Abstract
HTRF (Homogeneous Time Resolved Fluorescence) is the most frequently used generic assay technology to measure analytes in a homogenous format, which is the ideal platform used for drug target studies in high-throughput screening (HTS). This technology combines fluorescence resonance energy transfer technology (FRET) with time-resolved measurement (TR). In TR-FRET assays, a signal is generated through fluorescent resonance energy transfer between a donor and an acceptor molecule when in close proximity to each other. Buffer and media interference is dramatically reduced by dual-wavelength detection, and the final signal is proportional to the extent of product formation. The HTRF assay is usually sensitive and robust that can be miniaturized into the 384 and 1536-well plate formats. This assay technology has been applied to many antibody-based assays including GPCR signaling (cAMP and IP-One), kinases, cytokines and biomarkers, bioprocess (antibody and protein production), as well as the assays for protein-protein, proteinpeptide, and protein-DNA/RNA interactions.
Since its introduction to the drug-screening world over ten years ago, researchers have used HTRF to expedite the study of GPCRs, kinases, new biomarkers, protein-protein interactions, and other targets of interest. HTRF has also been utilized as an alternative method for bioprocess monitoring. The first-generation HTRF technology, which uses Europium cryptate as a fluorescence donor to monitor reactions between biomolecules, was extended in 2008 through the introduction of a second-generation donor, Terbium cryptate (Tb), enhancing screening performance. Terbium cryptate possesses different photophysical properties compared to Europium, including increased quantum yield and a higher molar extinction coefficient. In addition to being compatible with the same acceptor fluorophors used with Europium, it can serve as a donor fluorophore to green-emitting fluors because it has multiple emission peaks including one at 490 nm. Moreover, all Terbium HTRF assays can be read on the same HTRF-compatible instruments as Europium HTRF assays.
Overall, HTRF is a highly sensitive, robust technology for the detection of molecular interactions in vitro and is widely used for primary and secondary screening phases of drug development. This review addresses the general principles of HTRF and its current applications in drug discovery.
Keywords: HTRF, TR-FRET, GPCR, Kinase, Biomarker, Bioprocess.
INTRODUCTION
Utilizing rare-earth lanthanides with long emission half-lives as donor fluorophores, HTRF technology combines standard FRET with the time resolved measurement (TR) of fluorescence [1,2]. This powerful combination provides significant benefits to drug discovery researchers in high throughput screening (HTS), including assay flexibility, reliability, increased assay sensitivity, higher throughput, and fewer false positive/false negative results. These features, allied with automated liquid handling and robust detection instrumentation, allow for a broad range of applications in HTS.
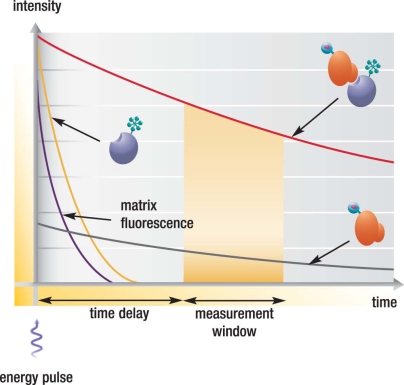
The principle behind FRET is based on the transfer of energy between two fluorophores, a donor (long-lived fluorescence) and an acceptor (short-lived fluorescence), when in close proximity [3, 4]. Molecular interactions between biomolecules can be assessed by coupling each partner with a fluorescent label and detecting the level of energy transfer (Fig. 1). In the past, organic fluorescent compounds such as fluorescein and rhodamine have been widely used in the regular fluorescence assay. However, these bioassays have great disadvantages in that fluorescent detection because it is dramatically inhibited by noise in the back ground derived from scattered excitation light and significantly interfered by fluorescence from coexisting material in the sample (fluorescent compounds and dust/line), making it difficult to obtain a highly sensitive measurement. Through time-resolved measurement of fluorescence, HTRF allows the elimination of short-lived background fluorescence. Introducing a time delay (50-150 microseconds) between the initial light excitation and fluorescence measurement minimizes the contribution of all non-specific short-lived fluorescence emissions. In contrast, HTRF acceptor fluorophores emit long-lived fluorescence when engaged in the TR-FRET process with a long-lived donor fluorophore, signifying energy transfer through proximity of the labeled biomolecules (Fig. 2).
Fig. (1).
The general principle of fluorescence resonance energy transfer technology (FRET). When the donor and acceptor apart, there is no FRET signal. Once they are brought in proximity, the FRET signals are generated (for example, the emission spectrum at 665 nm).
Fig. (2).
The energy pulse from the excitation source (flash lamp, laser) is followed by a time delay, allowing interfering short-lived fluorescence (compounds, proteins, medium etc.) to decay. The red line: FRET signal intensity generated at 665 nm; black line, emission of donor cryptate at 620 nm; orange line, fluorescent signal generated from acceptor fluorophores.
Four specific fluorophores are used in HTRF forming different TR-FRET systems. The central element, the energy donor, is either Europium cryptate (Eu3+ cryptate) (Fig. 3a) [5] or Lumi4-Tb (Tb2+ cryptate) (unpublished structure) (Fig. 3b). These rare earth complexes, based on the work from Nobel Prize laureate, Professor Jean-Marie Lehn, and from Professor Raymond respectively, have a macrocycle within which a Eu3+ ion or Tb2+ is tightly embedded. These ions are not fluorescent on their own; they need a light-collection device (i.e. the cage) to be excited. Similar to functions of the chelate in other luminescent lanthanide technologies, this cage acts as an antenna and allows both energy collection and transfer to the ions, which ultimately releases this energy with a specific fluorescent pattern. In particular, these cryptates are not subject to the photo-bleaching that affects a number of more conventional fluorophores, and the ions are almost inseparable from their macrocycles [5]. However, due to the unique caged structure, the kinetics stability of the cryptate is extremely higher than the one of those lanthanide chelates [3]. Rare earth chelates would be dissociated and unstable in acidic media or in the presence of divalent ions like Mn2+, while rare earth cryptates are extraordinarily stable under wide range of chemical conditions, like reverse phase chromatography in the presence of trifluoroacetic acid, and are not affected by the presence of divalent ions in the media [4]. It therefore allows for enhanced assay performance in terms of sensitivity, assay window, and robustness, without compromising the features and benefits that HTRF brings to assays: a ‘mix and measure’ non-radioactive format, miniaturizable to uHTS formats, showing excellent robustness and a low compound interference rate (Fig. 3). Lumi4-Tb’s structure, a lanthanide tightly embedded in a surrounding macrocycle, remains very much in line with that of previous HTRF cryptates and shows superior stability compared to other Terbium complexes. Terbium is exceptionally bright – 10 to 20 times brighter than Europium that significantly increases the detection sensitivity in assays such as the exploration of cell surface receptors.
Fig. (3).
Features of donor cryptate. a. Structure of cryptate trisbipyridine (TBP). Lambda max absorption: 305nm, molar extinction coefficient at 305nm: 30000 M-1 cm-1, molar extinction coefficient 337nm: 4500 M-1 cm-1. b. Detection wavelengths for Europium and Terbium cryptate. Both Europium and Terbium have emission spectra at 620 nm so that these signals can be used as reference for data analysis.
The first acceptor developed for HTRF was XL665, a phycobiliprotein pigment purified from red algae [6]. XL665 is a large heterohexameric edifice of 105 kDa, cross-linked after isolation for better stability and preservation of its photophysical properties in HTRF assays [6]. Unlike fluorescein, it is fully compatible with Eu cryptates. It is red-shifted and its emission is more likely to be away from possible medium and compound interference. The second-generation acceptor, d2, possesses a series of photophysical properties very similar to those of XL665 but is characterized by organic structures 100 times smaller than XL665. As a much smaller entity, d2 limits the steric hindrance problems sometimes suspected in XL665 based TR-FRET systems. These near-infrared acceptors are also particularly suited for homogeneous assays since their emission is less likely to be disturbed by intrinsic medium or compound autofluorescence arise in the typical compound screen process. The properties of these red acceptors also make them suitable for coupling with Terbium cryptate. Moreover, due to additional peaks in its emission spectrum, Terbium cryptate can be coupled with green acceptors such as fluorescein, emitting in the 520 nm range that may for instance allow designing multiplex assays with two readouts.
HTRF emissions are measured at two different wavelengths, 620nm (donor) and 665nm (acceptor). This feature is extremely advantageous, particularly for reducing well-to-well variations that may arise in homogeneous assay formats. Because wells contain a range of compounds and/or medium additives, each well will have different photophysical properties and cause varying degrees of signal interference and therefore altered signal intensities. This well-to-well signal variation is not due to true differences in light transition, but it can lead to misleading results if only a single emission wavelength is measured. A unique ratio-metric measurement of two emission wavelengths (patent US 5,527,684 and foreign equivalents) corrects for well-to-well variability and signal quenching from assay components and medium variability. Emissions at 620nm (donor fluor) are used as an internal reference while emissions at 665nm (acceptor fluor) are used as an indicator of the biological reaction being assessed. Because both the 620nm and 665nm emissions are decreased by sample interferences, the ratio remains unchanged (Fig. 4). In addition, the ratiometric readout can minimize the system errors caused by liquid handling instruments and detectors.
Fig. (4). Ratiometric reduction of the signal output data.
The specific signal at 665 nm may be affected by light transmission. The ratio (i.e. 665nm/620 nm) will normalize the signal measured and generate a variable that is independent of the optical properties of the medium in which the interaction is studied. In the case shown above, the 665 nm fluorescence decreases proportionally to the transmission, whereas the ratio corrects this interference and is equivalent in both situations.
HTRF ASSAY APPLICATIONS
GPCR Investigation and Screening
GPCRs have offered a broad playfield to HTRF development and the assays that have been described may be segmented into two categories: functional assays and mechanistic assays. Functional assays have been based on the quantification of secondary messengers, whose concentration correlates tightly to events occurring at the cell surface receptor level, such as ligand binding. They are part of a signaling cascade that may be blocked by specific inhibitors to enable their accumulation in the cell.
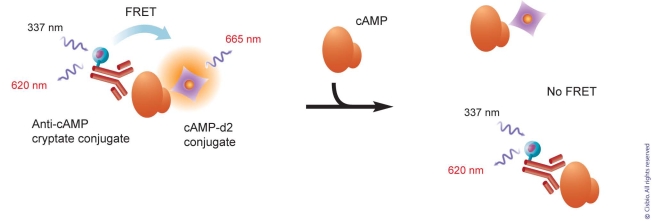
Cyclic AMP has long been used as a secondary messenger, as its production results from the up or down regulation of adenylate cyclase upon Gs and Gi-coupled receptor activity. The HTRF assay was developed based on the detection on cAMP (Fig. 5). On the other hand, Gq-coupled receptors activate the phospholipase C pathway (PLC), which acts upstream to the inositol phosphate cascade. Thus far, the main readout consisted of the detection of calcium release triggered by IP3, one on the cascade’s metabolites. The assessment of IP3 as a potential secondary messenger has been hampered by its extreme instability, as no inhibitor of the cascade is known to block its degradation. The measurement of total inositol phosphate metabolites by radioactive uptake translates receptor activity well, however, these methods are far from being HTS-friendly. Recently, Trinquet et al. demonstrated that inositol-1-phosphate, a sub-metabolite of the same pathway, could be accumulated in the cell in the presence of Lithium Chloride, an agent inhibiting downstream phosphatases [7]. Similarly to cAMP assay development, the generation of specific high affinity monoclonal antibodies have enabled the optimization of the HTRF IP-One assay [7, 8].
Fig. (5).
cAMP assay principle. In this competitive assay, an anti-cAMP antibody is labeled with a cryptate donor and cAMP is labeled with acceptor d2. Once both of them are place together, the binding of antibody to cAMP brings donor and acceptor into proximity range. Upon the excitation of donor at 337 nm, the energy is transferred from donor to acceptor. Donor and acceptor generate emission at 620 nm and 665 nm, respectively. In the cAMP assay, the level of free cAMP generated by the signal cascade can be measured by competing with the cAMP d2 for antibody binding. IP-One assay from HTRF has the similar principle.
The application of HTRF to functional detection of GPCRs has been illustrated for many receptors including 5HT4 and 5HT6 [9, 10], BLT1 and cysLT1 leukotriene receptors [11], canabinoid receptors [12, 13], H1, 2 and 3 receptors [14, 15], PTH receptor [16], PACAP and PAR1 [17, 18] and a number of orphan receptors such as GPR22, 30 or 109 [19-21]. The assay system allows several advantages over existing methodologies. First, the robustness of the chemistry makes it suitable for most cell-based experimental procedures, allowing the direct measurement of the cell lysate in the presence of cell culture medium [8, 22, 23]. Second, these assays are capable of being highly miniaturized, making them particularly adaptable to HTS [22, 24]. Third, the cellular accumulation of secondary messengers has proven advantages over more transient readouts such as calcium flux for instance. As recently demonstrated, the identification of slow-acting compounds on Gq-coupled targets via IP1 assessment (unpublished data) confirm more general previous findings suggesting the potential for the IP-One assay to be used as an orthogonal screening method [25]. Finally, Jean et al. have recently shown that the sensitivity of these methodologies could enable the detection of these secondary messengers in rat native cerebral tissues [9]. In a recent communication, Trinquet et al. have also demonstrated how the detection of two secondary messenger could be multiplexed (unpublished data) thereby offering a way to detect simultaneously different signal mediations from the same GPCR. Taking advantage of the fact that the new Lumi4-Tb donor can be associated with different acceptors, and using V2 and M1 receptors, cAMP was run as a fluorescein assay while IP1 detection was performed with the standard d2 near-infrared dye. Both assays could be run and detected in the same well using the same cellular material and confirmed receptor responses for both pathways upon agonist stimulation.
The second field of application of HTRF methods was developed with the emergence of new concepts around receptor organization at the cell surface. Upon increasing evidence that a large number of receptors oligomerize at the cell surface to mediate intracellular signaling, researchers have started to develop methodologies that enable the detection of such rearrangements. These mechanistic studies have first been undertaken with antibodies specific to receptors or to encoded tags, and several groups have been able to investigate, among others, the way A1-A2 receptors assemble and permit adenosine to modulate glutamatergic neurotransmission [26], how BILF1 and chemokine receptor oligomerization may contribute to the alteration of the immune response [27], or mGlu and GABA receptors organization at the cell surface [28, 29]. We will further discuss the receptor oligomerization in the section of Protein-Protein/Protein-Peptide/Protein-DNA Interaction Assays. Because the use of conjugated antibodies is sometimes limiting, new labeling techniques based on suicide enzyme technologies like SNAP-tag were applied to address the same receptor mechanistics. Maurel et al. [30] have used specific HTRF fluorophore-labeled benzyl guanine substrates to covalently label SNAP-encoded GABA receptors and investigate their organization at the cell surface. These recent optimizations and findings preclude a new field of application for HTRF applied to receptor investigation, particularly for ligand binding and receptor trafficking.
Kinase Assays
HTRF technology has been applied to a number of enzymatic activity assays, such as kinases, proteases, ubiquitination, etc. There are two major formats for HTRF in these assays, a “sandwich” format to measure the signals generated from the status change of a labeled substrate, such as phosphorylation and cleavage by proteases; and a competition format to monitor the other products generated in the reactions, such as ADP production in the kinase assays (Fig. 6).
Fig. (6).
The comparison of “sandwich” assay and competition assay. a. “Sandwich” assay. HTRF signals are generated through the energy transfer from donor to acceptor that are labeled on the antibodies recognizing the target at different regions. b. Competition assay. Targets generated through biological events compete binding activity to the donor labeled antibody with the target labeled with acceptor so that the decrease of the HTRF signals are measured.
Serine/Threonine Kinase Assays
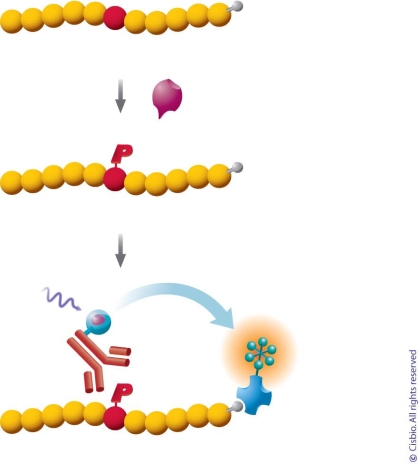
Protein kinases are key elements in intracellular signaling pathways that control many physiological processes and the activities of protein kinases are altered in a number of human diseases such as cancer and autoimmune disorders [31-33]. The rapidly growing interest in kinases as potential targets for therapeutic intervention has prompted the development of many kinase assay technologies. HTRF-based tools for HTS could apply to all kinases, with specific applications for Tyrosine (Tyr) and Serine/Threonine (Ser/Thr) kinases. This kinase screening platform offers the ability to use a single technology for both universal and specific kinase assays, saving valuable development time and cost (Fig. 7). A comprehensive review summarized the recent literatures and meeting reports on HTRF kinase assay [34].
Fig. (7).
Kinase assay principle. The substrates used in HTRF kinase assays are tagged by fusion proteins, peptides, or biotin. An antibody to these proteins or peptides, or streptavidin (for biotin in this case) is labeled with acceptor. An antibody against phosphorylated substrate is labeled by donor cryptate. In the enzymatic reaction, once substrate is phosphorylated, donor and acceptor can be brought into proximity by affinity interactions of antibodies/substrate or streptavidin/biotin.
HTRF KinEASE is a universal and sensitive assay allowing for profiling and HTS of a large range of Ser/Thr kinases. This platform consists of three different biotinylated substrates and a single proprietary monoclonal anti-phospho antibody labeled with Eu3+ cryptate recognizing only the phosphorylated substrates, and streptavidin-XL665. Each peptide has a common C-terminal phospho-serine epitope with three different N-terminal sequences allowing access to a large range of kinases. FRET signals are generated once the monoclonal antibody binds to the phosphorylated serine in the biotinylated peptide that in turn is bound to streptavidin-XL665. It is highly likely that many more kinases will phosphorylate one or more of the three peptides. Using this universal kinase activity detection tool and other more specific substrates, numerous assay systems have been successfully built to monitor protein kinase activity with purified enzymes, as well as with cellular lysates and membrane preparations, including COT [35], AKT [36], and Rock II [37]. Recently a cellular Ser/Thr-kinase platform was developed by co-expression of targeted kinases with a generic peptide substrates followed by quantification of phosphorylation on intracellular substrate by HTRF [38]. Although the development of this platform is still in the early stage, the proof-of principle assays were shown that activity of cellular AKT, B-RAF and CamK2δ could be detected and IC50 could be measured [38]. To date, this KinEASE screening platform has been validated on over 160 kinases.
Tyrosine Kinase Assays
Based on the same monoclonal antibody principle as HTRF KinEASE, HTRF KinEASE TK was developed to measure tyrosine kinase activity [39]. Compared to the well-known universal tyrosine kinase substrates, poly(GT) and poly (GAT), the TK substrate has shown vast flexibility in assay design without sacrificing assay performance and its single phosphorylation site makes kinetic determinations such as Km apparent easily investigated [40]. The application of HTRF on tyrosine kinases have been demonstrated in many reports, including Lck [41], FGF receptor [42], c-kit [43], c-Met [44], IGF-1R [45], KDR (kinase insert domain receptor (a type III receptor tyrosine kinase)) [46], and Tie2 [47]. So far, at least 59 unique tyrosine kinases have been validated by the HTRF KinEASE TK platform.
In contrast to the above sandwich format, HTRF Transcreener ADP is a universal and flexible assay for profiling and HTS of all kinases and ATPases using a competition format. This homogeneous, non-radioactive competition assay measures the generation of ADP (Adenosine 5’-diphosphate), which is directly detected by a specific anti-ADP antibody labeled with Eu3+ cryptate, and correlates tightly with the amount of phosphorylated substrate. Because the assay measures ADP and not the substrate itself, it is compatible with any substrate. The enzyme-free detection system also provides an alternative with less interference and a lower false positive rate than assays whose detection systems involve the use an enzyme to measure ATP depletion. The HTRF Transcreener ADP assay is particularly suitable when measuring difficult kinases for which no anti-phospho-specific antibody is available.
Other Enzyme Assays
HTRF has been used to measure the activity of heparanase [48]. Heparanase functions as an enzyme cleaving heparan sulfate proteoglycan to release growth factors and plays an important role in tissue invasion by malignant tumor cells and inflammatory cells. In this assay, biotinylated heparan sulfate proteoglycan labeled with Europium cryptate served as the substrate for heparanase. Heparanase activity resulting in substrate cleavage by murine melanoma cell extracts was detected by measuring the HTRF signal after addition of streptavidin-XL665. Subsequently, a rapid process for screening large numbers of samples can be established by the HTRF Heparanase assay.
Ubiquitination is one of the major post-translational modifications that proteins regularly undergo prior to degradation in the proteosome. The ubiquitin pathway is associated with many pathologies such as cancer, muscle atrophy, inflammation, viral infection, and metabolic and neurodegenerative disorders. Ubiquitination is carried out by the conjugation of ubiquitin to target proteins through the sequential activity of three enzymes: E1, E2 and E3. Poly-ubiquitinated proteins are then subject to degradation by proteasomes. In order to monitor p53 poly-ubiquitination, a HTS assay system using HTRF has been established [49]. In the ubiquitination reaction, Europium cryptate-labeled ubiquitin is incorporated as poly-ubiquitin chains onto the biotinylated p53 target protein. The assay is stopped by the addition of streptavidin-XL665 in a buffer containing EDTA before being read. The HTRF Ubiquitin assay is robust enough to be used in synthetic compound screening and has almost the same level of sensitivity as that monitored by the scintillation proximity assay (SPA) using 125I-labeled ubiquitin. The HTRF Ubiquitin assay was recently used as a research tool to measure self-ubiquitination of POSH (plenty of SH3s) [50].
Protein-Protein/Protein-Peptide/Protein-DNA Interaction Assays
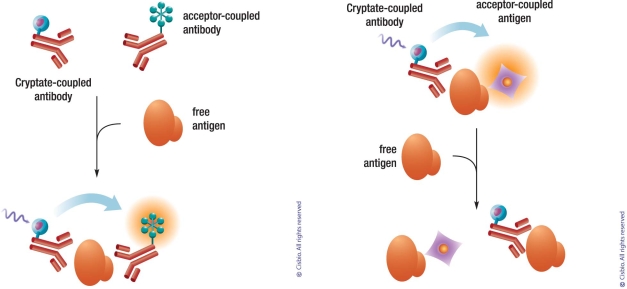
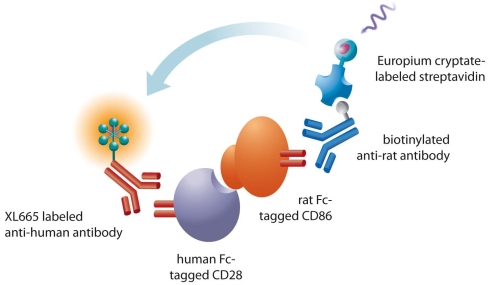
Protein-protein, protein-peptide, and protein/DNA interactions are involved at almost every level of cell function, in surface protein communication (e.g. GPCR dimerization, surface molecule interaction, and receptor-ligand binding), in signal transduction machinery across various biological membranes (e.g. G-protein cascades, kinase/substrate interactions), the regulation of gene expression (e.g. interaction of transcription factors with co-activators or co-repressors and transcription factors with DNA), the packaging of chromatin, etc. Several applications using HTRF in these interactions have been demonstrated to date. The general principle of these assays is very similar. A pair of antibodies against the target proteins or the tags that they are fused to is labeled with the HTRF donor and acceptor fluorophore, respectively. The interaction of target proteins brings the antibodies into close proximity, generating the FRET signal (Fig. 8).
Fig. (8).
Principle of protein-protein interaction. Antibodies against the desired proteins (CD28 and CD 86 as examples) for interaction or the fused tags on these proteins are labeled with donor and acceptor, respectively. The FRET signals are generated once these proteins interact with each other to bring the labeled antibodies in into proximity.
Cell Surface Protein Interaction
It is well established that membrane receptors may dimerize to regulate the activity of each member of the dimer or to work cooperatively to bind ligand on one receptor while mediating intracellular signaling through the other. In 2004, the Pin lab pioneered the work demonstrating the application of HTRF in studying dimerization of glutamate and γ-aminobutyric acid (GABA) receptors [51]. In this study, anti-tag (Flag, HA, and 6-His) antibodies labeled with donor Eu cryptate and acceptor AlexaFluor 647 or XL665 were used to generate fluorescent signals. The mechanism of this dimerization was further pursued by HTRF technology [29]. Recently, the application of HTRF on GPCR oligomerization and receptor-ligand binding was further expanded with SNAP-Tag technology as mentioned above [30] (and data not shown). The Pin lab later conducted a series of studies using HTRF to demonstrate the dimerization of other GPCRs, such as metabotropic glutamate-like receptors (mGluR) [28, 52], vasopressin and oxytocin receptors [53]. Ciruela et al. developed an HTRF assay to measure the heterodimerization of adenosine A1 receptors (A1Rs) and A2A receptors (A2ARs), revealing a more precise modulation of glutamatergic neurotransmission by adenosine [26]. The human hemagglutinin (HA)-A2AR and Flag-A1R constructs were generated and transiently transfected into HEK cells. The level of heterodimerization of the receptors was reflected through the FRET generated when Eu-labeled anti-Flag antibody and XL665-labeled anti-HA antibody were incubated with these transfected cells [26].
In addition to utilizing HTRF to study GPCR dimerization such as the dimerization of orphan GPR50 receptor with MT1 and MT2 melatonin receptors [54], many groups have used HTRF technology to study higher order oligomer formation of the Dopamine D2 receptor [55], oligomerization of human histamine H4 receptor [56], and to determine the function of C-terminal ends of mGluR1 in subunit association [57]. It has also been used to study chemokine receptors (reviewed by Wang and Norcross [58]) and the interaction of surface molecules, such as Trail receptor 4 and OX40 [59], CD80 and CD28 [60], and adhesive molecule Chemokine (C-X3-C motif) ligand 1 (CX3CL1) aggregation [61]. Moreover, the protein – peptide interaction was demonstrated with HTRF technology in cell surface calreticulin (CRT) and Shared Epitope (SE) from HLA-DR chain [62].
Viral Protein Interactions with Human Proteins
Viruses interact with protein machinery in intact cells to propagate in order to maintain functionality in infectious disease and oncology. For example, reverse transcription complex (RTC) is formed by viral genomic RNA associated with viral proteins (including RT) and, presumably, as yet uncharacterized cellular proteins from human immunodeficiency virus type 1 (HIV-1) infected cells. Lemay et al. reported an HTRF assay to confirm the yeast two-hybrid results that nucleocytoplasmic shuttling protein – the RNA-binding protein HuR is the cellular protein in RTC to interact with viral RT [63]. In 2002, Leblanc et al. showed that SV40-LTAg interacts with p53 by using the antibodies directly against them labeled by Eu cryptate and XL665, respectively [64]. Another HTRF assay showed that HIV integrase (IN) interacts with SNF5, a subunit of the SWI/SNF chromatin remodeling complex [65]. Interestingly, Epstein-Barr virus (EBV) BILF1 gene, which is a GPCR, has been shown with HTRF assays to form heterodimers with various chemokine receptors, altering the responsiveness of B lymphocytes to chemokines that is essential for EBV dissemination and/or involved in EBV-induced pathogenesis [27].
Protein/DNA Binding Assay
HTRF technology has also been useful in monitoring the interaction of protein/DNA, such as transcription factors binding with their regulatory elements on DNA or RNA. The MutS protein is part of the dam-directed MutHLS mismatch repair pathway in Escherichia coli. By using directly labeled MutS with europium cryptate, Lopez-Crapez et al. have developed an HTRF assay to measure interactions between MutS and mismatched DNA or DNA containing the most common lesion of the anticancer drug cisplatin [66].
Biomarker Detection
Monitoring the production of biomarkers is a useful tool for examining aberrant cell signaling processes often associated with diseases in therapeutic areas such as inflammation, metabolism and neuroscience. Understanding the relationship between the production of these downstream signaling products and the upstream effectors of signaling cascades is not only important for mapping the signaling pathways, but is also useful in identifying specific agents of therapeutic value for a particular disease state.
In this light, HTRF has proven a viable technology for constructing assays that can be used to examine the in-vivo production of biomarkers measured in either whole cells or cell supernatants, as well as serve as a platform for monitoring the in-vitro production of biomarkers via the enzymatic modifications of proteins. Such assays typically involve a sandwich or competition format.
Cell based assays employing HTRF to the direct measurement of protein biomarkers have allowed researchers to gain a better understanding of the biology behind signaling processes. Elucidation of signaling pathways and their regulatory mechanisms allows researchers to better identify targets for therapeutic intervention. By applying HTRF assays to directly measure IL-6 and IL-8 production by cells resulting from TNF stimulation, as well as stimulation by other immune stimuli, researchers were able to establish the essential functional role of a regulatory protein, called TMEM9B, in the production of these cytokines [67].
HTRF biomarker detection assays can also be used to monitor cellular responses to novel pharmacological substances and screen for potential therapeutic agents in many disease pathways. Two such biomarker detection assays include the measurement of insulin and cortisone. Insulin has been well established as a key regulator in glucose homeostasis. Deficiency of insulin leads to insulin-dependent (type 1) diabetes, whereas abnormal insulin secretion and resistance to insulin action is common in non-insulin-dependent (type 2) diabetes. An HTRF assay for the measurement of insulin has been developed that can be used to quantify both secreted and intracellular insulin. Such an assay has clear utility for the investigation of novel anti-diabetic agents. Researchers used HTRF to measure the insulin response of cells treated with various amounts glucose as well as an agent of potential therapeutic value to ascertain its effectiveness at promoting insulin production [68]. Other recently published applications of HTRF in cell-based assays have highlighted its utility for monitoring cortisol production in cell cultures. This application was used to characterize and identify inhibitors of the enzyme 11beta-Hydroxysteroid Dehydrogenase Type 1 (11β-HSD1). Inhibition of this target is believed to have therapeutic value in the treatment of type 2 diabetes as well as in metabolic syndrome. Cell-based assays directly measuring cortisol production have been used to monitor the activity of 11β-HSD1 in screening for inhibitors of the cellular enzyme [69,70].
Utility of these same HTRF biomarker assays has extended into the development of biochemical-based enzyme screens as well, including targets such as prostaglandin E2 synthase and 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1). Recently, the development of an assay to measure the activity of microsomal prostaglandin E2 synthase-1 (mPGES-1) was described in the literature. The authors used an HTRF assay to directly detect prostaglandin E2 formed by the enzyme. This assay proved useful in assessing the activity of the enzyme as well as determining the potency of inhibitors [71]. Previous methods used for measuring the activity of this enzyme have proven labor intensive. Also, an article was recently published that described a biochemical assay for measuring human 11β-HSD1 activity. In this instance, the authors have applied the HTRF detection of cortisol production to screening for inhibitors of the enzyme contained in human liver microsome fractions [72]. In another publication, researchers describe the use of the same cortisol measurement kit to monitor 11β-HSD1 activity in order to elucidate the relationship of the cofactor NADPH/NAD+ ratio to the reductase activity of the enzyme [73]. Understanding the enzyme and cofactor relationship is important for constructing screens to correctly measure for modulators of the different activities.
The assays described above highlight just some of the quantitative biomarker assays that have been developed using HTRF. Other examples of HTRF biomarker assays developed include: cGMP, histamine, IL-2, IL-17, estradiol, Leukotriene B4 (LTB4) and Amyloid β 1-40.
In addition to drug discovery applications, investigative applications of the quantification of biomarkers with HTRF have allowed researchers to examine the clinicopathology of diseases such as vascular dementia. By measuring the levels of Amyloid-β in brain samples from diseased individuals vs. age-matched controls, researchers were able to identify a link between the presence of these proteins and vascular dementia [74].
Bioprocess
In addition to its broad applications in investigation of new drug targets, HTRF shows other niches, such as monitoring the manufacturing of biotherapeutics to insure safety and quality. Antibodies are of interest in their potential for therapeutic use and a significant portion of new drugs stem from the biopharmaceutical industry. The compatibility of HTRF technology with cell culture media and other biological materials makes it applicable in the quantification of human antibodies and Fc fragments/human Fc-tagged chimeras. HTRF bioprocess assays are a highly sensitive and cost-effective alternative to automated systems and other methods used to screen for positive clones and high-producing clones during the antibody engineering and selection process [75]. The Human MAb assay is a competitive immunoassay for the measurement of all isotypes of human IgGs and Fc-tagged proteins. The assay utilizes an antibody specific to human Fc IgGs conjugated to europium cryptate and human IgG conjugated to XL665. The assay is run with a minimal number of additions: a sample of the antibody being assayed is titred into 96-, 384-, or 1536-well assay plates followed by the addition of detection reagents and then allowed to incubate at room temperature for 2 hours before being read.
Additionally, HTRF may be used to quantify and/or detect the presence of tagged protein during protein production and through each step of the purification process. The assay for GST- or 6HIS-tagged proteins is similar to that of the Human MAb assay. It is a competitive immunoassay in which an anti-tag antibody is coupled to europium cryptate and GST or 6HIS-peptide is coupled to XL665. The tagged fusion protein competes with the XL665 conjugate for binding to the anti-tag-cryptate conjugate and the signal is inversely proportional to the concentration of tagged fusion protein.
Most bioprocess ELISA assays can be easily converted to a no-wash, one-step HTRF assay. HTRF can be applied in several other ways to increase productivity in a bioprocess research facility, for instance, through monitoring cellular impurities which play a role in immunotoxicity [76].
CONCLUSION
HTRF is widely used in drug discovery and HTS. Even though this technology is extremely sensitive, versatile, and well adapted to automation for HTS, intrinsic limitations also exist, for example, quenching generated by external interactions with the intramolecular excitation process (electron transfer, FRET, and bleaching) and fluorescence of library compounds or biological proteins. It is our experience that the results obtained by using fluorescent techniques that do not use ratiometric measurement and/or time resolution should be carried out and validated with great care in order to avoid false positive or negative results.
Recently alternative methods were developed to measure the second messenger accumulation. However, classical HTRF applications on other cell signaling process such as phosphorylation, proteolysis, ubiquitination, and glycosylation, remain irreplaceable. The different photophysical properties of Terbium cryptate and its compatibility with not only red acceptors but also green acceptors open new doors to multiplexing with HTRF. Moreover, ligand-binding assays that are selective for cell-surface receptors run more efficiently with Terbium than Europium cryptates (unpublished data). Most recently, the introduction of Terbium cryptate and SNAP-Tag into the HTRF portfolio broadens HTRF applications, for instance, by allowing to study receptor dimerization, receptor mechanistics (internalization and dimerization), and by providing a non-radioactive alternative to ligand binding assays on whole, live cells [30].
Footnotes
Additional information for HTRF assays and other related reagents can be found on the web at www.HTRF.com.
Lumi4® is a trademark of Lumiphore Inc.
HTRF KinEASETM is developed with Millipore.
HTRF TranscreenerTM ADP assay is licensed from BellBrook Lab.
REFERENCES
- 1.Mathis G. Probing molecular interactions with homogeneous techniques based on rare earth cryptates and fluorescence energy transfer. Clin Chem. 1995;41(9):1391–7. [PubMed] [Google Scholar]
- 2.Selvin PR. Principles and biophysical applications of lanthanide-based probes. Annu Rev Biophys Biomol Struct. 2002;31:275–302. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 3.Mathis G. HTRF(R) Technology. J Biomol Screen. 1999;4(6):309–14. doi: 10.1177/108705719900400605. [DOI] [PubMed] [Google Scholar]
- 4.Bazin H, Trinquet E, Mathis G. Time resolved amplification of cryptate emission: a versatile technology to trace biomolecular interactions. J Biotechnol. 2002;82(3):233–50. doi: 10.1016/s1389-0352(01)00040-x. [DOI] [PubMed] [Google Scholar]
- 5.Alpha B, Ballardini R, Balzani V, Lehn JM, Perathoner S, Sabbatini N. Antenna effect in luminescent lanthanide cryptate: a photophysical study. Photochem Photobiol. 1990;52(2):299–306. [Google Scholar]
- 6.Mathis G. Rare earth cryptates and homogeneous fluoroim-munoassays with human sera. Clin Chem. 1993;39(9):1953–9. [PubMed] [Google Scholar]
- 7.Trinquet E, Fink M, Bazin H, et al. D-myo-inositol 1-phosphate as a surrogate of D-myo-inositol 1,4,5-tris phosphate to monitor G protein-coupled receptor activation. Anal Biochem. 2006;358(1):126–35. doi: 10.1016/j.ab.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Degorce F, Achard S, Preaudat M, et al. A new HTRF assay for the direct assessment of cyclic AMP in high throughput screening. SBS in St Louis, MO, USA. 2008.
- 9.Jean A, Conductier G, Manrique C, et al. Anorexia induced by activation of serotonin 5-HT4 receptors is mediated by increases in CART in the nucleus accumbens. Proc Natl Acad Sci USA. 2007;104(41):16335–40. doi: 10.1073/pnas.0701471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero G, Sanchez E, Pujol M, et al. Efficacy of selective 5-HT6 receptor ligands determined by monitoring 5-HT6 receptor-mediated cAMP signaling pathways. Br J Pharmacol. 2006;148(8):1133–43. doi: 10.1038/sj.bjp.0706827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peres CM, Aronoff DM, Serezani CH, Flamand N, Faccioli LH, Peters-Golden M. Specific leukotriene receptors couple to distinct G proteins to effect stimulation of alveolar macrophage host defense functions. J Immunol. 2007;179(8):5454–61. doi: 10.4049/jimmunol.179.8.5454. [DOI] [PubMed] [Google Scholar]
- 12.Cascio MG, Bisogno T, Palazzo E, et al. In vitro and in vivo pharmacology of synthetic olivetol- or resorcinol-derived cannabinoid receptor ligands. Br J Pharmacol. 2006;149(4):431–40. doi: 10.1038/sj.bjp.0706888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto W, Mikami T, Iwamura H. Involvement of central cannabinoid CB2 receptor in reducing mechanical allodynia in a mouse model of neuropathic pain. Eur J Pharmacol. 2008;583(1):56–61. doi: 10.1016/j.ejphar.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Pos Z, Wiener Z, Pocza P, et al. Histamine suppresses fibulin-5 and insulin-like growth factor-II receptor expression in melanoma. Cancer Res. 2008;68(6):1997–2005. doi: 10.1158/0008-5472.CAN-07-2816. [DOI] [PubMed] [Google Scholar]
- 15.Ito S, Yoshimoto R, Miyamoto Y, et al. Detailed pharmacological characterization of GT-2331 for the rat histamine H3 receptor. Eur J Pharmacol. 2006;529(1-3):40–6. doi: 10.1016/j.ejphar.2005.10.066. [DOI] [PubMed] [Google Scholar]
- 16.Hsiao EC, Boudignon BM, Chang WC, et al. Osteoblast expression of an engineered Gs-coupled receptor dramatically increases bone mass. Proc Natl Acad Sci USA. 2008;105(4):1209–14. doi: 10.1073/pnas.0707457105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delcourt N, Thouvenot E, Chanrion B, et al. PACAP type I receptor transactivation is essential for IGF-1 receptor signalling and antiapoptotic activity in neurons. EMBO J. 2007;26(6):1542–51. doi: 10.1038/sj.emboj.7601608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayoub MA, Maurel D, Binet V, et al. Real-time analysis of agonist-induced activation of protease-activated receptor 1/Galphai1 protein complex measured by bioluminescence resonance energy transfer in living cells. Mol Pharmacol. 2007;71(5):1329–40. doi: 10.1124/mol.106.030304. [DOI] [PubMed] [Google Scholar]
- 19.Adams JW, Wang J, Davis JR, et al. Myocardial expression, signaling, and function of GPR22: a protective role for an orphan G protein-coupled receptor. Am J Physiol Heart Circ Physiol. 2008;295(2):H509–21. doi: 10.1152/ajpheart.00368.2008. [DOI] [PubMed] [Google Scholar]
- 20.Otto C, Fuchs I, Kauselmann G, et al. GPR30 does not mediate estrogenic responses in reproductive organs in mice. Biol Reprod. 2009;80(1):34–41. doi: 10.1095/biolreprod.108.071175. [DOI] [PubMed] [Google Scholar]
- 21.Skinner PJ, Cherrier MC, Webb PJ, et al. Fluorinated pyrazole acids are agonists of the high affinity niacin receptor GPR109a. Bioorg Med Chem Lett. 2007;17(20):5620–3. doi: 10.1016/j.bmcl.2007.07.101. [DOI] [PubMed] [Google Scholar]
- 22.Bergsdorf C, Kropp-Goerkis C, Kaehler I, et al. A one-day, dispense-only IP-One HTRF assay for high-throughput screening of Galphaq protein-coupled receptors: towards cells as reagents. Assay Drug Dev Technol. 2008;6(1):39–53. doi: 10.1089/adt.2007.108. [DOI] [PubMed] [Google Scholar]
- 23.Amoravain M, Lyotard S, Jaga D, Lebreton ML, Servent F, Bomer U. Introduction of a new HTRF acceptor, d2: Development of a complete GPCR platform for a Gs, Gi and Gq screening. SBS; 11th Annual Conference; Geneva. 2005. [Google Scholar]
- 24.Liu K, Titus S, Southall N, et al. Comparison on functional assays for Gq-coupled GPCRs by measuring inositol monophospate and intracellular calcium in 1536-well plate format. Curr Chem Genomics. 2008;1:70–78. doi: 10.2174/1875397300801010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassutt KJ, Orsini MJ, Abousleiman M, Colone D, Tang W. Identifying nonselective hits from a homogeneous calcium assay screen. J Biomol Screen. 2007;12(2):285–7. doi: 10.1177/1087057106298538. [DOI] [PubMed] [Google Scholar]
- 26.Ciruela F, Casado V, Rodrigues RJ, et al. Presynaptic control of striatal glutamatergic neurotransmission by adenosine A1-A2A receptor heteromers. J Neurosci. 2006;26(7):2080–7. doi: 10.1523/JNEUROSCI.3574-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vischer HF, Nijmeijer S, Smit MJ, Leurs R. Viral hijacking of human receptors through heterodimerization. Biochem Biophys Res Commun. 2008;377(1):93–7. doi: 10.1016/j.bbrc.2008.09.082. [DOI] [PubMed] [Google Scholar]
- 28.Rondard P, Liu J, Huang S, et al. Coupling of agonist binding to effector domain activation in metabotropic glutamate-like receptors. J Biol Chem. 2006;281(34):24653–61. doi: 10.1074/jbc.M602277200. [DOI] [PubMed] [Google Scholar]
- 29.Rondard P, Huang S, Monnier C, et al. Functioning of the dimeric GABA(B) receptor extracellular domain revealed by glycan wedge scanning. EMBO J. 2008;27(9):1321–32. doi: 10.1038/emboj.2008.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurel D, Comps-Agrar L, Brock C, et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat Methods. 2008;5(6):561–7. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–65. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 32.Fantl WJ, Johnson DE, Williams LT. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–81. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 33.Edelman AM, Blumenthal DK, Krebs EG. Protein serine/threonine kinases. Annu Rev Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- 34.Jia Y. Current status of HTRF technology in kinase assays. Expert Opin Drug Discov. 2008;3(12):1461–74. doi: 10.1517/17460440802518171. [DOI] [PubMed] [Google Scholar]
- 35.Jia Y, Quinn CM, Clabbers A, et al. Comparative analysis of various in vitro COT kinase assay formats and their applications in inhibitor identification and characterization. Anal Biochem. 2006;350(2):268–76. doi: 10.1016/j.ab.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Jia Y, Quinn CM, Gagnon AI, Talanian R. Homogeneous time-resolved fluorescence and its applications for kinase assays in drug discovery. Anal Biochem. 2006;356(2):273–81. doi: 10.1016/j.ab.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Schroter T, Minond D, Weiser A, et al. Comparison of miniaturized time-resolved fluorescence resonance energy transfer and enzyme-coupled luciferase high-throughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II) J Biomol Screen. 2008;13(1):17–28. doi: 10.1177/1087057107310806. [DOI] [PubMed] [Google Scholar]
- 38.Adams DG, Wang Y, Mak PA, et al. Cellular Ser/Thr-kinase assays using generic peptide substrates. Curr Chem Genomics. 2008;1:54–64. doi: 10.2174/1875397300801010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tardieu J-L. HTRF KinEASE(TM) TK: a new solution for tyrosine kinase screening. Nat Methods. 2007;4:i–ii. [Google Scholar]
- 40.Harbert C, Marshall J, Soh S, Steger K. Development of a HTRF kinase assay for determination of syk activity. Curr Chem Genomics. 2008;1:20–6. doi: 10.2174/1875397300801010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbott L, Betschmann P, Burchat A, et al. Discovery of thienopyridines as Src-family selective Lck inhibitors. Bioorg Med Chem Lett. 2007;17(5):1167–71. doi: 10.1016/j.bmcl.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Kammasud N, Boonyarat C, Tsunoda S, et al. Novel inhibitor for fibroblast growth factor receptor tyrosine kinase. Bioorg Med Chem Lett. 2007;17(17):4812–8. doi: 10.1016/j.bmcl.2007.06.058. [DOI] [PubMed] [Google Scholar]
- 43.Gracias V, Ji Z, Akritopoulou-Zanze I, et al. Scaffold oriented synthesis. Part 2: design synthesis and biological evaluation of pyrimido-diazepines as receptor tyrosine kinase inhibitors. Bioorg Med Chem Lett. 2008;18(8):2691–5. doi: 10.1016/j.bmcl.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 44.Bellon SF, Kaplan-Lefko P, Yang Y, et al. c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations. J Biol Chem. 2008;283(5):2675–83. doi: 10.1074/jbc.M705774200. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Malkowski M, Hailey J, Turek-Etienne T, Tripodi T, Pachter JA. Screening for small molecule inhibitors of insulin-like growth factor receptor (IGF-1R) kinase: comparison of homogeneous time-resolved fluorescence and 33P-ATP plate assay formats. J Exp Ther Oncol. 2004;4(2):111–9. [PubMed] [Google Scholar]
- 46.Dominguez C, Smith L, Huang Q, et al. Discovery of N-phenyl nicotinamides as potent inhibitors of Kdr. Bioorg Med Chem Lett. 2007;17(21):6003–8. doi: 10.1016/j.bmcl.2007.07.077. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Lin TH, Cole AG, et al. Identification and characterization of small-molecule inhibitors of Tie2 kinase. FEBS Lett. 2008;582(5):785–91. doi: 10.1016/j.febslet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Enomoto K, Okamoto H, Numata Y, Takemoto H. A simple and rapid assay for heparanase activity using homogeneous time-resolved fluorescence. J Pharm Biomed Anal. 2006;41(3):912–7. doi: 10.1016/j.jpba.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 49.Yabuki N, Watanabe S, Kudoh T, Nihira S, Miyamato C. Application of homogeneous time-resolved fluorescence (HTRFTM) to monitor poly-ubiquitination of wild-type p53. Comb Chem High Throughput Screen. 1999;2(5):279–87. [PubMed] [Google Scholar]
- 50.Tuvia S, Taglicht D, Erez O, et al. The ubiquitin E3 ligase POSH regulates calcium homeostasis through spatial control of Herp. J Cell Biol. 2007;177(1):51–61. doi: 10.1083/jcb.200611036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurel D, Kniazeff J, Mathis G, Trinquet E, Pin JP, Ansanay H. Cell surface detection of membrane protein interaction with homogeneous time-resolved fluorescence resonance energy transfer technology. Anal Biochem. 2004;329(2):253–62. doi: 10.1016/j.ab.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Brock C, Oueslati N, Soler S, Boudier L, Rondard P, Pin JP. Activation of a dimeric metabotropic glutamate receptor by intersubunit rearrangement. J Biol Chem. 2007;282(45):33000–8. doi: 10.1074/jbc.M702542200. [DOI] [PubMed] [Google Scholar]
- 53.Albizu L, Balestre MN, Breton C, et al. Probing the existence of G protein-coupled receptor dimers by positive and negative ligand-dependent cooperative binding. Mol Pharmacol. 2006;70(5):1783–91. doi: 10.1124/mol.106.025684. [DOI] [PubMed] [Google Scholar]
- 54.Levoye A, Dam J, Ayoub MA, et al. The orphan GPR50 receptor specifically inhibits MT1 melatonin receptor function through hetero-dimerization. EMBO J. 2006;25(13):3012–23. doi: 10.1038/sj.emboj.7601193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo W, Urizar E, Kralikova M, Mobarec JC, Shi L, Filizola M, Javitch JA. Dopamine D2 receptors form higher order oligomers at physiological expression levels. EMBO J. 2008;27(17):2293–304. doi: 10.1038/emboj.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Rijn RM, Chazot PL, Shenton FC, Sansuk K, Bakker RA, Leurs R. Oligomerization of recombinant and endogenously expressed human histamine H(4) receptors. Mol Pharmacol. 2006;70(2):604–15. doi: 10.1124/mol.105.020818. [DOI] [PubMed] [Google Scholar]
- 57.Kumpost J, Syrova Z, Kulihova L, et al. Surface expression of metabotropic glutamate receptor variants mGluR1a and mGluR1b in transfected HEK293 cells. Neuropharmacology. 2008;55(4):409–18. doi: 10.1016/j.neuropharm.2008.06.073. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Norcross M. Dimerization of chemokine receptors in living cells: key to receptor function and novel targets for therapy. Drug Discov Today. 2008;13(13-14):625–32. doi: 10.1016/j.drudis.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Newton P, Harrison P, Clulow S. A novel method for determination of the affinity of protein: protein interactions in homogeneous assays. J Biomol Screen. 2008;13(7):674–82. doi: 10.1177/1087057108321086. [DOI] [PubMed] [Google Scholar]
- 60.Uvebrant K, da Graca Thrige D, Rosen A, et al. Discovery of selective small-molecule CD80 inhibitors. J Biomol Screen. 2007;12(4):464–72. doi: 10.1177/1087057107300464. [DOI] [PubMed] [Google Scholar]
- 61.Hermand P, Pincet F, Carvalho S, et al. Functional adhesiveness of the CX3CL1 chemokine requires its aggregation. Role of the transmembrane domain. J Biol Chem. 2008;283(44):30225–34. doi: 10.1074/jbc.M802638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling S, Pi X, Holoshitz J. The rheumatoid arthritis shared epitope triggers innate immune signaling via cell surface calreticulin. J Immunol. 2007;179(9):6359–67. doi: 10.4049/jimmunol.179.9.6359. [DOI] [PubMed] [Google Scholar]
- 63.Lemay J, Maidou-Peindara P, Bader T, Ennifar E, Rain JC, Benarous R, Liu LX. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leblanc V, Delaunay V, Claude Lelong J, et al. Homogeneous time-resolved fluorescence assay for identifying p53 interactions with its protein partners, directly in a cellular extract. Anal Biochem. 2002;308(2):247–54. doi: 10.1016/s0003-2697(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 65.Maroun M, Delelis O, Coadou G, et al. Inhibition of early steps of HIV-1 replication by SNF5/Ini1. J Biol Chem. 2006;281(32):22736–43. doi: 10.1074/jbc.M604849200. [DOI] [PubMed] [Google Scholar]
- 66.Lopez-Crapez E, Malinge JM, Gatchitch F, et al. A homogeneous resonance energy transfer-based assay to monitor MutS/DNA interactions. Anal Biochem. 2008;383(2):301–6. doi: 10.1016/j.ab.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 67.Dodeller F, Gottar M, Huesken D, Iourgenko V, Cenni B. The lysosomal transmembrane protein 9B regulates the activity of inflammatory signaling pathways. J Biol Chem. 2008;283(31):21487–94. doi: 10.1074/jbc.M801908200. [DOI] [PubMed] [Google Scholar]
- 68.Tousch D, Lajoix AD, Hosy E, et al. Chicoric acid, a new compound able to enhance insulin release and glucose uptake. Biochem Biophys Res Commun. 2008;377(1):131–5. doi: 10.1016/j.bbrc.2008.09.088. [DOI] [PubMed] [Google Scholar]
- 69.Vicker N, Su X, Ganeshapillai D, et al. Novel non-steroidal inhibitors of human 11beta-hydroxysteroid dehydrogenase type 1. J Steroid Biochem Mol Biol. 2007;104(3-5):123–9. doi: 10.1016/j.jsbmb.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 70.Wang DY, Lu Q, Walsh SL, et al. Development of a high-throughput cell-based assay for 11beta-hydroxysteroid dehydro-genase type 1 using BacMam technology. Mol Biotechnol. 2008;39(2):127–34. doi: 10.1007/s12033-008-9050-y. [DOI] [PubMed] [Google Scholar]
- 71.Goedken ER, Gagnon AI, Overmeyer GT, et al. HTRF-based assay for microsomal prostaglandin E2 synthase-1 activity. J Biomol Screen. 2008;13(7):619–25. doi: 10.1177/1087057108321145. [DOI] [PubMed] [Google Scholar]
- 72.Su X, Vicker N, Lawrence H, et al. Inhibition of human and rat 11beta-hydroxysteroid dehydro-genase type 1 by 18beta-glycyrrhetinic acid derivatives. J Steroid Biochem Mol Biol. 2007;104(3-5):312–20. doi: 10.1016/j.jsbmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 73.Dzyakanchuk AA, Balazs Z, Nashev LG, Amrein KE, Odermatt A. 11beta-Hydroxysteroid dehydrogenase 1 reductase activity is dependent on a high ratio of NADPH/NADP(+) and is stimulated by extracellular glucose. Mol Cell Endocrinol. 2009;301(1-2):137–41. doi: 10.1016/j.mce.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 74.Lewis H, Beher D, Cookson N, et al. Quantification of Alzheimer pathology in ageing and dementia: age-related accumulation of amyloid-beta(42) peptide in vascular dementia. Neuropathol Appl Neurobiol. 2006;32(2):103–18. doi: 10.1111/j.1365-2990.2006.00696.x. [DOI] [PubMed] [Google Scholar]
- 75.Idusogie EE, Castro JM, Casipit C, Sato A, Terasawa Y, Mulkerrin MG. Development of an antibody screening assay for selection of production cell lines. BioProcess Int. 2008;6(4):20–33. [Google Scholar]
- 76.Potera C. Methodology garners researchers' interest. Genet Eng Biotechnol News. 2008;28(20):15–6. [Google Scholar]