Abstract
The formation and maintenance of prions in the yeast Saccharomyces cerevisiae is highly regulated by the cellular chaperone machinery. The most important player in this regulation is Hsp104p, which is required for the maintenance of all known prions. The requirements for other chaperones, such as members of the Hsp40 or Hsp70 families, vary with each individual prion. [RNQ+] cells do not have a phenotype that is amenable to genetic screens to identify cellular factors important in prion propagation. Therefore, we used a chimeric construct that reports the [RNQ+] status of cells to perform a screen for mutants that are unable to maintain [RNQ+]. We found eight separate mutations in Hsp104p that caused [RNQ+] cells to become [rnq−]. These mutations also caused the loss of the [PSI+] prion. The expression of one of these mutants, Hsp104p-E190K, showed differential loss of the [RNQ+] and [PSI+] prions in the presence of wild type Hsp104p. Hsp104p-E190K inefficiently propagated [RNQ+] and was unable to maintain [PSI+]. The mutant was unable to act on other in vivo substrates, as strains carrying it were not thermotolerant. Purified recombinant Hsp104p-E190K showed a reduced level of ATP hydrolysis as compared to wild type protein. This is likely the cause of both prion loss and lack of in vivo function. Furthermore, it suggests that [RNQ+] requires less Hsp104p activity to maintain transmissible protein aggregates than Sup35p. Additionally, we show that the L94A mutation in Rnq1p, which reduces its interaction with Sis1p, prevents Rnq1p from maintaining a prion and inducing [PSI+].
Key words: [RNQ+], [PSI+], Hsp104p, Sis1p, mutagenesis
Introduction
Prions are proteins that can form a self-propagating aggregated state. In mammals, prions are a causative agent of neurodegenerative disease.1 In yeast, however, prions act as an epigenetic mode of inheritance. The prion-forming proteins in yeast are involved in a variety of cellular processes, including nitrogen catabolism, translation termination and chromatin modification.2–4 One of the yeast prions, [PSI+], provides a potential mechanism to respond to environmental stresses.5–8 Interestingly, [PSI+] is regulated by another prion, [RNQ+], whose presence increases the rate of [PSI+] appearance.9–12 However, [RNQ+] is not required for the maintenance of [PSI+]. Thus, the maintenance of [RNQ+] is critical for the formation of [PSI+] and its adaptive effect on the cells.
The maintenance of yeast prions is regulated by the cellular chaperone machinery.13,14 A crucial player in this regulation is the AAA+ ATPase Hsp104p that is essential for the survival of acute heat shock.15,16 Hsp104p is a disaggregase and has been shown to dissociate aggregated proteins both in vivo and in vitro.15,17,18 Like its bacterial homolog ClpB, the active form of Hsp104p is a ring-shaped hexamer with a pore in the center.19 Interestingly, Hsp104p can be divided up into five functionally distinct domains. Although the N terminus is generally dispensable for function, the amino and carboxy termini are likely involved in substrate binding.20,21 The two nucleotide binding domains (NBDs) coordinate to hydrolyze ATP to ADP and this enzymatic activity is required for Hsp104p function in prion maintenance.22–25 Finally, the M domain lies between the two NBDs and is thought to coordinate the activities of the NBDs by propagating a conformational change in response to binding or hydrolysis of ATP.23,26,27 Due to the discrete nature of the domains, a variety of mutations have been described in each domain of Hsp104p that affect prion propagation.20,25,28–31
One model of Hsp104p function in prion biology posits that Hsp104p has two roles in prion maintenance: generation of transmissible material (seeds) from prion aggregates and conversion of monomer into a prion-competent form.17,32–34 A complete loss of Hsp104p activity eliminates (cures) all known yeast prions.3,4,11,25,35 However, increasing Hsp104p activity by overexpressing the disaggregase cures only the [PSI+] prion and does not affect [RNQ+] or any other yeast prion.3,4,9,11,25,35 This indicates that Hsp104p has both general effects on all known yeast prions and specific effects on the [PSI+] prion, at least when overexpressed.
Other members of the cellular chaperone machinery also show differential effects on prion propagation. For example, overexpression of the Hsp70 family member Ssa1p cures [URE3] but not [PSI+].36 Excess Ssa1p does, however, reduce the curing of [PSI+] by Hsp104p overexpression and promote [PSI+] formation.37,38 Overexpression of the Hsp40 member Ydj1p cures the [URE3] prion but affects only certain variants of [PSI+] and [RNQ+].35,39,40 Deletion of either of two different Ssa1p nucleotide exchange factors, SSE1 or FES1, cures [URE3] but not [PSI+], though the deletion of SSE1 weakens [PSI+]-mediated nonsense suppression.41,42 Moreover, Hsf1p, which regulates the expression of heat shock proteins, influences [PSI+] formation. Hsf1p contains two activation domains and deletion of the N-terminal activation domain inhibits [PSI+] formation while deletion of the C-terminal activation domain promotes [PSI+] formation.43 This suggests two classes of interaction with [PSI+] by proteins regulated by Hsf1p.
Rnq1p has been shown to interact with both Ssa1p and the Hsp40 family member Sis1p.44,45 The propagation of [RNQ+] can be abolished by mutations within Sis1p.33,44,46,47 Sis1p depletion cures [RNQ+], [PSI+] and [URE3], albeit with varying efficiency, which may indicate different requirements for its activity in prion maintenance.48
Here, we describe a screen for cellular factors that affect the propagation of the [RNQ+] prion. Using a chimeric reporter for the [RNQ+] status of the cell, we found several novel alleles of HSP104 that are unable to propagate both [RNQ+] and [PSI+]. Interestingly, one of the alleles shows differential rates of curing of [RNQ+] and [PSI+] in the presence of wild type Hsp104. Additionally, we show that Rnq1p-L94A, which has a decreased interaction with Sis1p, aggregates non-specifically and cannot support [PSI+] induction.
Results
EMS mutagenesis reveals genes necessary for the maintenance of [RNQ+].
We developed a system to identify cellular factors important in the propagation of [RNQ+]. The phenotypes associated with [RNQ+], insolubility of the Rnq1 protein and an increase in the induction rate of [PSI+], are not amenable for use in a high-throughput screen. Therefore, we used the RRP reporter to assay the [RNQ+] status of the cells. As described previously,50 RRP consists of a fusion of the Rnq1p prion forming domain (PrD) and the C-terminal translation termination domain of Sup35p. In [rnq−] cells, RRP is soluble and able to promote faithful termination of translation. In [RNQ+] cells, RRP aggregates and is unable to promote faithful translation termination. We used a strain that has the ade1-14 allele for a sensitive readout of nonsense suppression. This allele harbors a premature stop codon that is read through when RRP is aggregated. Therefore, [rnq−] cells expressing RRP are adenine auxotrophs and appear red on rich medium (YPD) due to the accumulation of an intermediate in the adenine biosynthesis pathway. [RNQ+] cells expressing RRP are adenine prototrophs and appear light pink on YPD.
We used EMS to create an unbiased set of mutants that are unable to maintain [RNQ+]. A strain that was light pink due to expression of RRP in a [RNQ+] background was mutagenized with EMS to approximately 14% viability and plated on YPD. Following plating on YPD, some of the colonies turned red, indicating a deficiency in adenine biosynthesis, possibly as a consequence of the loss of RRP-associated nonsense suppression (Fig. 1A). Of the approximately 150,000 colonies screened, 312 colonies turned red on YPD. These 312 colonies were re-plated onto YPD to determine if the red phenotype was stable. 40 of these colonies remained red following two passages on YPD, indicating a stable phenotype.
Figure 1.
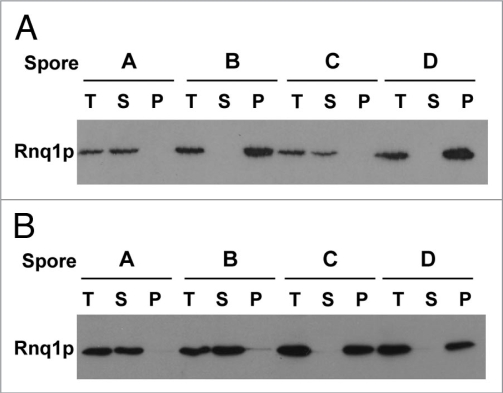
RRP reporter reveals [rnq−] cells after EMS mutagenesis. (A) [RNQ+] cells carrying the RRP reporter were mutagenized with EMS to approximately 14% viability and plated on YPD to assess color nonsense suppression of ade1-14. Left panel shows colonies prior to EMS treatment while right panel shows colonies after EMS treatment. (B) Solubility of Rnq1p in mutagenized cells. Following EMS mutagenesis, the red colonies were lysed and the lysate was fractionated into soluble and insoluble components by ultracentrifugation. The total (T), soluble (S) and insoluble pellet (P) were subjected to SDS-PAGE, transferred to PVDF and probed with an anti-Rnq1p antibody. The strain is indicated above the blot with [RNQ+] and [rnq−] controls shown on the left. An example of a mutagenized strain that remained [RNQ+], Red4, and one that converted to [rnq−], Red8, are shown.
The red phenotype created by the EMS mutagenesis could result not only from the loss of the [RNQ+] prion but also from a variety of different mutations affecting adenine biosynthesis or translation termination. Therefore, we wanted to select those red mutants that had, in fact, converted from [RNQ+] to [rnq−]. The solubility of the Rnq1 protein was analyzed by western blot following high speed centrifugation of cell lysates. In [rnq−] cells, Rnq1p remains in the soluble fraction while in [RNQ+] cells Rnq1p is found in the insoluble fraction. The solubility of Rnq1p in the 40 stable red colonies was assayed. Of these 40 mutants, 14 showed soluble Rnq1p, indicating that [RNQ+] had been cured (Fig. 1B and data not shown).
Genetic linkage indicates the [rnq−] phenotype is caused by single mutations.
Next, we assessed the genetic properties of the red mutants. We mated the 14 [rnq−] mutants to a wild type [RNQ+] strain. We then assayed the solubility of Rnq1p in the resulting diploids as described above. Three of the diploids showed soluble Rnq1p, indicating that the diploids were [rnq−] and that the mutant phenotype was dominant over wild type (data not shown). The remaining eleven mutants had insoluble Rnq1p in the diploids, indicating that [RNQ+] was propagating in the cells and that the mutations were recessive (data not shown).
To determine if the loss of [RNQ+] was caused by a single genetic lesion we employed a genetic test. The [RNQ+] diploids generated above were sporulated to obtain haploid progeny. Single mutations should segregate in a 2:2 ratio and produce two [rnq−] cells and two [RNQ+] cells. Multiple mutations would create a variety of ratios due to random segregation. The haploid progeny acquired from the sporulation of the recessive mutants were analyzed for their [RNQ+] status using the solubility assay described above. Eight of the recessive mutants generated the 2:2 ratio of [rnq−] to [RNQ+] cells, indicating a single mutation (Fig. 2A and data not shown). The remaining three mutants showed other ratios of [RNQ+] to [rnq−] haploid progeny, suggesting that multiple mutations had possibly been acquired.
Figure 2.
Curing of the [RNQ+] prion is caused by mutation at a single genetic locus. (A) Biochemical analysis of recessive mutations that cure [RNQ+]. The red [rnq−] cells carrying recessive mutations created by EMS were mated to a strain carrying wild type Rnq1p in the [RNQ+] state. The resulting diploids were sporulated and the solubility of the Rnq1 protein from individual haploids was analyzed as described in the legend of Figure 1. A single representative tetrad is shown. A minimum of eight tetrads were tested for each mutant. (B) Analysis of dominant mutation that cure [RNQ+]. The red [rnq−] cells carrying dominant mutations created by EMS were mated to a strain carrying wild type Rnq1p in the [RNQ+] state. The resulting diploid was sporulated and haploid progeny of this cross were then backcrossed to strains carrying wild type Rnq1p in the [RNQ+] state. The resulting diploids from these matings were lysed and fractionated as above. Three tetrads were tested and a single representative tetrad is shown.
Due to the ability of the three dominant mutations to cure [RNQ+] in the diploid, all four haploid progeny will be [rnq−], regardless of whether the phenotype is caused by one or more loci. However, the haploids can then be backcrossed to a wild type, [RNQ+], haploid strain and the [RNQ+] status of the resulting diploids analyzed. Single gene traits should produce a 2:2 ratio of [RNQ+] diploids to [rnq−] diploids. The haploid progeny from the dominant mutants were analyzed in this manner. Only one of the dominant mutations produced the 2:2 ratio indicative of a single gene trait (Fig. 2B). One of the dominant mutants failed to sporulate and, therefore, could not be analyzed in this manner, while the third produced variable ratios indicative of a multi-locus trait. We chose to focus on the mutants whose phenotypes were caused by alteration of a single locus.
Missense mutations in HSP104 cure [RNQ+].
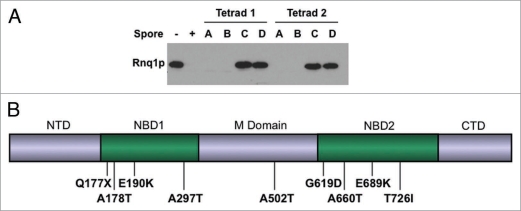
We set out to identify the specific mutations that cured the [RNQ+] prion. Since inactivation of the cellular disaggregase Hsp104p is known to cure all yeast prions it was an obvious candidate for our mutants.3,4,11,25,35 To determine if our mutants were in Hsp104p, we deleted the HSP104 gene in each of our mutants. The resulting Δhsp104 mutants were then mated to a [RNQ+] strain. These diploids were sporulated and the [RNQ+] status of the haploid progeny was assessed by a Rnq1p well-trap assay (Fig. 3A and data not shown). For this assay, lysates from the haploid cells were resuspended in sample buffer containing 1% SDS and incubated at room-temperature. Insoluble Rnq1p in [RNQ+] strains resists solubilization by SDS and does not enter the separating gel when subjected to electrophoresis, but soluble Rnq1p enters the gel and can be detected by western blot. All of the HSP104 deletions produced a 2:2 ratio of [RNQ+] to [rnq−] haploids, indicating that the mutations caused by EMS were in HSP104.
Figure 3.
The EMS-induced mutations that cure [RNQ+] are in HSP104. (A) HSP104 was deleted in the mutant strains as indicated in the methods. The resulting Δhsp104 strains were mated to a strain carrying wild type Rnq1p in the [RNQ+] state. The resulting diploids were sporulated and the solubility of Rnq1p from individual haploids determined by well trap assay as described in Materials and Methods, transferred to PVDF and probed with an anti-Rnq1p antibody. Two representative tetrads are shown. A minimum of four tetrads were tested for each mutant. (B) Diagram of mutations in Hsp104p that cure [RNQ+]. The general domain structure of Hsp104p is indicated above the diagram. NTD, N-terminal domain; NBD1, nucleotide binding domain one; M domain, middle domain; NBD2, nucleotide binding domain two; CTD, C-terminal domain. Mutations found in strains cured of [RNQ+] are indicated below the diagram.
We sequenced HSP104 from each of the mutant strains and found that eight of the single gene mutants carried a single missense mutation in the gene while one carried a nonsense mutation (Fig. 3B). The missense mutations found were as follows: A178T, E190K, A297T, A502T, G619D, A660T, E689K and T726I. The G619D mutant was dominant for the curing of [RNQ+]. Of these mutations, T726I, has been previously described.26 Additionally, a different mutation in G619, G619V, has also been described and both T726I and G619V have been shown to affect prion maintenance.26,28,54 The other mutations found in this screen are novel.
HSP104 missense mutations cure [PSI+].
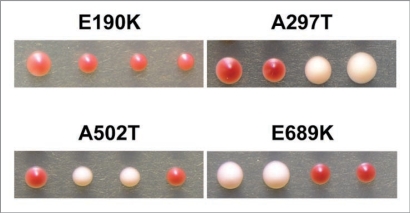
Because Hsp104p is also required for the propagation of [PSI+], we tested whether the mutations also affected the [PSI+] prion. In order to isolate haploids containing both the HSP104 mutation and SUP35, we mated the original mutant strain which contained the mutant HSP104 and RRP to a [RNQ+] strain that contained wild type HSP104 and SUP35. We used western blot analysis to identify spores containing wild type Sup35p that had become [rnq−] due to the presence of the mutant HSP104 (data not shown). RRP is recognized by polyclonal antibodies raised against Sup35p and can easily be distinguished from wild type Sup35p by its larger size. The [rnq−] strains carrying wild type Sup35p will also carry the HSP104 mutation. We then backcrossed these strains to a [PSI+] [RNQ+] strain. We dissected the resulting diploids and analyzed the degree of nonsense suppression of their haploid progeny in order to determine their [PSI+] status (Fig. 4 and data not shown). Each of the mutants that cured [RNQ+] was also able to cure [PSI+], indicating that the mechanism of curing by the mutants is not specific to [RNQ+] but likely affects prion propagation in general. Surprisingly, Hsp104p-E190K, which was recessive for the curing of [RNQ+], appeared to be dominant for the curing of [PSI+] (Fig. 4). All of the other mutations showed the same dominant or recessive effects on the curing of both [RNQ+] and [PSI+].
Figure 4.
The identified mutations in Hsp104p cure [PSI+]. Haploid [rnq−] progeny from a cross between the mutant strains and a wild type strain (SUP35) were mated to a wild type [PSI+] strain. The resulting diploids were sporulated and the haploid progeny plated on YPD. Representative tetrads from four mutants are shown. Hsp104-E689K shows a 2:2 inheritance of [RNQ+] to [rnq−] colonies which is indicative of a recessive trait. Hsp104p-A297T and Hsp104p-A502T also show a 2:2 inheritance. Hsp104p-E190K shows a 0:4 inheritance of [RNQ+] to [rnq−] colonies which indicates a dominant trait. A minimum of eight tetrads were analyzed for each mutant.
Hsp104p-E190K cures [RNQ+] slowly in the presence of wild-type Hsp104p.
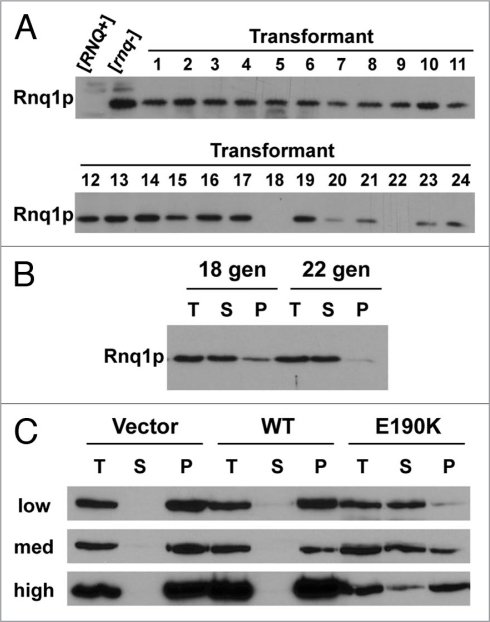
To further investigate the E190K mutation, the mutant strain was recreated by the pop-in/pop-out method in an unmutagenized version of 74-D694. Interestingly, initial results indicated that HSP104-E190K was dominant for curing [PSI+] but recessive for curing [RNQ+]. Further analysis, however, suggested that [RNQ+] was cured in a dominant fashion as well. We hypothesized that Hsp104p-E190K was less efficient at curing [RNQ+] than [PSI+] in the presence of wild type Hsp104p. Two other mutations of Hsp104, P557L and L462R, are known to cure [PSI+] but not [RNQ+].29 Of these two, Hsp104p-L462R looked cured by the lack of Rnq1p-GFP fluorescent foci but was confirmed [RNQ+] by SDD-AGE and mating to [rnq−]. Therefore, Hsp104p-L462R was suggested to weaken or destabilize [RNQ+] but not cure it.29 To determine if Hsp104p-E190K was similar to either of these two mutants, we transformed haploid [RNQ+] cells expressing wild type Rnq1p with a plasmid that expresses Hsp104p Hsp104p-E190K. We then tested the transformants for the [RNQ+] prion by well-trap assay. Of the 24 transformants tested, all but two had become [rnq−] (Fig. 5A). The two remaining [RNQ+] transformants were grown in selective medium for 22 generations and then retested for soluble Rnq1p (Fig. 5B). Both had become [rnq−]. This is consistent with our hypothesis that Hsp104-E190K shows inefficient curing of [RNQ+] in the presence of wild type Hsp104p.
Figure 5.
E190K cures [RNQ+] slowly in the presence of wild type Hsp104p. (A) Well-trap assays of cells containing both wild type Hsp104p and the E190K mutant. A strain carrying wild type Hsp104p was transformed with a plasmid expressing the E190K mutant. Lysate from individual transformants was subjected to a well trap assay as detailed in Materials and Methods, transferred to PVDF membrane, and blotted with an anti-Rnq1p antibody. (B) Strains carrying both wild type Hsp104p and the Hsp104p-E190K mutant lose [RNQ+] over time. The strains from (A) that remained [RNQ+] were grown in selective media for the number of generations indicated (# gen), lysed, and the [RNQ+] status was determined by [RNQ+] solubility assay as described in Materials and Methods. (C) The rate of [RNQ+] curing differs among [PIN+] prion strain variants. The strain variants of [RNQ+] (high, med, low [PIN+]) which have endogenous wild type Hsp104p were transformed with an empty plasmid (Vector) or a plasmid containing either wild type Hsp104p (WT) or Hsp104p-E190K (E190K). Transformants were scraped off plates, lysed and subjected to a full solubility assay as described in Materials and Methods, transferred to PVDF membrane and blotted with an anti-Rnq1p antibody. Three separate transformations were analyzed and all showed similar results. Total, T; soluble, S; insoluble pellet, P.
In order to gain more insight into the slow curing of [RNQ+] by Hsp104p-E190K, we looked at the effect of Hsp104p-E190K on prion strain variants of [RNQ+]. Three strain variants of [RNQ+] have been identified that induce [PSI+] with differing frequencies.40 The three [RNQ+] variants we used were previously characterized as low, medium and high [PIN+] ([PSI+] inducibility) and are so named for their increasing ability to induce [PSI+] in the presence of moderate overexpression of Sup35p. We transformed haploid cells of low, medium and high [PIN+] with a plasmid expressing Hsp104p, Hsp104p-E190K, or a vector control. We then immediately analyzed the transformants that grew for the state of [RNQ+] by a solubility assay (Fig. 5C). Contrary to a previous report that showed differences in the amount of soluble Rnq1p in these variants, our vector control revealed that all three variants had only insoluble Rnq1p.40 This discrepancy could be due to the difference in the solubility assays with which Rnq1p was measured but our assay consistently shows no soluble Rnq1 protein in the lysates from these strain variants. As expected, overexpression of Hsp104p did not cure [RNQ+] in any of the variants. Interestingly, the expression of Hsp104p-E190K in the presence of wild type Hsp104p resulted in significant Rnq1 protein in the soluble pools of the three variants. In the low [PIN+] strain, the majority of the protein had shifted to the soluble pool while in the high [PIN+] strain, the majority of Rnq1p stayed in the insoluble fraction. In the medium [PIN+] strain, the distribution between soluble and insoluble was approximately equal. Thus, the efficiency of [RNQ+] curing by Hsp104p-E190K is dependent on the strain variant of [RNQ+].
Hsp104p-E190K does not support thermotolerance.
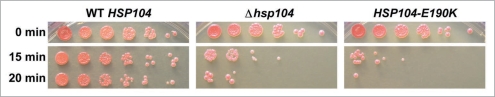
Since our data suggest that there is a difference in handling various substrates by Hsp104p-E190K, we next asked whether the mutant was defective in recognition and processing of other substrates in vivo. As Hsp104p is required for resistance to lethal heat shock, we tested whether the strain carrying the Hsp104p-E190K mutant was thermotolerant. Strains expressing either wild type Hsp104p, Hsp104p-E190K, or no Hsp104p at all were first exposed to a sublethal heat stress (37°C) to induce HSP104 expression and then exposed to a lethal heat stress (50°C). Strains expressing wild type Hsp104p were able to recover from a 20 minute heat shock while the Δhsp104 strains showed significant cell death (Fig. 6). The strain expressing Hsp104p-E190K was also unable to survive lethal heat stress and appeared similar to the deletion (Fig. 6). This indicates that the in vivo function of Hsp104p in resolubilizing essential proteins aggregated by heat stress is compromised by the E190K mutation.
Figure 6.
Hsp104p-E190K does not allow for the yeast to recover from acute heat shock. Following 30 minutes of pre-treatment at 37°C, liquid cultures containing identical numbers of cells of the indicated genotypes were heat-shocked at 50°C for the time indicated. Following heat shock, the cells were serially diluted and spotted onto YPD.
E190K forms hexamers but has a defect in ATP hydrolysis.
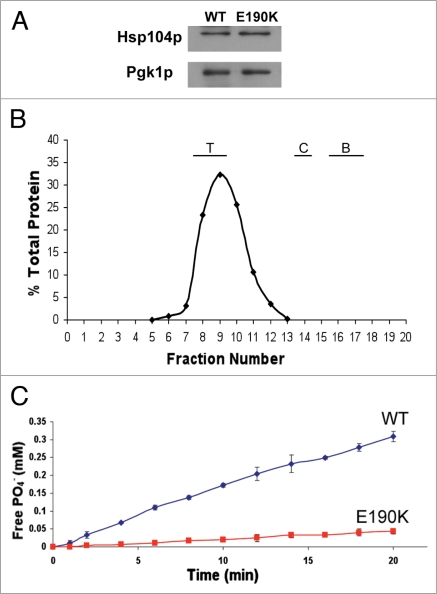
As the maintenance of prions is closely tied to the levels of Hsp104p, we wanted to determine if the steady state expression of the Hsp104p-E190K mutant is similar to that of wild type. Lysates from logarithmically growing cells were analyzed by western blot using an antibody against Hsp104p. Hsp104p-E190K was expressed to a similar level as wild type Hsp104p (Fig. 7A).
Figure 7.
Hsp104-E190K is deficient in hydrolyzing ATP but not in forming hexamers. (A) Expression analysis of Hsp104p-E190K. Lysate from strains grown to an OD600 of 1.0 were normalized by Bradford analysis, subjected to SDS-PAGE, transferred to PVDF and blotted with an anti-Hsp104p antibody. Equal loading was confirmed by probing the same blot with anti-Pgk1p antibody. (B) Hsp104p-E190K forms hexamers. Recombinant purified Hsp104-E190K was run through a Sephacryl S-300 column in the presence of ATP. Fractions were collected and analyzed by SDS-PAGE followed by western blot with anti-Hsp104p and the bands quantified by densitometry. The percent of total protein in each fraction was calculated. The positions of eluted standards are indicated above the graph. The molecular weight of the standards is thyroglobulin-670 kDa (T), catalase-250 kDa (C), bovine serum albumin-66 kDa (B). (C) ATP hydrolysis by Hsp104p-E190K. The assays were performed in buffer A at 37°C. At various time points the amount of free Pi in the reaction of either Hsp104p (circles) or Hsp104p-E190K (squares) was calculated as compared to a standard of KH2PO4 concentrations.
To determine the reason why Hsp104p-E190K is unable to support prion propagation and is defective in protecting the cells from heat stress, we examined two biochemical properties of Hsp104p, hexamer formation and ATPase activity. To test the ability of Hsp104p-E190K to form hexamers, we expressed and purified recombinant wild type Hsp104p and Hsp104p-E190K from E. coli. Purified protein was analyzed by size exclusion chromatography under conditions that promote hexamer formation.54 The mutant protein eluted in a similar volume as both the wild type Hsp104 protein and a 670 kDa standard, indicating that under these conditions, the mutant is able to form hexamers (Fig. 7B and data not shown).
Given that Hsp104p-E190K forms hexamers, we wanted to see whether it is able to hydrolyze ATP as efficiently as the wild type protein. To test this, the purified protein was incubated with a fixed concentration of ATP. At various time points, the amount of free Pi produced by the hydrolysis of ATP to ADP was measured. While Hsp104p-E190K did show hydrolysis of ATP, it did not hydrolyze ATP as well as wild type Hsp104p (Fig. 7C). This provides a likely explanation for its defect in both thermotolerance and prion propagation.
Rnq1p-L94A is unable to propagate the [RNQ+] prion.
Our screen uncovered novel mutations in Hsp104p but failed to uncover other novel factors required for the maintenance of the [RNQ+] prion. Had our screen been to saturation, we would have predicted it to reveal point mutations in SIS1, which is required for [RNQ+] propagation, as well as mutations in RNQ1 itself that abolish prion propagation. The domains of Sis1p required for the propagation of [RNQ+] have been described.44 We recently performed a screen for mutations in Rnq1p that disrupt prion propagation with the same RRP reporter and [RNQ+] dependent phenotype used here.50 Interestingly, we identified mutations that allow [RNQ+] to propagate but affect the ability of [RNQ+] to induce [PSI+]. None of those mutations affected the interaction with Sis1p, however, as detected by co-immunoprecipitation (Bardill JP and True HL, unpublished data). Recently, one binding site for Sis1p on Rnq1p was identified and a mutation in Rnq1p that severely reduces the interaction with Sis1p has been described.58 We created this mutation in Rnq1p, L94A, to ask whether the alteration in the interaction with Sis1p at this site would affect the propagation of the [RNQ+] prion.
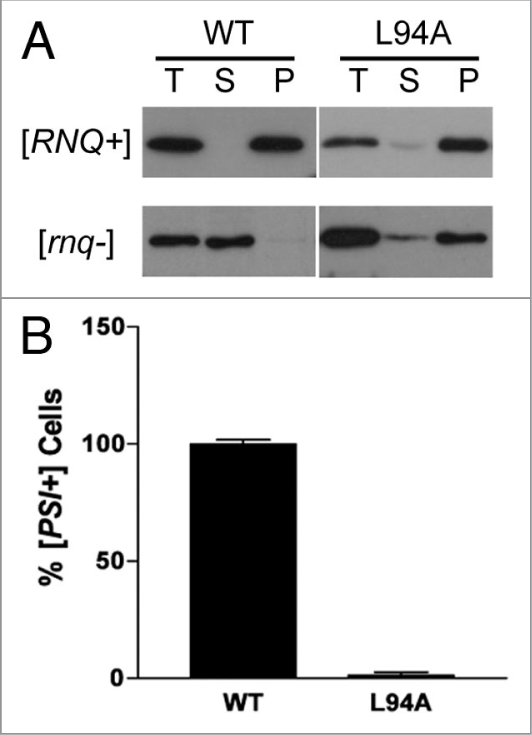
We created the L94A mutation in a Rnq1p-expressing plasmid and analyzed its ability to maintain the [RNQ+] prion. Expression from the native RNQ1 promoter was undetectable by western blot, so we used a GPD promoter which produced Rnq1p-L94A at steady state levels approximately equal to wild type Rnq1p (data not shown). This plasmid was used to replace wild type Rnq1p by plasmid shuffle in either a [RNQ+] or [rnq−] cell. The solubility of the Rnq1p-L94A mutant was then assayed. Wild type Rnq1p fractionates into either the insoluble or soluble fraction when shuffled through a [RNQ+] or [rnq−] strain, respectively (Fig. 8A). Rnq1p-L94A also fractionated into the insoluble fraction when shuffled through a [RNQ+] strain. However, the mutant was also insoluble when shuffled through a [rnq−] strain, indicating that the aggregation of Rnq1p-L94A is non-specific (Fig. 8A). Furthermore, we crossed the strain carrying Rnq1p-L94A to a [rnq−] strain and found that the haploid progeny carrying the wild type RNQ1 remained [rnq−] (data not shown). This suggests that the aggregates from the mutant L94A strain were unable to transmit the aggregate structure and are thus not prion-like. We also tested the ability of Rnq1p-L94A to induce [PSI+]. Following overexpression of Sup35p, cells containing Rnq1p-L94A showed a severe reduction in the ability to induce [PSI+] (Fig. 8B). These data indicate that the interaction of Rnq1p with Sis1p is important for the maintenance of the [RNQ+] prion as well as [PSI+] induction.
Figure 8.
Rnq1p-L94A aggregates but does not maintain [RNQ+] or facilitate [PSI+] induction. (A) Rnq1p-L94A was expressed as the only copy of Rnq1p in either a [RNQ+] or [rnq−] strain by plasmid shuffle. Lysate from these strains were fractionated into total (T), soluble (S) and insoluble pellet (P) by high speed centrifugation and subjected to SDS-PAGE. A western blot was performed with anti-Rnq1p antibody. [RNQ+] status of original plasmid shuffle strain is indicated to the left of the blot. (B) Wild type Rnq1p was replaced with Rnq1p-L94A by plasmid shuffle in a [RNQ+] strain. The plasmid-shuffled strain was transformed with a plasmid that overexpresses Sup35p and plated on YPD. [PSI+] was scored by the appearance of light pink sectoring on the colonies. Three independent transformants were scored for [PSI+] induction. Approximately 200 colonies were analyzed for each and a total of four [PSI+] colonies were obtained. The graph indicates the percentage of sectoring cells with wild type set to 100%. Error bars indicate standard error of at least three separate replicates.
Discussion
Here we describe six novel mutations within HSP104 that cure both the [RNQ+] and [PSI+] prions. Five of these mutations are located in one of the nucleotide binding domains and one was found in the M domain. One of the Hsp104p mutants, E190K, displays differential kinetics for curing [PSI+] and [RNQ+] in the presence of wild type Hsp104p. Furthermore, Hsp104p-E190K shows varying kinetics within [RNQ+] variants that suggests Hsp104 may function differently in different prion strain variants. Hsp104p-E190K is unable to support thermotolerance on its own and shows reduced ATPase activity, suggesting that [RNQ+] requires less Hsp104p activity to propagate the prion state than [PSI+].
To our knowledge this represents the first screen for cellular factors that affect [RNQ+] propagation. The phenotypes typically associated with the [RNQ+] prion, aggregation of Rnq1p and [PSI+] inductions, are not readily amenable to large scale screens. However, as the nonsense suppression of RRP is dependent on the cells being [RNQ+],50 it constitutes an excellent system to screen for factors that affect [RNQ+] propagation.
Our screen for mutants that are unable to propagate [RNQ+] only uncovered mutations within HSP104. Hsp104p is the only known protein required for the propagation of all known yeast prions.3,4,11,25,35 Furthermore, over thirty different mutations have been discovered in Hsp104p that affect prion propagation.20,25,28–31 These mutations are found throughout the protein and may interfere with Hsp104p activity at a variety of different steps including substrate recognition, hexamer formation and nucleotide hydrolysis. Thus, there are many targets within Hsp104p that can be altered and cause the loss of prion propagation. Generally, the mutations that cure [PSI+] cause the cells to lose the thermotolerance phenotype. However, this is not always a complete loss of thermotolerance.28 This indicates that thermotolerance and prion propagation are genetically separable.
The majority of the mutants found in this screen were in either NBD1 (A178T, E190K, A297T) or NBD2 (G619D, A660T, E689K, T726I). A loss of ATPase activity from either of these domains or the inability to form hexamers generally results in defects in the propagation of [PSI+]. Of the two domains, NBD1 appears to provide the majority of the ATPase activity required for disaggregation activity.23,59 While NBD2 has a low rate of hydrolysis, nucleotide binding to NBD2 regulates the formation of hexamers as well as the rate of hydrolysis of NBD1.22–24 As an example of the importance of these two domains, the K218T and K620T mutations which lie within the Walker A motifs of NBD1 and NBD2, respectively, have been shown previously to impair ATPase activity.54 Thus, in our screen the mutants in NBD1 and NBD2 likely alter the ATPase activity or oligomerization ability of the protein to produce the loss of prion phenotypes. This conclusion is supported by the decreased rate of ATP hydrolysis of Hsp104p-E190K as compared to wild type Hsp104p.
The final mutation we identified was in the M domain (A502T). Deletions within this middle domain abrogate the function of Hsp104p27 as this domain likely plays a role in transducing the allosteric signal from NBD2 to NBD1. Thus, the A502T mutation we identified likely disrupts the coordination between the domains, reducing the ATPase activity and inhibiting the overall disaggregation ability of the protein. Interestingly, the mutation A503V has differential effects on [PSI+] and [RNQ+].60 It causes toxicity in [PSI+] strains and affects aggregate size but does not affect [RNQ+] propagation.60
Due to its differential effects on curing [RNQ+] and [PSI+] in the presence of wild type Hsp104, we extended our analysis of the E190K mutant. While it is competent to form hexamers, it shows significantly decreased ATPase activity in vitro as compared to wild type Hsp104. The in vivo function of the mutant is also compromised as it cannot support thermotolerance. Given that it appeared to have no defect in hexamer formation, the mutant likely forms mixed hexamers with wild type Hsp104p. This mixed hexamer may have enough activity to temporarily propagate [RNQ+] but not [PSI+]. Our data indicate that aggregates of Rnq1p may require less Hsp104p activity to propagate than aggregates of Sup35p. Interestingly, too much Hsp104p activity cures [PSI+] but not [RNQ+].11,25 Thus, [RNQ+] appears to persist through a broader range of Hsp104 activities. This may be one reason why [RNQ+] cells are found in nature while [PSI+] cells are not.61 Allowing [RNQ+] to persist over a broader range of conditions may also benefit the yeast as the presence of the [RNQ+] prion dramatically increases the appearance of [PSI+].10–12,62 Further investigations into the different regulation of yeast prions will lead to a better understanding of their biological function.
In further characterizing the effect of E190K on [RNQ+] by looking at the high, medium and low [PIN+] variants, we found that the mixed hexamers of wild type and Hsp104p-E190K differ in their ability to recognize and/or propagate the [RNQ+] variants. Previous data suggests that medium and low [PIN+] have different aggregate structures than high [PIN+].63 While Hsp104p-E190K is not the first Hsp104p mutant to show differential curing of [PSI+] and [RNQ+],29 it is the first Hsp104 mutant that demonstrates a difference in recognition between the variants of [RNQ+]. Interestingly, the overexpression of the Hsp40 co-chaperone Ydj1p showed differential curing of the [RNQ+] strain variants, suggesting that the whole chaperone network is involved in recognizing distinct structures.40
Another Hsp40, Sis1p, is essential for the propagation of [RNQ+] and interactions between Sis1p and Rnq1p have been proposed to generate the infectious seeds required for [RNQ+] propagation.33,44 The Rnq1p-L94A mutation dramatically reduces the ability of Rnq1p to bind Sis1p but does not completely abolish binding.58 Our data indicate that the reduction in Sis1p binding prevents the protein from effectively maintaining a prion. It appears that Rnq1p-L94A aggregates non-specifically, indicating that the interaction with Sis1p promotes the formation of ordered aggregates. These non-specific aggregates are unable to induce [PSI+], suggesting the aggregation of Rnq1p alone is not sufficient to promote [PSI+] induction.
Materials and Methods
Strain and plasmid construction.
All S. cerevisiae strains were derived from 74-D694. Standard culturing and manipulation techniques were used for both yeast and Escherichia coli.49 Yeast strains were grown in either a rich medium, YPD (1% yeast extract, 2% peptone, 2% glucose), or a synthetic medium (0.67% yeast nitrogen base, 2% glucose) lacking the appropriate amino acids to select for plasmids. Haploid spores were generated from diploid parents and isolated by micromanipulation.
RRP has been described previously.50 pPROEx Htb Hsp104 has previously been described.21 A fragment containing the E190K mutation was amplified using the oligonucleotides 5′-TTC TTT CAA AGG CAC CAT CC and 5′-CGG GAT CCA CCC TTG AAT CGA ATC AGC A. This product was digested with BglII and EcoRI and ligated into a BglII/EcoRI fragment of pPROEx Htb Hsp104 to generate pPROEx Htb Hsp104-E190K. HSP104 was cloned into pRS316,51 by digesting pPROEx Htb Hsp104 with BamHI and XhoI and ligated into a BamHI/XhoI fragment of pRS316. Sequence carrying the E190K mutation was excised from pPROEx Htb Hsp104-E190K on a BglII and EcoRI fragment and ligated into a cognate fragment of pRS316-Hsp104 or pRS306-Hsp104 to create pRS316-Hsp104-E190K and pRS306-Hsp104-E190K, respectively. Single mutants were also cloned with bridge PCR. The N-terminus of RNQ1 was amplified with the oligonucleotide 5′-GGG GAT ATC ATG GAT ACG GAT AAG TTA ATC TCA GAG G-3′ and an oligonucleotide specific to the mutant. The reverse complement of the mutant specific oligonucleotide was then used along with the oligonucleotide 5′-CCC GTC GAC TCA GTA GCG GTT CTG GTT GCC G-3′ to amplify the C-terminus of RNQ1. This produced full length RNQ1 carrying the desired mutation which was digested with EcoRV and SalI and ligated into pRS313 that contained the RNQ1 promoter on an EcoRV/EcoRI fragment and the RNQ1 terminator on a SalI/XhoI fragment. For GPD-Rnq1-L94A, an EcoRV/SalI fragment was removed from the pRS313-Rnq1-L94A mutant and cloned into a EcoRV/SalI fragment of p413-GPD.
EMS mutagenesis.
Overnight cultures of [RNQ+] cells expressing RRP were washed twice in 50 mM potassium phosphate buffer pH 7.0 and then resuspended in buffer. The cells were normalized to 5 × 107 cells/ml and incubated with ethyl methyl sulfonate (EMS) at a final concentration of 3% (v/v). Mutagenesis was halted at various time points by the addition of an equal volume of 10% Na2S2O3. The treated cells were extensively washed with H2O and their viability was determined by plating assay. A culture with approximately 14% viable cells was plated on YPD to assess color.
Analysis of [RNQ+] status.
The [RNQ+] status of the cells was biochemically assessed in one of two ways.52 For a full solubility assay, yeast cells were lysed with glass beads in buffer containing 100 mM Tris-HCl pH 7, 200 mM NaCl, 1 mM ethylendiaminetetraacetic acid (EDTA), 5% glycerol, 0.5 mM dithiothreitol (DTT), 50 mM N-ethylmaleimide (NEM), 3 mM phenylmethanesulphonylfluoride (PMSF) and complete Mini protease inhibitor cocktail (Roche). Following lysis, an equal volume of RIPA buffer (50 mM Tris-HCl pH 7, 200 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)) was added to the lysate and cell debris was cleared by a brief centrifugation. This cleared lysate is the total protein. Insoluble protein was pelleted by centrifugation at 80,000 RPM (27,000 xg) in a Beckman TLA-100 rotor for 30 minutes. The supernatant containing the soluble protein was removed and the pellet was resuspended in a 1:1 mix of lysis buffer and RIPA buffer. Total, supernatant and pellet fractions were subjected to SDS-PAGE, transferred to PVDF membranes and probed with an anti-Rnq1p antibody.
Well trap assays were performed as previously described.52 Cells were lysed with glass beads in buffer containing 50 mM Tris-HCL pH 7.5, 50 mM KCl, 10 mM MgCl2 and 5% glycerol supplemented with an anti-protease solution (Sigma), 10 mM PMSF, 50 mM NEM. Lysate was incubated in sample buffer (100 mM Tris-Cl pH 6.8, 1% SDS, 10% glycerol, 0.1% bromophenol blue) for seven minutes at room temperature. Treated lysates were subjected to SDS-PAGE, transferred to PVDF and blotted with an anti-Rnq1p antibody.
Thermotolerance assays.
Thermotolerance assays were performed as previously described.53 Equal numbers of yeast cells in logarithmic growth were resuspended in liquid medium. The resuspended cells were pretreated at 37°C for 30 minutes and then shifted to 50°C for various periods of time. Following heat shock, the cells were incubated on ice for 2 minutes. The cells were then serially diluted and spotted onto YPD.
Hsp104p purification.
His6-Hsp104p and His6-Hsp104p-E190K were expressed and purified from E. coli BL21(DE3) as previously described.21 Polyhistidine-tagged Hsp104p and Hsp104-E190K were isolated by affinity chromatography on Ni-NTA sepharose. The histidine tag was cleaved with TEV protease and the untagged protein further purified by anion exchange chromatography. The fractions were analyzed by SDS-PAGE, pooled and frozen at −80°C.
ATPase activity assay.
Characterization of Hsp104 ATPase activity was performed in Buffer A (40 mM Tris, pH 7.5, 175 mM NaCl, 5 mM MgCl2, 0.02% Triton X-100) as reported.54 The reactions were performed at 37°C with either 1 µg Hsp104p or 5 µg Hsp104-E190K and 5 mM ATP (Sigma). At various time points during the reaction, Malachite Green Reagent was added to the reaction tube to quantify the amount of free Pi and the color development was stopped by the addition of citric acid. The A650 of the sample was determined and the absorbance units calibrated against a standard of known concentrations of KH2PO4. Each time point was done three independent times and the mean ± standard deviation was calculated.
Gel filtration chromatography.
Gel filtration was performed as previously described.55 Two milligrams of purified Hsp104p or Hsp104p-E190K were incubated for 5 minutes on ice with Buffer B (40 mM Tris-HCL, pH 8.0, 15 mM NaCl, 5 mM MgCl2, 10 mM ATP) before loading onto Sephacryl S-300 High Resolution resin (Amersham Biosciences). The column was equilibrated in Buffer B and was run at 4°C at a flow rate of 0.05 mL/min. Fractions (5 mL) were collected and analyzed for the presence of Hsp104 by western blot using an antibody probe against Hsp104p. Resolved bands were quantified using Image J. Molecular weights of Hsp104p and Hsp104p-E190K were estimated by comparison of their elution profiles with the following standards: thyroglobulin Mr 670,000; catalase Mr 250,000; bovine serum albumin Mr 66,000.
[PSI+] induction assays.
[PSI+] induction assays were performed as described previously.50 Plasmid shuffled strains containing the Rnq1p mutants were transformed with a Ura-marked plasmid carrying SUP35 (pSUP2,56) and plated on selective media for both plasmids. Individual transformants were grown in selective media to OD600 ∼1.6 and plated on YPD. After five days of growth, [PSI+] colonies were counted as any colony with a light pink sector. Representative colonies were checked for curing on plates containing 3 mM GdnHCl. The vast majority (>95%) of colonies with light pink sectors were curable on 3 mM GdnHCl (data not shown). Overexpression of Sup35p in a [RNQ+] strain has also been shown to create non-heritable amyloids of Sup35p that cause nonsense suppression similar to [PSI+].57 Since this nonsense suppression is dependent on the overexpression of Sup35p, we selected cells with light pink sectors and spotted them onto medium containing 5-FOA. Cells that converted to [PSI+] remained light pink on 5-FOA medium, while cells with non-heritable amyloids reverted back to red. Experiments with both wild type Rnq1p and the mutants revealed that, on average, about 12% of colonies with light pink sectors were the result of non-heritable amyloids while the rest were bona fide [PSI+] (data not shown). This proportion is in line with the frequency of non-heritable amyloid induction previously reported.57
Acknowledgements
The authors would like to thank Dr. John Glover for plasmids, protocols and advice. We thank Dr. Susan Lindquist for providing the anti-Rnq1p antibody. We thank Dr. Lisa Underwood and Rachel Bouttenot for critical reading of this manuscript. This research was supported by National Institutes of Health grant GM072228 (H.L.T.).
Abbreviations
- RRP
[RNQ+] reporter protein
- NBD
nucleotide binding domain
- M
middle domain
- EMS
ethyl methyl sulfonate
- GdnHCl
guanidine hydrochloride
- PrD
prion-forming domain
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/9662
References
- 1.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Wickner RB, Masison DC, Edskes HK. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 3.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009 doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 7.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 8.Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 12.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 13.Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- 14.True HL. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 18.Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 20.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tkach JM, Glover JR. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J Biol Chem. 2004;279:35692–35701. doi: 10.1074/jbc.M400782200. [DOI] [PubMed] [Google Scholar]
- 22.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 23.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattendorf DA, Lindquist SL. Analysis of the AAA sensor-2 motif in the C-terminal ATPase domain of Hsp104 with a site-specific fluorescent probe of nucleotide binding. Proc Natl Acad Sci USA. 2002;99:2732–2737. doi: 10.1073/pnas.261693199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 26.Schirmer EC, Homann OR, Kowal AS, Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol Biol Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cashikar AG, Schirmer EC, Hattendorf DA, Glover JR, Ramakrishnan MS, Ware DM, et al. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–760. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi A, Hara H, Kurahashi H, Nakamura Y. A systematic evaluation of the function of the protein-remodeling factor Hsp104 in [PSI+] prion propagation in S. cerevisiae by comprehensive chromosomal mutations. Prion. 2007;1:69–77. doi: 10.4161/pri.1.1.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurahashi H, Nakamura Y. Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol Microbiol. 2007;63:1669–1683. doi: 10.1111/j.1365-2958.2007.05629.x. [DOI] [PubMed] [Google Scholar]
- 30.Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation and thermotolerance. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 32.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 33.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Molecular & Cellular Biology. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 40.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;30:30. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andreasson C, Lindquist S, et al. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 2008;3:1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park KW, Hahn JS, Fan Q, Thiele DJ, Li L. De Novo Appearance and “Strain” Formation of Yeast Prion [PSI+] Are Regulated by the Heat-Shock Transcription Factor. Genetics. 2006;173:35–47. doi: 10.1534/genetics.105.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian X, Hou W, Zhengang L, Sha B. Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp40: yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1. Biochem J. 2002;361:27–34. doi: 10.1042/0264-6021:3610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aron R, Lopez N, Walter W, Craig EA, Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169:1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego: Academic Press, Inc; 1991. [Google Scholar]
- 50.Bardill JP, True HL. Heterologous prion interactions are altered by mutations in the prion protein Rnq1p. J Mol Biol. 2009;388:583–596. doi: 10.1016/j.jmb.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liebman SW, Bagriantsev SN, Derkatch IL. Biochemical and genetic methods for characterization of [PIN+] prions in yeast. Methods. 2006;39:23–34. doi: 10.1016/j.ymeth.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 54.Schirmer EC, Queitsch C, Kowal AS, Parsell DA, Lindquist S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J Biol Chem. 1998;273:15546–15552. doi: 10.1074/jbc.273.25.15546. [DOI] [PubMed] [Google Scholar]
- 55.Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- 56.Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, et al. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 57.Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- 58.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, et al. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schirmer EC, Ware DM, Queitsch C, Kowal AS, Lindquist SL. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc Natl Acad Sci USA. 2001;98:914–919. doi: 10.1073/pnas.031568098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 61.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. Epub 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 63.Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]