 12.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 13.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 16.Stefanovic S, Puceat M. Oct-3/4: not just a gatekeeper of pluripotency for embryonic stem cell, a cell fate instructor through a gene dosage effect. Cell Cycle. 2007;6:8–10. doi: 10.4161/cc.6.1.3633. [DOI] [PubMed] [Google Scholar]
- 17.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 18.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 19.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 20.Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 21.Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 23.Boer B, Kopp J, Mallanna S, Desler M, Chakravarthy H, Wilder PJ, et al. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 2007;35:1773–1786. doi: 10.1093/nar/gkm059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 25.Kalani MY, Cheshier SH, Cord BJ, Bababeygy SR, Vogel H, Weissman IL, et al. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc Natl Acad Sci USA. 2008;105:16970–16975. doi: 10.1073/pnas.0808616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, et al. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 29.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 30.Zechner D, Fujita Y, Hulsken J, Muller T, Walther I, Taketo MM, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 31.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 32.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 33.Israsena N, Hu M, Fu W, Kan L, Kessler JA. The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev Biol. 2004;268:220–231. doi: 10.1016/j.ydbio.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 34.Gridley T. Notch signaling in vertebrate development and disease. Mol Cell Neurosci. 1997;9:103–108. doi: 10.1006/mcne.1997.0610. [DOI] [PubMed] [Google Scholar]
- 35.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 37.Ohtsuka T, Sakamoto M, Guillemot F, Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278:44808–44815. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- 39.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 40.Gao F, Zhang Q, Zheng MH, Liu HL, Hu YY, Zhang P, et al. Transcription factor RBP-J-mediated signaling represses the differentiation of neural stem cells into intermediate neural progenitors. Mol Cell Neurosci. 2009;40:442–450. doi: 10.1016/j.mcn.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 43.Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M, et al. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 44.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 45.Stecca B, Ruiz I, Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009 doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palma V, Ruiz I, Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- 47.Takanaga H, Tsuchida-Straeten N, Nishide K, Watanabe A, Aburatani H, Kondo T. Gli2 Is A Novel Regulator of Sox2 Expression In Telencephalic Neuroepithelial Cells. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0580. [DOI] [PubMed] [Google Scholar]
- 48.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 49.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 50.Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, et al. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 51.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- 52.Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 53.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 54.Miyagi S, Masui S, Niwa H, Saito T, Shimazaki T, Okano H, et al. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582:2811–2815. doi: 10.1016/j.febslet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 56.Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- 57.Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, et al. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- 58.Miyagi S, Nishimoto M, Saito T, Ninomiya M, Sawamoto K, Okano H, et al. The Sox2 regulatory region 2 functions as a neural stem cell-specific enhancer in the telencephalon. J Biol Chem. 2006;281:13374–13381. doi: 10.1074/jbc.M512669200. [DOI] [PubMed] [Google Scholar]
- 59.Alvarez-Bolado G, Rosenfeld MG, Swanson LW. Model of forebrain regionalization based on spatiotemporal patterns of POU-III homeobox gene expression, birthdates and morphological features. J Comp Neurol. 1995;355:237–295. doi: 10.1002/cne.903550207. [DOI] [PubMed] [Google Scholar]
- 60.Hara Y, Rovescalli AC, Kim Y, Nirenberg M. Structure and evolution of four POU domain genes expressed in mouse brain. Proc Natl Acad Sci USA. 1992;89:3280–3284. doi: 10.1073/pnas.89.8.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathis JM, Simmons DM, He X, Swanson LW, Rosenfeld MG. Brain 4: a novel mammalian POU domain transcription factor exhibiting restricted brain-specific expression. EMBO J. 1992;11:2551–2561. doi: 10.1002/j.1460-2075.1992.tb05320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hudson CD, Podesta J, Henderson D, Latchman DS, Budhram-Mahadeo V. Coexpression of Brn-3a POU protein with p53 in a population of neuronal progenitor cells is associated with differentiation and protection against apoptosis. J Neurosci Res. 2004;78:803–814. doi: 10.1002/jnr.20299. [DOI] [PubMed] [Google Scholar]
- 64.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 66.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 68.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malas S, Postlethwaite M, Ekonomou A, Whalley B, Nishiguchi S, Wood H, et al. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience. 2003;119:421–432. doi: 10.1016/s0306-4522(03)00158-1. [DOI] [PubMed] [Google Scholar]
- 70.Rizzoti K, Brunelli S, Carmignac D, Thomas P, Robinson I, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–255. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- 71.Kan L, Jalali A, Zhao L, Zhou X, McGuire T, Kazanis I, et al. Dual function of Sox1 in telencephalic progenitor cells. Dev Biol. 2007;310:85–98. doi: 10.1016/j.ydbio.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kan L, Israsena N, Zhang Z, Hu M, Zhao L, Jalali A, et al. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269:580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Wang TW, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J Comp Neurol. 2006;497:88–100. doi: 10.1002/cne.20984. [DOI] [PubMed] [Google Scholar]
- 74.Fuccillo M, Joyner A, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 75.Matise M, Epstein D, Park H, Platt K, Joyner A. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 76.Theil T, Alvarez-Bolado G, Walter A, Rüther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- 77.Park H, Bai C, Platt K, Matise M, Beeghly A, Hui C, et al. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 78.Galvin KE, Ye H, Erstad DJ, Feddersen R, Wetmore C. Gli1 induces G2/M arrest and apoptosis in hippocampal but not tumor-derived neural stem cells. Stem Cells. 2008;26:1027–1036. doi: 10.1634/stemcells.2007-0879. [DOI] [PubMed] [Google Scholar]
- 79.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267:21879–21885. [PubMed] [Google Scholar]
- 81.Kabos P, Kabosova A, Neuman T. Blocking HES1 expression initiates GABAergic differentiation and induces the expression of p21(CIP1/WAF1) in human neural stem cells. J Biol Chem. 2002;277:8763–8766. doi: 10.1074/jbc.C100758200. [DOI] [PubMed] [Google Scholar]
- 82.Solecki D, Liu X, Tomoda T, Fang Y, Hatten M. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 83.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizo A, Dontje B, Vellenga E, de Haan G, Schuringa J. Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood. 2008;111:2621–2630. doi: 10.1182/blood-2007-08-106666. [DOI] [PubMed] [Google Scholar]
- 85.Moon JH, Yoon BS, Kim B, Park G, Jung HY, Maeng I, et al. Induction of neural stem cell-like cells (NSCLCs) from mouse astrocytes by Bmi1. Biochem Biophys Res Commun. 2008;371:267–272. doi: 10.1016/j.bbrc.2008.04.068. [DOI] [PubMed] [Google Scholar]
- 86.Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, et al. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenway DJ, Street M, Jeffries A, Buckley NJ. RE1 Silencing transcription factor maintains a repressive chromatin environment in embryonic hippocampal neural stem cells. Stem Cells. 2007;25:354–363. doi: 10.1634/stemcells.2006-0207. [DOI] [PubMed] [Google Scholar]
- 89.Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 91.Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- 92.Bani-Yaghoub M, Kubu CJ, Cowling R, Rochira J, Nikopoulos GN, Bellum S, et al. A switch in numb isoforms is a critical step in cortical development. Dev Dyn. 2007;236:696–705. doi: 10.1002/dvdy.21072. [DOI] [PubMed] [Google Scholar]
- 93.Elmi M, Faigle R, Yang W, Matsumoto Y, Rosenqvist E, Funa K. Mechanism of MASH1 induction by ASK1 and ATRA in adult neural progenitors. Mol Cell Neurosci. 2007;36:248–259. doi: 10.1016/j.mcn.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 94.Lo L, Sommer L, Anderson DJ. MASH1 maintains competence for BMP2-induced neuronal differentiation in post-migratory neural crest cells. Curr Biol. 1997;7:440–450. doi: 10.1016/s0960-9822(06)00191-6. [DOI] [PubMed] [Google Scholar]
- 95.Paris M, Wang WH, Shin MH, Franklin DS, Andrisani OM. Homeodomain transcription factor Phox2a, via cyclic AMP-mediated activation, induces p27Kip1 transcription, coordinating neural progenitor cell cycle exit and differentiation. Mol Cell Biol. 2006;26:8826–8839. doi: 10.1128/MCB.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- 97.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.El Wakil A, Francius C, Wolff A, Pleau-Varet J, Nardelli J. The GATA2 transcription factor negatively regulates the proliferation of neuronal progenitors. Development. 2006;133:2155–2165. doi: 10.1242/dev.02377. [DOI] [PubMed] [Google Scholar]
- 99.Bel-Vialar S, Medevielle F, Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 100.Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev Biol. 2005;281:318–333. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 101.Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 102.Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 103.Satoh M, Sugino H, Yoshida T. Activin promotes astrocytic differentiation of a multipotent neural stem cell line and an astrocyte progenitor cell line from murine central nervous system. Neurosci Lett. 2000;284:143–146. doi: 10.1016/s0304-3940(00)00981-2. [DOI] [PubMed] [Google Scholar]
- 104.Taga T, Fukuda S. Role of IL-6 in the neural stem cell differentiation. Clin Rev Allergy Immunol. 2005;28:249–256. doi: 10.1385/CRIAI:28:3:249. [DOI] [PubMed] [Google Scholar]
- 105.Kahn MA, Huang CJ, Caruso A, Barresi V, Nazarian R, Condorelli DF, et al. Ciliary neurotrophic factor activates JAK/Stat signal transduction cascade and induces transcriptional expression of glial fibrillary acidic protein in glial cells. J Neurochem. 1997;68:1413–1423. doi: 10.1046/j.1471-4159.1997.68041413.x. [DOI] [PubMed] [Google Scholar]
- 106.Ge W, Martinowich K, Wu X, He F, Miyamoto A, Fan G, et al. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J Neurosci Res. 2002;69:848–860. doi: 10.1002/jnr.10364. [DOI] [PubMed] [Google Scholar]
- 107.Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, et al. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134:285–293. doi: 10.1242/dev.02727. [DOI] [PubMed] [Google Scholar]
- 108.Alberta JA, Park SK, Mora J, Yuk D, Pawlitzky I, Iannarelli P, et al. Sonic hedgehog is required during an early phase of oligodendrocyte development in mammalian brain. Mol Cell Neurosci. 2001;18:434–441. doi: 10.1006/mcne.2001.1026. [DOI] [PubMed] [Google Scholar]
- 109.Abematsu M, Kagawa T, Fukuda S, Inoue T, Takebayashi H, Komiya S, et al. Basic fibroblast growth factor endows dorsal telencephalic neural progenitors with the ability to differentiate into oligodendrocytes but not gamma-aminobutyric acidergic neurons. J Neurosci Res. 2006;83:731–743. doi: 10.1002/jnr.20762. [DOI] [PubMed] [Google Scholar]
- 110.Ligon KL, Kesari S, Kitada M, Sun T, Arnett HA, Alberta JA, et al. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci USA. 2006;103:7853–7858. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 112.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 115.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 116.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 117.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 118.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 119.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 120.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 121.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 122.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 124.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 125.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 128.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 130.Smirnova L, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of microRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 131.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 133.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 134.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 135.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 136.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 139.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 140.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, et al. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 141.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, et al. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- 143.Havrda MC, Harris BT, Mantani A, Ward NM, Paolella BR, Cuzon VC, et al. Id2 is required for specification of dopaminergic neurons during adult olfactory neurogenesis. J Neurosci. 2008;28:14074–14086. doi: 10.1523/JNEUROSCI.3188-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 145.Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zencak D, Lingbeek M, Kostic C, Tekaya M, Tanger E, Hornfeld D, et al. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, et al. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suh H, Consiglio A, Ray J, Sawai T, D'Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 151.Josephson R, Muller T, Pickel J, Okabe S, Reynolds K, Turner PA, et al. POU transcription factors control expression of CNS stem cell-specific genes. Development. 1998;125:3087–3100. doi: 10.1242/dev.125.16.3087. [DOI] [PubMed] [Google Scholar]
- 152.Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136:531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- 153.Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, et al. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- 154.Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, et al. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur J Neurosci. 2004;19:3129–3141. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- 155.Kala K, Haugas M, Lillevali K, Guimera J, Wurst W, Salminen M, et al. Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development. 2009;136:253–262. doi: 10.1242/dev.029900. [DOI] [PubMed] [Google Scholar]
- 156.Batista MF, Lewis KE. Pax2/8 act redundantly to specify glycinergic and GABAergic fates of multiple spinal interneurons. Dev Biol. 2008;323:88–97. doi: 10.1016/j.ydbio.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jo AY, Park CH, Aizawa S, Lee SH. Contrasting and brain region-specific roles of neurogenin2 and mash1 in GABAergic neuron differentiation in vitro. Exp Cell Res. 2007;313:4066–4081. doi: 10.1016/j.yexcr.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 158.Dullin JP, Locker M, Robach M, Henningfeld KA, Parain K, Afelik S, et al. Ptf1a triggers GABAergic neuronal cell fates in the retina. BMC Dev Biol. 2007;7:110. doi: 10.1186/1471-213X-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Krueger KC, Deneris ES. Serotonergic transcription of human FEV reveals direct GATA factor interactions and fate of Pet-1-deficient serotonin neuron precursors. J Neurosci. 2008;28:12748–12758. doi: 10.1523/JNEUROSCI.4349-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Couch JA, Chen J, Rieff HI, Uri EM, Condron BG. robo2 and robo3 interact with eagle to regulate serotonergic neuron differentiation. Development. 2004;131:997–1006. doi: 10.1242/dev.00962. [DOI] [PubMed] [Google Scholar]
- 161.Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, et al. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- 163.Zhao ZQ, Scott M, Chiechio S, Wang JS, Renner KJ, Gereau RWt, et al. Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. J Neurosci. 2006;26:12781–12788. doi: 10.1523/JNEUROSCI.4143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, et al. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- 165.Fukuda S, Kondo T, Takebayashi H, Taga T. Negative regulatory effect of an oligodendrocytic bHLH factor OLIG2 on the astrocytic differentiation pathway. Cell Death Differ. 2004;11:196–202. doi: 10.1038/sj.cdd.4401332. [DOI] [PubMed] [Google Scholar]
- 166.Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963–968. doi: 10.1083/jcb.200404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438:360–363. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- 168.Sakurai K, Osumi N. The neurogenesis-controlling factor, Pax6, inhibits proliferation and promotes maturation in murine astrocytes. J Neurosci. 2008;28:4604–4612. doi: 10.1523/JNEUROSCI.5074-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vrotsos EG, Kolattukudy PE, Sugaya K. MCP-1 involvement in glial differentiation of neuroprogenitor cells through APP signaling. Brain Res Bull. 2009 doi: 10.1016/j.brainresbull.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cebolla B, Vallejo M. Nuclear factor-I regulates glial fibrillary acidic protein gene expression in astrocytes differentiated from cortical precursor cells. J Neurochem. 2006;97:1057–1070. doi: 10.1111/j.1471-4159.2006.03804.x. [DOI] [PubMed] [Google Scholar]
- 171.Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 172.Yanagisawa M, Nakashima K, Arakawa H, Ikenaka K, Yoshida K, Kishimoto T, et al. Astrocyte differentiation of fetal neuroepithelial cells by interleukin-11 via activation of a common cytokine signal transducer, gp130, and a transcription factor, STAT3. J Neurochem. 2000;74:1498–1504. doi: 10.1046/j.1471-4159.2000.0741498.x. [DOI] [PubMed] [Google Scholar]
- 173.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 174.Potzner MR, Griffel C, Lutjen-Drecoll E, Bosl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27:5316–5326. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 176.Stolt CC, Schmitt S, Lommes P, Sock E, Wegner M. Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev Biol. 2005;281:309–317. doi: 10.1016/j.ydbio.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 177.Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Inoue K, Tanabe Y, Lupski JR. Myelin deficiencies in both the central and the peripheral nervous systems associated with a SOX10 mutation. Ann Neurol. 1999;46:313–318. doi: 10.1002/1531-8249(199909)46:3<313::aid-ana6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 179.Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sohn J, Natale J, Chew LJ, Belachew S, Cheng Y, Aguirre A, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26:9722–9735. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Parras CM, Hunt C, Sugimori M, Nakafuku M, Rowitch D, Guillemot F. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci. 2007;27:4233–4242. doi: 10.1523/JNEUROSCI.0126-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lee J, Wu Y, Qi Y, Xue H, Liu Y, Scheel D, et al. Neurogenin3 participates in gliogenesis in the developing vertebrate spinal cord. Dev Biol. 2003;253:84–98. doi: 10.1006/dbio.2002.0868. [DOI] [PubMed] [Google Scholar]
- 183.Balasubramaniyan V, Timmer N, Kust B, Boddeke E, Copray S. Transient expression of Olig1 initiates the differentiation of neural stem cells into oligodendrocyte progenitor cells. Stem Cells. 2004;22:878–882. doi: 10.1634/stemcells.22-6-878. [DOI] [PubMed] [Google Scholar]
- 184.Gong X, Lin T, Sun Z, Fu M, Zuo H, Xie Z. Olig1 is downregulated in oligodendrocyte progenitor cell differentiation. Neuroreport. 2008;19:1203–1207. doi: 10.1097/WNR.0b013e328308b322. [DOI] [PubMed] [Google Scholar]
- 185.Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Copray S, Balasubramaniyan V, Levenga J, de Bruijn J, Liem R, Boddeke E. Olig2 overexpression induces the in vitro differentiation of neural stem cells into mature oligodendrocytes. Stem Cells. 2006;24:1001–1010. doi: 10.1634/stemcells.2005-0239. [DOI] [PubMed] [Google Scholar]
- 187.Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- 188.Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- 189.Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, et al. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 191.Sock E, Leger H, Kuhlbrodt K, Schreiber J, Enderich J, Richter-Landsberg C, et al. Expression of Krox proteins during differentiation of the O-2A progenitor cell line CG-4. J Neurochem. 1997;68:1911–1919. doi: 10.1046/j.1471-4159.1997.68051911.x. [DOI] [PubMed] [Google Scholar]
- 192.Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol Cell Neurosci. 2004;25:111–123. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 193.Schreiber J, Enderich J, Sock E, Schmidt C, Richter-Landsberg C, Wegner M. Redundancy of class III POU proteins in the oligodendrocyte lineage. J Biol Chem. 1997;272:32286–32293. doi: 10.1074/jbc.272.51.32286. [DOI] [PubMed] [Google Scholar]
- 194.Berndt JA, Kim JG, Tosic M, Kim C, Hudson LD. The transcriptional regulator Yin Yang 1 activates the myelin PLP gene. J Neurochem. 2001;77:935–942. doi: 10.1046/j.1471-4159.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- 195.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mason JL, Angelastro JM, Ignatova TN, Kukekov VG, Lin G, Greene LA, et al. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol Cell Neurosci. 2005;29:372–380. doi: 10.1016/j.mcn.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 197.Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, et al. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- 198.Awatramani R, Beesley J, Yang H, Jiang H, Cambi F, Grinspan J, et al. Gtx, an oligodendrocyte-specific homeodomain protein, has repressor activity. J Neurosci Res. 2000;61:376–387. doi: 10.1002/1097-4547(20000815)61:4<376::AID-JNR4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 199.Nicolay DJ, Doucette JR, Nazarali AJ. Hoxb4 in oligodendrogenesis. Cell Mol Neurobiol. 2004;24:357–366. doi: 10.1023/B:CEMN.0000022768.34389.4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Roybon L, Hjalt T, Christophersen NS, Li JY, Brundin P. Effects on differentiation of embryonic ventral midbrain progenitors by Lmx1a, Msx1, Ngn2 and Pitx3. J Neurosci. 2008;28:3644–3656. doi: 10.1523/JNEUROSCI.0311-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]