Abstract
Chronic wasting disease (CWD) is the only known transmissible spongiform encephalopathy affecting free-ranging wildlife. Although the exact mode of natural transmission remains unknown, substantial evidence suggests that prions can persist in the environment, implicating components thereof as potential prion reservoirs and transmission vehicles.1–4 CWD-positive animals may contribute to environmental prion load via decomposing carcasses and biological materials including saliva, blood, urine and feces.5–7 Sensitivity limitations of conventional assays hamper evaluation of environmental prion loads in soil and water. Here we show the ability of serial protein misfolding cyclic amplification (sPMCA) to amplify a 1.3 × 10−7 dilution of CWD-infected brain homogenate spiked into water samples, equivalent to approximately 5 × 107 protease resistant cervid prion protein (PrPCWD) monomers. We also detected PrPCWD in one of two environmental water samples from a CWD endemic area collected at a time of increased water runoff from melting winter snow pack, as well as in water samples obtained concurrently from the flocculation stage of water processing by the municipal water treatment facility. Bioassays indicated that the PrPCWD detected was below infectious levels. These data demonstrate detection of very low levels of PrPCWD in the environment by sPMCA and suggest persistence and accumulation of prions in the environment that may promote CWD transmission.
Key words: prions, chronic wasting disease, water, environment, serial protein misfolding cyclic amplification
Introduction
Transmissible spongiform encephalopathies (TSEs) are a group of unusual infectious diseases that cause progressive neurodegeneration and certain death that include bovine spongiform encephalopathy (BSE) in cattle, scrapie in sheep and goats, chronic wasting disease (CWD) in deer, elk and moose and kuru and Creutzfeldt-Jakob disease in humans. CWD is endemic to Northeastern Colorado and was first described in mule deer in 1967 in Fort Collins,8 located in the South Platte River Basin, which has the highest incidence of CWD in Colorado.9 Considerable evidence supports the hypothesis that PrPSc, an aberrant, pathological form of the normal, host-encoded cellular prion protein (PrPC), is the major constituent of prions, the causative agent of TSEs.10–12 PrPSc autocatalyzes conversion of PrPC into more PrPSc in a wide variety of tissues such as tonsils, spleen, retina, skeletal muscle, peripheral nervous system, endocrine organs, blood, saliva, lymph nodes, spinal cord and most robustly in the brain.5,13–16
Prions have been shown to strongly adsorb to mineral and organic soil components1,3,17 and persist in aquatic environments.4,18 Adsorption of hamster scrapie prions to soil increased the infectious titer 680-fold compared to unbound prions, enhancing oral transmission of scrapie.17 Prions may contaminate the natural environment via potentially infectious decomposing carcasses and biological materials such as saliva, urine and feces from CWD-positive animals.5–7,13,15,16 Carcass decomposition and shedding of CWD PrPSc (PrPCWD) in saliva, urine and feces into soil and water, which can be consumed during foraging and drinking, may contribute to efficient transmission of CWD in endemic areas like northern Colorado where prevalence estimates range up to thirty percent.2,9,14 Considering the high number of infected cervids in the South Platte River Basin and the possibility of environmental CWD contamination, it is conceivable that surface water sources draining the basin, including the Cache la Poudre River, may contain minute quantities of CWD prions. Rainfall and mountain snowmelt runoff wash soil and organic materials into surface water,19 increasing its total dissolved solids (TDS) and total suspended solids (TSS) content that includes total organic carbon (TOC), inorganic soil and mineral components, and colloids.20 The city of Fort Collins water treatment facility measures TOC levels to estimate runoff intensity.21 Environmental PrPCWD load in surface water could increase during spring melting of the winter mountain snow pack that increases TSS, TDC and TOC levels.
Detecting extremely small quantities of prions in the environment requires a very sensitive assay. Commonly used detection methods such as western blotting and immunohistochemistry detect PK-resistant forms of PrPSc (PrPRES) only in animal tissues harboring relatively large quantities of prions, typically in the central nervous and lymphoid systems. Their low sensitivity prohibits detection of small amounts of infectious prions in other biological and environmental samples. Serial protein misfolding cyclic amplification (sPMCA) is an in vitro technique that amplifies minute amounts of PrPRES to detectable levels by expediting conversion of PrPC to PrPRES.22–25 sPMCA has been used to efficiently and specifically amplify CWD prions26,27 and detect as few as 26 Hamster 263K PrPSc molecules,22 equivalent to approximately one infectious particle.28
We used sPMCA in this study to determine the detection limit of PrPCWD spiked into water samples and examine raw surface water for PrPCWD before, during and after processing at the Fort Collins water treatment facility (FCWTF). We detected a 1.3 × 10−7 dilution of CWD-infected brain homogenate in water after sPMCA with 78.57% sensitivity and 99.59% specificity. We also detected PrPCWD in one of two raw surface water samples from a CWD-endemic area at a time of increased water runoff from melting winter snow pack, as well as in the flocculation stage of processing this water. Aliquots of raw water samples testing positive for PrPCWD by sPMCA failed to cause diseases in susceptible transgenic mice.
Results
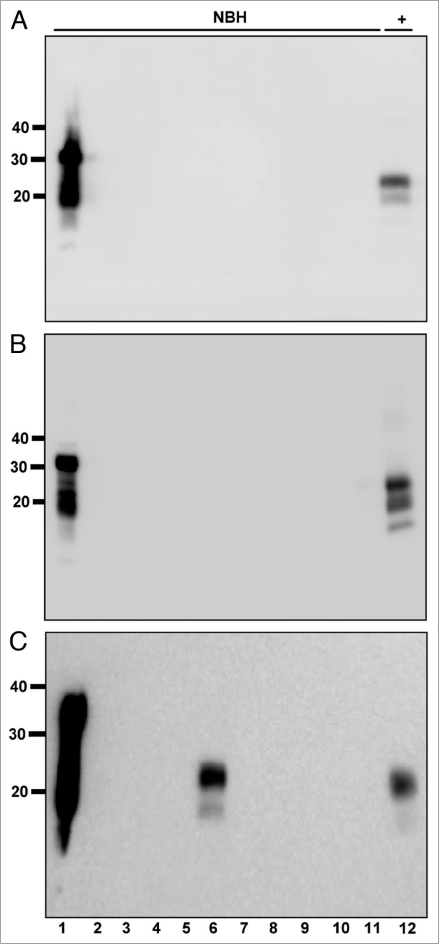
We optimized our sPMCA protocol to avoid false positive results that have recently been reported with this assay,29 achieving 99.59% specificity in our normal brain controls after sPMCA (n = 241, Fig. 1, Table 1 and Materials and Methods).
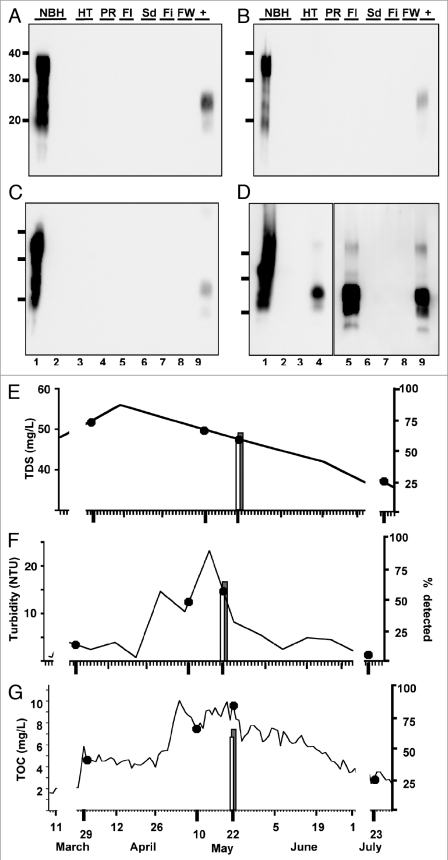
Figure 1.
Normal brain homogenate negative controls. All samples were digested with Proteinase K except normal brain homogenate (NBH) in lane 1. NBH, Normal brain homogenate control. +, 1:100,000 positive amplification control. Molecular weight markers in kilodaltons are shown to the left of the blots, which show thirty samples representative of 241 NBH negative controls. All samples were negative after six rounds of sPMCA except the sample in Gel C, lane 6.
Table 1.
Summary of sPMCA and bioassay results
| Sample | Dilutions | sPMCA1 | Bioassay2 | |||||||||
| cerPrP3 | moPrP3 | Tg1536 | Tg5037 | wild type | ||||||||
| PrPCWD dilutions | none4 | D104 | E14 | E24 | RML5 | none4 | E24 | RML5 | ||||
| no spike | 1/241 | ND6 | ND | ND | ND | ND | ND | ND | 0/20 (550) | 0/207 | ND | |
| 1.0 − 10−2 | ND | 10/10 | 10/10 | 10/10 | ND | ND | 0/10 | ND | 30/30 (251 ± 3) | 25/308 | 0/10 | |
| 1.0 × 10−3 | ND | 10/10 | 10/10 | 10/10 | ND | ND | 0/10 | ND | 8/8 (323 ± 14)9 | ND | ND | |
| 1.0 × 10−4 | ND | 16/16 | 10/10 | 10/10 | ND | ND | 0/10 | ND | 0/69 | ND | ND | |
| 1.0 × 10−5 | ND | 127/127 | 74/74 | 10/10 | ND | ND | 0/10 | ND | 1/8 (485)9 | ND | ND | |
| 2.0 × 10−5 | ND | 7/8 | ND | 8/8 | ND | ND | ND | ND | ND | ND | ND | |
| 4.1 × 10−5 | ND | 7/8 | ND | 8/8 | ND | ND | ND | ND | ND | ND | ND | |
| 8.2 × 10−5 | ND | 7/8 | ND | 8/8 | ND | ND | ND | ND | ND | ND | ND | |
| 1.0 × 10−6 | ND | 9/10 | 10/10 | 8/8 | ND | ND | 0/10 | ND | 0/10 (550) | 0/5 (400) | ND | |
| 3.3 × 10−6 | ND | 8/10 | ND | 7/8 | ND | ND | ND | ND | ND | ND | ND | |
| 6.6 × 10−6 | ND | 9/10 | ND | 8/8 | ND | ND | ND | ND | ND | ND | ND | |
| 1.3 × 10−7 | ND | 7/10 | 8/10 | 7/8 | ND | ND | ND | ND | 0/89 | ND | ND | |
| 2.6 × 10−7 | ND | 1/10 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| 5.3 × 10−7 | ND | 1/10 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| 1.1 × 10−8 | ND | 0/10 | 0/10 | 1/8 | ND | ND | ND | ND | 0/79 | ND | ND | |
| PrPSc dilutions | ||||||||||||
| no spike | ND | ND | ND | ND | 0/9 | 0/9 | ND | ND | ND | ND | 0/10 | |
| 1.0 × 10−2 | ND | ND | ND | ND | ND | ND | ND | ND | 0/10 | 0/5 | 15/15 | |
| 1.0 × 10−3 | ND | ND | ND | ND | 0/9 | ND | ND | 27/27 | 0/6 | 0/5 | 5/5 | |
| 1.0 × 10−4 | ND | ND | ND | ND | 0/9 | ND | ND | 27/27 | ND | ND | ND | |
| 1.0 × 10−5 | ND | ND | ND | ND | 0/9 | ND | ND | 27/27 | ND | ND | ND | |
| Negative control water samples | ||||||||||||
| CWD non-endemic US raw | no spike | 0/80 | ND | ND | ND | ND | ND | 0/16 | ND | ND | ND | ND |
| 1.0 × 10−4 | ND | ND | ND | 16/16 | ND | ND | 0/48 | ND | ND | ND | ND | |
| 1.0 × 10−5 | ND | ND | ND | 16/16 | ND | ND | ND | ND | ND | ND | ND | |
| 1.0 × 10−6 | ND | ND | ND | 16/16 | ND | ND | ND | ND | ND | ND | ND | |
| 1.0 × 10−7 | ND | ND | ND | 12/16 | ND | ND | ND | ND | ND | ND | ND | |
| 1.0 × 10−8 | ND | ND | ND | 1/16 | ND | ND | ND | ND | ND | ND | ND | |
| CWD non-endemic US finished | no spike | 0/18 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CWD non-endemic bottled | no spike | 0/18 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Test water samples | ||||||||||||
| 3-29-2007 FCWTF10 | no spike | 0/18 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 5-10-2007 FCWTF | no spike | 0/18 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 7-23-2007 FCWTF | no spike | 0/18 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 5-22-2007 FCWTF raw HT | no spike | 0/18 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 5-22-2007 FCWTF raw PR | no spike | 24/42 | ND | ND | ND | ND | 0/40 | ND | ND | 0/10 (550) | ND | ND |
| 5-22-2007 FCWTF flocculant | no spike | 20/30 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 5-22-2007 FCWTF sediment | no spike | 0/6 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 5-22-2007 FCWTF filtered | no spike | 0/6 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 5-22-2007 FCWTF finished | no spike | 0/6 | ND | ND | ND | ND | ND | ND | ND | ND | 0/10 (400) | ND |
| 9-27-2007 raw HT/PR+ alum | no spike | 0/12 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 4.0 × 10−5 | ND | 12/12 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| 9-27-2007 raw HT/PR+ alum | no spike | 0/12 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 4.0 × 10−5 | ND | 12/12 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
number of positive samples/total samples
number of terminally sick mice/number of animals inoculated (DPI* ± SD)
normal brain homogenate used for sPMCA substrate and negative controls
CWD-infected brain source
RML, Rocky Mountain Lab strain of mouse-adapted scrapie
ND, no data
0/15 by intracerebral (i.c.) route, 0/5 per OS (p.o.) at 400 days post inoculation (*DPI)
20/20 by i.c. (225 ± 6), 5/10 p.o. at 400 DPI
data from Angers, et al.31
FCWTF, Fort Collins water treatment facility.
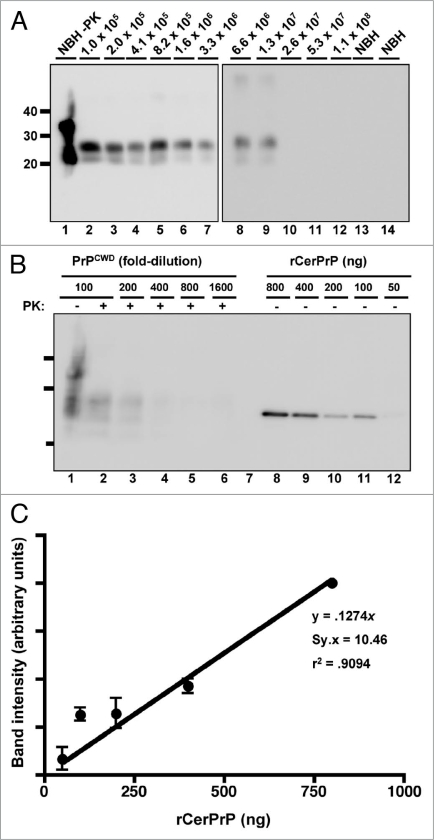
We next determined our prion detection limit using sPMCA by amplifying known CWD positive deer brain homogenates serially diluted into commercially purchased laboratory-grade water. We reproducibly detected a 1.3 × 10−7 dilution of brain homogenate by sPMCA with 78.87% sensitivity while all normal brain homogenate controls remained negative (Fig. 2A and Table 1). We quantified the amount of PrPCWD in this brain dilution by densitometric comparison of western blot signals of serially diluted, unamplified PrPCWD and known concentrations of recombinant cervid PrP (Fig. 2B). Using the standard curve generated from these data (Fig. 2C) and PrPCWD dilution and amplification factors (see Materials and Methods), we estimate that a 1.3 × 10−7 brain dilution contains approximately 101 ± 44 fg/µl of PrPCWD, or 5 ± 2.2 × 107 PrPCWD monomers. In addition to 241 normal brain homogenate negative controls, we tested numerous aliquots of raw water from non-CWD-endemic areas and detected no positive samples (Fig. 3 and Table 1). We also performed sPMCA on serial dilutions of PrPCWD spiked into each raw water sample to ensure that contaminants therein did not interfere with amplification. sPMCA amplified these dilutions similarly to those prepared in purified water, indicating little or no effect of raw water contaminants on PMCA efficiency in these samples. We also tested numerous aliquots of municipal and bottled water from non-CWD-endemic areas and detected no positive samples (Fig. S5 and Table 1).
Figure 2.
PrPCWD detection limit in water. (A) All samples were digested with Proteinase K except normal brain homogenate (NBH) in lane 1. Lanes 2 through 12 show amplified samples at the indicated starting dilution of CWD-positive brain into water. Lanes 13 and 14 show amplified NBH controls. The highest dilution reproducibly detected was 1:1.3 × 107 (lane 9). (B) Representative western blot showing serial 2-fold dilutions of PrPCWD (lanes 1–6) and rCerPrP (lanes 8–12). Samples were PK digested where indicated. Known quantities of rCerPrP were loaded in the indicated lanes to generate the standard curve in shown in (C). Densitometric analyses were performed on four unsaturated replicate blots. PRPCWD was estimated by interpolation of band intensities to the standard curve. The linear regression equation, the standard deviation of the residuals (Sy.x) and the goodness-of-fit (r2) of the curve are indicated. Molecular weight markers in kilodaltons are shown to the left of the blots, which are representative of four replicates using two distinct CWD prion isolates.
Figure 3.
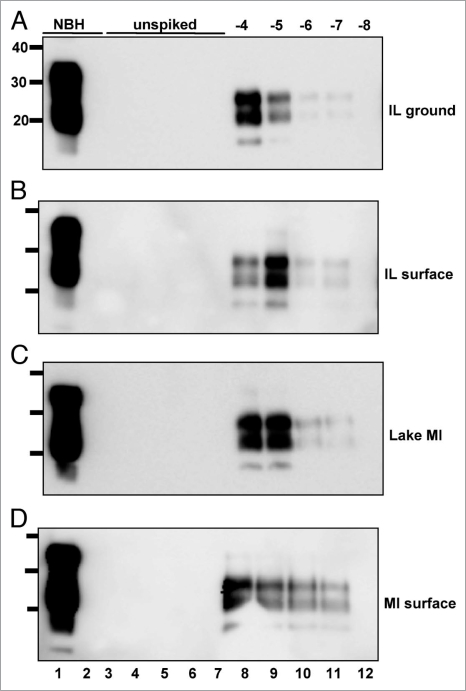
PrPCWD amplification in raw water samples from non-CWD-endemic areas. All samples were digested with Proteinase K except normal brain homogenate (NBH) in lane 1. Lane 2 shows amplified NBH control. sPMCA failed to amplify any PrPCWD from five replicate samples (lanes 3 to 7) of raw water from areas in IL (A, ground water and B, surface water) and Michigan (C, Lake Michigan and D, surface water) without any reported CWD cases, but amplified dilutions of PrPCWD spiked into these samples (log10, lanes 8 to 12) with similar efficiency as PrPCWD spiked into purified water. Data are representative of four separate experiments.
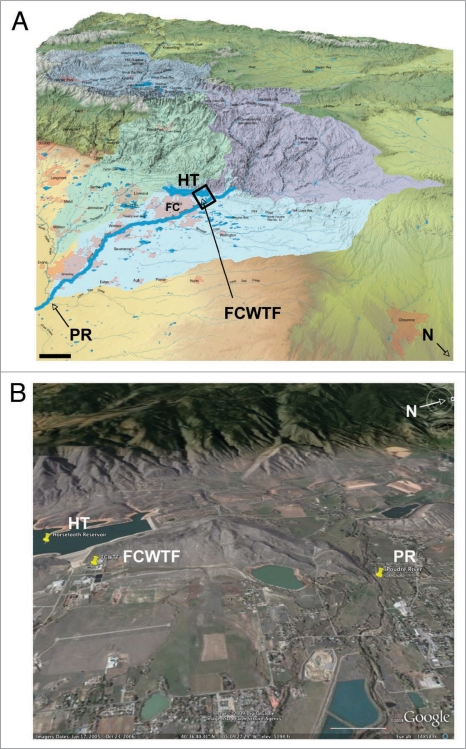
We then analyzed two discrete sources of raw surface water entering the Fort Collins water treatment facility. Water from the western slope of the Colorado Rocky Mountains pumped into Horsetooth reservoir (HT) represents runoff from an area of relatively low CWD prevalence compared to eastern slope runoff that drains into the South Platte River basin and the Cache la Poudre River (PR, Fig. 4), the other surface water source tested. We sampled raw water from designated sampling stations within the FCWTF (Fig. S4) collected at four different times of the year, 3-29-07, 5-10-07, 5-22-07 and 7-23-07, representing different runoff intensities and correspondingly different TDS, TSS (as measured by turbidity) and TOC levels. We also examined mixed HT and PR raw water at four different stages of processing at the FCWTF: flocculation, sedimentation, filtration and filter backwashing (Fig. S4 and Materials and Methods). The FCWTF facility treats an average of 1.13 × 108 L of water per day. Flocculation, in which alum is added to raw water, neutralizes charges on organic and inorganic settleable particles, colloids and dissolved constituents that precipitate to the bottom of sedimentation basins. The lowered-turbidity water is filtered through anthracite and sand, treated with calcium hydroxide, carbon dioxide for corrosion control, hydrofluorosilicic acid for fluoridation, and a chlorine solution for disinfection prior to distribution to the public. Water is periodically back flushed through the filters into a retention pond, mixed with raw water and recycled through the FCWTF.
Figure 4.
Locations of the Cache La Poudre river, Horsetooth reservoir and the Fort Collins Water Treatment Facility in the South Platte river basin watershed. (A) Map showing the South Platte river basin watershed. Cache La Poudre river (PR) and Horsetooth reservoir (HT) are shown 2X scale for clarity. The Colorado-Big Thompson western slope watershed, which feeds HT, is shown in dark blue (top left watershed on the map). The Cache la Poudre watershed is shown in purple (top right watershed). FC, city of Fort Collins; FCWTF, Fort Collins water treatment facility; HT, Horsetooth reservoir; N, north. Scale bar, 10 km. (B) Satellite map of the boxed area in (A) shows the relative locations of PR, HT and FCWTF in more detail.48 Scale bar, 1 km.
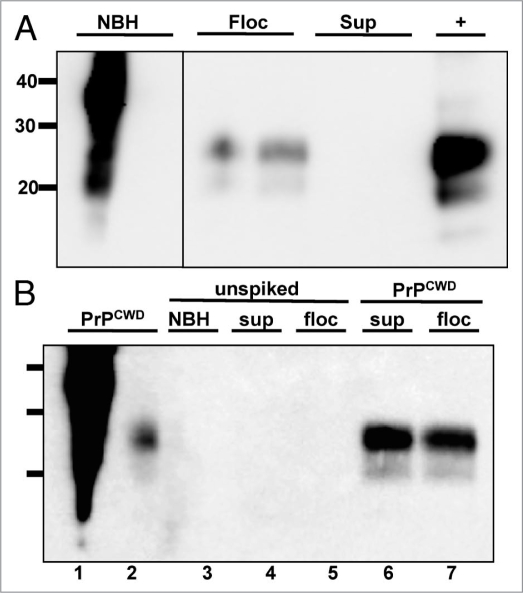
We detected no positive samples from raw or processed water samples collected on 3-29-07 (n = 18, Fig. 5A and Table 1), and 7-23-07 (n = 18, Fig. 5C and Table 1) corresponding to high to low TDS levels (52 and 36 mg/L, respectively, Fig. 5E), low turbidity (4.85 and 1.35 NTU, respectively, Fig. 5F) and moderate to low TOC levels (3.68 and 2.84 mg/L, respectively, Fig. 5G). We then examined water samples taken on 5-10-07 and 5-22-07, collected at times of peak snowmelt runoff and relatively high TDS (50 and 48 mg/L, respectively), turbidity (10.5 and 12.6 NTU, respectively) and TOC levels (7.54 and 9.50 mg/L, respectively) compared to earlier collections (Fig. 5E–G). While we detected no PrPCWD in water samples collected on 5-10-07 (n = 18, Fig. 5B and Table 1), we detected PrPCWD in 57% (24/42) of raw PR water and 67% (20/30) of flocculant subsamples from water collected on 5-22-07 (Fig. 5D and Table 1). We detected no PrPCWD in water from subsequent processing stages, including finished water, or raw HT water from the western slope (n = 36).
Figure 5.
PrPCWD detected in raw water samples collected at a time of increased snow-melt runoff. All samples were digested with Proteinase K except normal brain homogenate (NBH) in lane 1. Lane 2 shows amplified NBH control. Lanes 3–8 show Horsetooth reservoir (HT), Cache la Poudre river (PR), flocculant (Fl), post sedimentation (Sd), post filter (Fi) and finished (FW) water samples. No PrPCWD positive signals were detected in the three replicates for water samples collected on 3-29-2007 (A), 5-10-2007 (B) or 7-23-2007 (C). PrPCWD positive signals were detected in samples collected on 5-22-07 (D) from the Cache la Poudre river (lane 4) and flocculant (lane 5). Lane 9 shows a 1:100,000 positive amplification control (+). (E–G) Total dissolved solids (TDS), turbidity and total organice carbon (TOC) data spanning the entire winter snowmelt runoff. Gaps in the graphs between March 11 and March 27 and July 2 and 22 reflect lack of data between these dates. Collection days for this study are shown bolded with corresponding TDS (E), turbidity (F) and TOC (G) readings (read from the left y-axis) indicated by dark circles. Vertical bars indicate the percentage of samples testing positive by sPMCA (read from the right y-axis). TDC, turbidity and TOC levels in raw Cache La Poudre river water corresponding to elevated mountain snowmelt are at (F and G) or near (E) their peaks in May, at which time PrPCWD was detected in Cache la Poudre river (white bar) and flocculent (grey bar) samples collected on 5-22-07. NTU, nephelometric turbidity units.
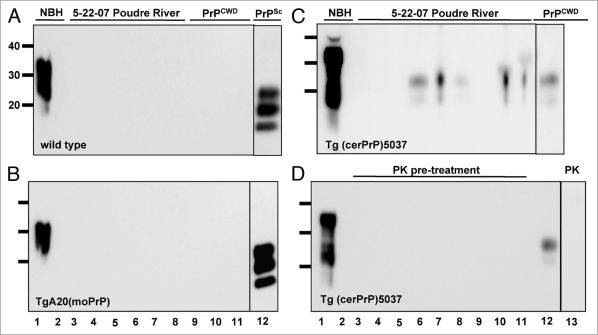
Although raw water contaminants did not impair PrPCWD spike amplification, they may affect the propensity of cerPrPC to misfold into PrPCWD in our sPMCA reactions, which could explain the detection of PrPCWD in samples with increased TDS, TSS and TOC. If this were true, we reasoned that other PrPC substrates would also be prone to misfolding during sPMCA. We therefore used moPrPC substrate from two NBH sources to attempt to amplify PrPSc from 5-22-07 raw PR water samples. We prepared NBH from wild type FVB and TgA20 mice that express 4–5 fold more moPrPC than wild type mice, similar to Tg(cerPrP) mice that overexpress cerPrP used in our previous sPMCA experiments. sPMCA failed to amplify PrPSc from six 5-22-07 PR raw water subsamples using either wild type or TgA20 NBH as substrate, strongly suggesting that PR raw water contaminants did not induce nonspecific conversion of the PrPC substrate into PrPSc in these reactions (Fig. 6A and B, lanes 3 to 8). Furthermore, moPrPC substrate did not support amplification of a 1:10,000 dilution of PrPCWD spiked into either NBH (lanes 9–11), but efficiently supported amplification of a 1:100,000 dilution of PrPSc (lane 12). These data corroborate previous studies demonstrating specific amplification of PrPCWD using cerPrPC substrate and inefficient PrPCWD amplification using murine substrate27 and indicate that sPMCA specifically detected PrPCWD in 5-22-07 PR samples. To verify the presence of PrPCWD prior to amplification, we performed sPMCA on nine additional 5-22-07 PR subsamples with or without pre-treatment with 25 µg/ml proteinase-K (PK), which digests a 1:100,000 dilution of PrPCWD in water such that it cannot be amplified (Fig. 6D, lane 13). These subsamples represent nine individual aliquots that were stored frozen at −70°C immediately after collection and not handled or opened until testing by sPMCA. We detected PrPCWD in 0/9 and 5/9 samples with or without PK treatment, respectively, confirming low yet demonstrable levels of PrPCWD in these raw water samples (Fig. 6C and D, lanes 3 to 11). Considering our previously determined PrPCWD detection limit in spiked dilutions, similar detection rates of this spike and 5-22-07 PR raw water samples and quantification using standard curves generated from recombinant cervid PrP, we estimate the contamination level of CWD prions in this raw water sample to be near our detection limit of 101 ± 44 fg/µl, or approximately 101 ± 44 parts per trillion.
Figure 6.
Abrogation of PrPCWD amplification from 5-22-07 raw PR water samples by PMCA using murine PrPC substrate or Proteinase K pre-treatment. All western blot samples were digested with Proteinase K except normal brain homogenate (NBH) in lane 1. Lane 2 shows amplified NBH controls. NBH from wild type (A) and TgA20 (B) mice failed to amplify PrPCWD from six subsamples of raw Poudre River water collected on 5-22-07 (lanes 3–8) and from 1:10,000 PrPCWD dilutions spiked into these NBHs (lanes 9–11), but efficiently amplified 1:100,000 PrPSc dilutions after six PMCA rounds (lane 12). (C) NBH from Tg(cerPrP)5037 mice amplified PrPCWD from 5/9 subsamples of 5-22-07 raw Poudre River water after six PMCA rounds (lanes 3 to 11). Lane 12 shows a 1:100,000 PrPCWD amplification control. (D) No PrPCWD signal was detected after PK pre-treatment and 6 rounds of sPMCA (lanes 3 to 11), indicating that the PrPCWD seed was present prior to sPMCA and can be reduced with protease digestion below sPMCA detection limit. Lanes 12 and 13 show amplification of a 1:100,000 dilution of PrPCWD pre-treated with heat-inactivated and active PK, respectively.
Inorganic soil components such as clay, quartz and montmorillonite have previously been reported to bind PrPRES,1,17 which may be precipitated with these components by flocculation. To determine which fraction of the 5-22-07 flocculation sample of mixed PR (58%) and HT (42%) water contained PrPCWD, we centrifuged this sample at 1,000 xg for five minutes and performed sPMCA on the pellet and supernatant fractions. PrPCWD was detected exclusively in the flocculant fraction, indicating that it was either bound to inorganic material in raw water or aggregated, possibly by the addition of alum (Fig. 7A). We next tested whether alum added to promote aggregation at the flocculation step of water processing could induce aggregation of PrPC substrate into PrPCWD that might explain the positive signal found in the flocculation water sample collected on 5-22-07. We modeled flocculation and sedimentation by adding 16 mg/L of alum to a mixture of PR (58%) and HT (42%) water collected on 9-27-07 according to a protocol used by the FCWTF (see Material and Methods). After sPMCA, we detected no PrPCWD in unspiked raw water control in either the supernatant or flocculant fraction and robust PrPCWD signal in raw water samples spiked with a 1:40,000 dilution of PrPCWD (Fig. 7B). Addition of alum had no apparent effect on sPMCA in these experiments.
Figure 7.
Effects of flocculation and alum on PrPCWD amplification. All samples were digested with Proteinase K except lane 1. (A) PrPCWD precipitates with flocculant water sample. Lanes 1 and 2 shows amplified NBH control. Lanes 3–6 show amplified samples after sPMCA. Lane 7 shows a 1:100,000 positive amplification control (+). PrPCWD signal was detected in the flocculant (lanes 3 and 4) but not supernatant (lanes 5 and 6) fraction, suggesting that the PrPCWD preferentially associates with the flocculant. (B) Addition of alum has no effect on PrPCWD amplification. Lanes 1 and 2 show PrPCWD amplification controls. Lane 3 shows NBH amplification control. Addition of Alum had no effect on sPMCA of unspiked NBH (lane 3) or mixed raw PR/HT water collected on 9-27-07 (lanes 4 and 5), nor on efficient amplification of PrPCWD diluted 1:40,000 into aliquots of the same raw water mixture by sPMCA (lanes 6 and 7).
We next used a cervid PrP transgenic mouse bioassay30,31 to estimate whether the PrPCWD levels detected in these raw water samples represent an infectious CWD prion dose. We used both Tg(cerPrP)1536 and Tg(cerPrP)5037 mice in our bioassays. Tg(cerPrP)1536 mice expressing mule deer PrP most prominently in nervous tissues have recently been used in an endpoint titration bioassay to determine the first infectious CWD prion titer.31 The infected brain preparation, D92, caused CWD in all inoculated Tg(cerPrP)1536 animals at a 10−3 dilution, no disease at 10−4, disease in one mouse at 10−5 and no mice at higher dilutions (Table 1).31 A 10−2 dilution of this inoculum caused terminal disease at similar DPI to D10, one of the CWD prion sources used in this study. Tg(cerPrP)5037 mice express elk PrP more abundantly in peripheral tissues including spleen, mesenteric and other lymph nodes, peyer's patches, tongue and intestine; and are susceptible to CWD by intraperitoneal and per os (p.o.) inoculation routes (Glenn Telling, personal communication and our unpublished data). Both transgenic lines were shown to be similarly susceptible to D92 and D10.31
To date, Tg(cerPrP)1536 mice acutely inoculated intracerebrally with 30 µl of UV-irradiated, antibiotic-treated 5-22-07 Cache la Poudre River, finished water, NBH or 10−6 dilution of D10 remain asymptomatic as far as 550 days post inoculation (DPI), whereas Tg(cerPrP)1536 mice inoculated with 30 µl of identically-treated 10−2 D10 succumbed to terminal CWD 251 ± 3 DPI (Table 1). To assess chronic low-level CWD prion infection in a more natural setting, we concurrently exposed additional cohorts of mice to CWD prions D10 and E2 diluted 10−6 daily in their drinking water. This dilution, which is thirteen times higher than our sPMCA detection limit, has caused no clinical disease in any animals (n = 15) as far as 550 DPI, while five of ten mice inoculated p.o. with a single 100 µl dose of 2% D10 or E2 succumbed to CWD 400 DPI. The remaining mice are currently under observation. Taken together, these data show that, while we have detected PrPCWD in 5-22-07 raw PR water by sPMCA, these samples have not proven to be infectious, even upon direct inoculation into the CNS.
Discussion
Indirect transmission of chronic wasting disease and scrapie through environmental exposure has been demonstrated in natural and experimental settings.2,7,32 Soil and surface water contaminated with urine, feces, saliva, blood and decomposing carcasses from prion infected animals have been implicated as possible transmission vehicles for CWD prions. Detecting vanishingly small traces of prions in environmental samples has not been possible with traditional prion detection techniques. sPMCA holds promise as a new technique for ultrasensitive prion detection, having been shown to be at least 4,000 times more sensitive than bioassay for detecting hamster prions.23 Similar to a recent report,33 modifications in our sPMCA reactions have enabled us to achieve highly sensitive and specific detection of PrPCWD without de novo prion generation previously reported for this assay.29
Here we show for the first time that sPMCA can detect minute concentrations of PrPCWD in experimental samples, and in environmental water samples from a single point source collection from the Cache la Poudre River, which accumulates water from melting winter snow pack draining through the South Platte River Basin, identified as a CWD-endemic area since the late 1960s. This sample, collected on 5-22-07 and representing one of two collected at a time of relatively high runoff, contained two to three-fold higher TOC levels than the other two samples collected, one before and one after peak runoff water flow. Increased water runoff also increases the concentration of inorganic environmental components, including minerals, clay and other soil components to which prions have been experimentally demonstrated to tightly adsorb and may carry prions into the Cache La Poudre river.1,3,17 Indeed, the 5-22-07 PR sample was two to ten-fold more turbid than early and late runoff samples, indicating higher TSS levels. While TDS levels did not correlate as tightly to runoff, exhibiting less variability compared to either turbidity or TOC levels, and with the 3-29-07 sample exhibiting the highest reading of all samples, the 5-22-07 PR samples still contained relatively high TDS levels. Environmental prions may be bound to both organic and inorganic material that comprise TDS and TSS. Recent studies have focused on competitive binding of PrPSc/CWD to organic and inorganic matrices that may influence prion adsorption to soil and its mobility in the environment.34 Because our study represents the first to identify PrPCWD in an environmental sample yet fails to definitively identify the matrix in which it resides, the nature of prion existence in the environment remains unclear. Too little data have been collected to definitively correlate PrPCWD with TDS, TSS or TOC levels in water draining into the South Platte River Basin. We continue to analyze water samples from this area for TDS, turbidity, TOC and PrPCWD content to more accurately define this putative relationship.
Other factors, such as temporal and spatial distribution of snow pack and rainfall, may influence PrPCWD levels, and its subsequent detectability, in water draining the basin. For example, a heavy snow pack or rainfall in an area highly contaminated with prions may promote increased movement of prions from that area, transiently increasing the prion load in the resulting runoff without increasing overall TDS, TSS or TOC levels in the Cache la Poudre River. Conversely, a heavy snow pack or rainfall in less contaminated areas may transiently dilute prion loads in the runoff below our detection limit. Kinetics of snowmelt may also influence prion loads and detection. An unusually warm or cold spring may accelerate or delay snowmelt and cause variations in prion deposition into the runoff that may not directly correlate with TDS, TSS or TOC levels.
Interaction of raw water components with PrPC substrate provides a plausible explanation for the positive PrPCWD signal arising from this lone sample in our sPMCA experiments. However, control sPMCA experiments using moPrPC as substrate failed to amplify PrPSc from this water sample, demonstrating that TOC or other raw water components did not induce spontaneous conversion of mouse PrPC to PrPSc. Only cerPrPC substrate supported PrPCWD amplification from the 5-22-07 raw PR water sample, consistent with the hypothesis that the sPMCA seed that it contains is of cervid origin. Significant species barriers for both CWD transmission to wild type mice and sPMCA of PrPCWD using NBH from moPrPC-expressing mice have been reported,27,30,35 further supporting this hypothesis.
Addition of alum, which neutralizes charges on particulates and colloids present in raw water to promote flocculation, may inadvertently predispose PrPC to aggregate and cause a false positive result in our sPMCA reactions. Control experiments in which alum was added to raw water samples produced no PrPCWD without the addition of a small amount of seed material, indicating little or no effect of alum in PrPCWD formation in our sPMCA reactions. Failure of all other flocculant samples to form PrPCWD by sPMCA, except that from the positive 5-22-07 raw water sample, predicted this outcome. Interestingly, although perhaps not surprisingly, PrPCWD detected in the flocculated raw water sample partitioned with the precipitate fraction upon low speed centrifugation. Flocculation may promote PrPCWD aggregation and precipitation directly or, more likely, sediments other soil components with which PrPCWD interacts, such as montmorillonite or other minerals previously shown to tightly bind prions.1
Finally, elimination of PrPCWD amplification upon pre-treatment of subsamples of the 5-22-07 raw PR water with PK serves as additional confirmation of the presence of PrPCWD prior to sPMCA that can be digested to a concentration below our 10−7 detection limit. We therefore conclude that this raw water sample contained extremely small but detectable amounts of PrPCWD.
We next sought to determine whether the 5-22-07 raw water sample in which we detected PrPCWD was infectious. Tg(cerPrP)1536 mice, which express 5–8-fold more cerPrPC per unit mass than white-tailed deer,26 have recently been used to detect infectious prions in urine from CWD-infected deer that were undetectable in natural host bioassays,36 and to determine the first infectious titer for a CWD prion source, D92. The bioassay revealed that a 10−2 D92 dilution caused terminal CWD in Tg(cerPrP)1536 mice at similar DPI to D10, one of the inocula used in this study, and no disease at 10−6 dilution.31 In this study, no mice inoculated i.c. with 5-22-07 raw water samples, NBH or finished water have contracted CWD after more than 500 DPI, while all 10−2 CWD prion controls have succumbed to disease. Since we estimated the prion concentration in the 5-22-07 sample to equate to a 10−7 dilution of D10 in water, we reasoned that this dose was not likely to cause disease in Tg(cerPrP)1536 mice even upon i.c. inoculation. We also exposed Tg(cerPrP)1536 mice and a second transgenic line, Tg(cerPrP)5037, with wider tissue distribution of cerPrPC and susceptibility to peripheral inoculation routes, to a 10−6 dilution of D10 and E2 CWD prions daily in their drinking water to mimic natural CWD prion exposure at a dose thirteen times higher than that estimated for the 5-22-07 sample. No Tg(cerPrP)1536 or Tg(cerPrP)5037 mice have contracted CWD at 500 and 400 DPI, respectively, while mice inoculated p.o. with 10−2 D10 have begun to succumb to disease. Again, these data are not surprising since no Tg(cerPrP)1536 mice contracted disease upon i.c. inoculation at 10−6 D10 dilution. We conclude from these data that the level of PrPCWD in the 5-22-07 raw PR water is not infectious in these bioassays. We also estimate that sPMCA of PrPCWD, which detected a 10−7 D10 dilution, is approximately 10,000 times more sensitive than the Tg(cerPrP)1536 mousebioassay, which detected a 10−3 D10 dilution, and approximates the fold increase in sensitivity of sPMCA of hamster prions compared to the corresponding hamster bioassay.6
It may initially seem highly unlikely that an infectious protein could be detected in a small sample of the sixty billion liters of water flowing through the South Platte River Basin at the time it was collected.37 However, given the extreme sensitivity that sPMCA affords, a very small amount of PrPCWD contaminant can be detected. At our detection limit established in this study, sixty billion liters of water need only contain six kilograms of PrPCWD to be successfully amplified by sPMCA. From the standard curve generated from densitometry of rCerPrP western blots, we estimate the PrPCWD concentration in the infected D10 deer brain to be 1 mg/ml, or roughly 1 mg PrPCWD/g of brain. Assuming a representative prion load in this brain and the average cervid brain weight to be 500 g, 1,200 infected deer contain six kilograms of PrPCWD in their brains, with lower but appreciable amounts in their spinal cords, spleen, lymph nodes, salivary glands, intestines and antlers. According to 2007 Colorado division of wildlife population estimates, there are 17,280 cervids in six game units surrounding the Cache la Poudre River.9 Ranges of prevalence estimates for each cervid population in these game units estimate that 819 to 1,465 infected cervids resided there in 2007 alone. CWD has been endemic in the area for forty years, and it remains unclear how long prions can persist in the environment. If persistent for at least several years, CWD prions deposited into the environment from thousands of infected carcasses may accumulate on soil and vegetation such that it can be washed into surface water draining the basin during snowmelt or rainstorms. Symptomatic and asymptomatic positive animals can also contribute to environmental CWD load via biological materials such as saliva, blood, urine and feces.5–7,32,36,38 Deer and elk defecate approximately 900,000 kg of feces and urinate approximately 14 million liters of urine in the area immediately surrounding the Cache la Poudre river per year.39–42 Although urine and feces likely contain much lower prion loads than blood or saliva, the sheer amount of excreta may contribute significantly to overall environmental prion contamination.
The data presented here demonstrate that sPMCA can detect low levels of PrPCWD in the environment, corroborate previous biological and experimental data suggesting long term persistence of prions in the environment2,3 and imply that PrPCWD accumulation over time may contribute to transmission of CWD in areas where it has been endemic for decades. This work demonstrates the utility of sPMCA to evaluate other environmental water sources for PrPCWD, including smaller bodies of water such as vernal pools and wallows, where large numbers of cervids congregate and into which prions from infected animals may be shed and concentrated to infectious levels.
Materials and Methods
Transgenic mice.
FVB mice were purchases from Charles River laboratories (Wilmington, MA). TgA20 mice overexpressing moPrP have been previously described.43 Tg(cerPrP)5037 mice were generated in the Telling laboratory in a similar fashion to Tg(cerPrP)1536 mice30 by injection of fertilized FVB/PrP null oocytes with the MoPrPXho expression vector containing elk PrP cDNA. This resulted in a wider tissue distribution of cerPrP expression, including spleen, mesenteric lymph nodes, peyer's patches, salivary glands, tongue and intestine (Glenn Telling, personal communication and our unpublished data) that was six times the expression of moPrP in wild type mice.31 Mice were housed and maintained at Lab Animal Resources, in accordance with protocols approved by the Institutional Animal Care and Use Committee at Colorado State University.
Sources and preparation of prion-infected brains.
The D10 isolate of CWD prions (D10) and the Rocky Mountain Lab strain of mouse-adapted scrapie prions passage 5 (RML) were previously described.30,44 The E1 and E2 isolates of CWD prions were prepared from brain hemispheres from terminally sick elk from game farms in northern Colorado. 10% brain homogenates were prepared in PBS or PMCA buffer 1 (4 mM EDTA, 150 mM NaCl in PBS). D10 and RML were homogenized using glass beads in a FastPrep machine (Biogene) three times for thirty seconds at 4.5 m/s with two minute intervals on ice. E1 and E2 10% brain homogenate was prepared using a commercial grade blender and homogenizing three times at maximum power for two minutes at 4°C with two minute intervals on ice. All brain homogenates were clarified by centrifugation at 1,000 rpm for five minutes and 50–1,000 µl aliquots stored at −70°C.
Environmental water samples.
Negative control raw water samples were collected from non-CWD-endemic areas (see Figs. S2–4) including Illinois surface water (41°31′49.87″N, 88°11′27.91″W), Illinois ground water (41°31′29.28″N, 88° 12′47.10″W approximate depth, 20 m), Michigan surface water (41°55′23.95″N, 84°4′50.68″W) and Lake Michigan water (43°44′31.03″N, 86°28′38.90″W). Four 50 ml aliquots of test water sample and one 50 ml aliquot field blank were collected at each site. Three samples were stored in Phosphoric acid at 4°C for subsequent TOC analyses. Samples were capped in 50 ml tubes that were sealed in individual double bags for transport to our laboratory. Upon arrival, the three aliquots stored in phosphoric acid were analyzed for TOC levels and the remaining aliquot was divided into 1 ml aliquots in a prion free room and stored at 4°C.
Thirty milliliter grab samples of the following water types were collected in 50 ml conical tubes in the FCWTF (40°35′33.35″N, 105° 9′32.41″W) with the assistance of FCWTF personnel (Fig. S4). Cache la Poudre River (PR) and Horsetooth Reservoir (HT) raw water was collected at designated sampling stations within the FCWTF before blending. After PR and HT water was blended, samples were collected from flocculation basin water, LaMella Sedimentation Basin water, filtered water, finished water and East and West backwash pond water prior to the backwash return pump station. Hydraulic residence times in the FCWTF range from 2.5 hours in summer to four hours in March. All samples were collected within one hour on the indicated days between 11 AM and 2 PM, ensuring that samples from individual collection days represent the same water source for that given collection. Samples were handled with clean gloves, placed in plastic bags for transport and aliquoted into 1–5 mL volumes on clean bench paper in a prion free room to avoid cross contamination, and stored at −80°C until use.
Preparation of normal brain homogenates.
Mice were perfused with 30 ml of 1x phosphate buffered saline (PBS) plus 5 mM EDTA, their brains removed and snap frozen in liquid nitrogen. Brain hemispheres were weighed and an appropriate volume of PMCA buffer 1 containing 2X Complete Protease Inhibitor Cocktail (Roche) and glass beads were added for FastPrep (Biogene) homogenizing for thirty seconds at 4.5 m/s to generate a 20% wt/vol solution. An equal volume of PMCA buffer 2 (PMCA buffer 1 plus 2.0% Triton X-100 without protease inhibitor) was added and samples were incubated on ice for 20 min then clarified by centrifugation at 300 xg for 5 min. The supernatant was carefully removed and stored at −80°C.
Serial protein misfolding cyclic amplification (sPMCA).
25 µl samples were added to 25 µl of NBH without addition of exogenous polyanions in individual 200 µl PCR tubes sealed shut with parafilm™. The tubes were then subjected to 40 sec pulses of sonication at 37°C, power setting 7, every 30 min for 24 hr using a 3000MP sonicator (Misonix) filled with 180 ml of deionized water. After 24 hrs each sample was mixed and 25 µl transferred to 25 µl of fresh NBH. The microplate horn was drained, washed thoroughly with 4% sodium dodecyl sulfate in 1% acetic acid45 and refilled with fresh water. Tubes were again sonicated as described above constituting two rounds of PMCA. This process was repeated four times for a total of six rounds unless otherwise indicated. A positive amplification control of 1:100,000 CWD+ brain homogenate was amplified with each sample group. Negative controls include normal brain homogenate alone (NBH) and raw, municipal and commercially purchased water samples from non CWD-endemic areas, where indicated. Two to three researchers blinded to sample identification independently performed sPMCA experiments.
CWD-spiked water detection.
Deionized water samples were spiked with a 10% solution of CWD-positive brain made from a terminally sick deer (D10) or elk (E1 and E2). 2-fold dilutions were made spanning 1:1,600 through 1:4.2 × 108 then amplified by sPMCA. Serial ten-fold dilutions of E2 were made in raw negative control and test water samples where indicated.
PK digestion and western blot (WB).
Amplified samples were treated with 50 µg/ml of Proteinase-K (Roche) for 30 min at 45°C and PK was inactivated by the addition of lithium dodecyl sulfate loading buffer (Invitrogen) and heat inactivation for five minutes at 99°C. Samples were electrophoresed through 12% polyacrylamide gels (Invitrogen) in running buffer containing sodium dodecyl sulfate, transferred to Immobilon PVDF membrane (Millipore), blocked with 5% nonfat milk in 0.2% Tween 20 in PBS and probed with a 1:20,000 dilution of HRP-conjugated Bar 224 anti-PrP monoclonal antibody (SPI bio) in Superblock (Pierce). Membranes were washed 6 × 10 min with 0.2% Tween 20 in PBS, incubated for five minutes with enhanced chemiluminescent substrate (Millipore) and visualized using the FujiDoc gel documentation system (Fuji).
Western blot quantification.
Densitometry was performed using Quantity One software (Bio-Rad, Hercules, CA). Band intensity values were plotted against known concentrations of rCerPrP to generate a standard curve using Prism software (La Jolla, CA). PrPCWD concentrations were estimated by interpolation of band intensities to this standard curve (see Suppl. methods). Statistical analyses were performed using Prism and Excel (Microsoft).
Water quality analyses.
Total Organic Carbon (TOC) analysis was performed at the FCWTF Process Laboratory using a GE/Sievers 900 series TOC Analyzer with an autosampler. The UV/Persulfate Oxidation Method is capable of a analytical range of 30 parts per trillion (ppt) to 50 parts per million (ppm) for USEPA compliance monitoring applications in water treatment facilities. Total dissolved solids (TDS) and turbidity analyses were performed at the FCWTF Process Laboratory by standard gravimetric and nephelometric methods, respectively. Routine QA/QC analysis and instrument calibration was performed according to Standard Methods procedures.46
PK pre-treatment of 5-22-07 Cache La Poudre raw water samples.
Aliqouts of Cache La Poudre river water samples collected on 5-22-07 were incubated with 25 µg/mL PK for thirty minutes at 45°C, heat inactivated for ten minutes at 95°C, then subjected to six rounds of PMCA as previously described. A 1:100,000 dilution of PrPCWD in water was treated with heat-inactivated or active PK, then amplified to demonstrate the amplification competency of the NBH and removal of amplifiable PrPCWD, respectively.
Jar test.
A jar test was conducted using 500 ml of raw water collected at the FCWTF on 9-27-07. 58% Cache la Poudre River was mixed with 42% Horsetooth Reservoir water, producing a final TOC level of 3.25 mg/L. The combination of raw water and subsequent chemical additive concentrations were based on those in use by the Fort Collins water treatment plant the day of collection. Water was added to either a control chamber spiked with 1:40,000 normal brain homogenate from CWD negative Tg(cerPrP) mice or a treatment chamber containing a 1:40,000 dilution of CWD positive D10 deer brain homogenate. To simulate full scale flocculation, samples were stirred at 300 rotations per minute (RPM) in a Minimix laboratory mixer (model MLM4, Nova-Tech International, Houston, TX) and liquid alum (General Chemical Co., Parsippany, NJ) was quickly added to final concentration of 16 mg/L and mixed for 2 min. Stir speed was reduced to 65 RPM and Optimer 8110 PULV polymer (Nalco, Napervile, Il) was quickly added to a final concentration of 0.12 mg/L and mixed for 15 min. Mixing speed was sequentially reduced to 38 RPM for 15 min, then to 18 RPM for an additional 15 min. Mixing was stopped and flocculant allowed to settle for 15 min then centrifuged at 1,000 RPM for 5 min. The supernatant and flocculant were separated and amplified via PMCA for a total of six rounds.
Mouse bioassay.
All inocula were treated with UV irradiation, 100 units/mL Penicillin and 100 µg/mL Streptomycin (Gibco) in PBS for thirty minutes, then sonicated for forty seconds prior to intracerebral inoculations. NBH and infected brain homogenates (IBH) were diluted 1:10 in 320 mM Sucrose. Thirty microliters of each inoculum was injected intracerebrally 3 mm deep through the coronal suture 3–5 mm lateral of the sagittal suture. Tg(cerPrP)1536 and Tg(cerPrP)5037 mice were chronically exposed to 1 × 10−6 D10 in drinking water to mimic environmental exposure and inoculated with 100 µl of 2% IBH p.o. as positive controls. At 150 DPI mice were monitored daily for clinical symptoms of prion disease, including tail rigidity, impaired extensor reflex, akinesia, tremors, ataxia and weight loss. Mice exhibiting any four of these symptoms or paralysis were scored terminally sick and euthanized.
Cloning, expression and purification of recombinant PrP (rPrP).
PCR reagents, spin column kits and Ni-NTA Superflow resin were from Qiagen; restriction enzymes from New England Biolabs; protease inhibitor (Complete EDTA-free) from Roche Applied Science; Snakeskin pleated 7K MWCO dialysis tubing from Pierce Biotechnology; Ultracel 10K filters from Amicon; DNA Ligation kit, pET-30a(+) vector DNA, E. coli NovaBlue and BL21(DE3) competent cells, kanamycin sulfate, BugBuster protein extraction reagent, and lysonase bioprocessing reagent were from EMD Biosciences.
Standard cloning procedures were used to construct plasmids. To avoid co-amplification of mule deer PrP pseudogene sequences with the functional PrP gene, PCR was carried out first on a mule deer genomic DNA template using primers MD582F and 1454RC that flank the functional PrP open reading frame only. Diluted product from that PCR was used as template with PCR primers 5′MD NdeI and 3′MD H3 (5′CCC ATA TGA AGA AGC GAC CAA AAC CTG GAG 3′ and 5′ GCA AGC TTT ACT AAC TTG CCC CTC TTT GGT 3′, respectively) designed to produce a DNA fragment that encodes the deduced mule deer mature peptide composed of amino acids 25 through 234, flanked upstream by an NdeI recognition sequence containing an in-frame methionine translation initiation codon, and downstream by two in-frame stop codons followed by the Hind III recognition sequence.
Following digestion with Hind III and Nde I and spin column purification, fragments were ligated into the Nde I and Hind III sites of plasmid vector pET-30a(+), and NovaBlue competent cells were transformed with ligation reaction for acquisition of kanamycin resistance. Plasmids containing correctly sized inserts released by Nde I-Hind III double digestion from ten isolates were purified and submitted to the University of Wyoming Nucleic Acid Sequence Facility for DNA sequencing. Clone MD29, containing an insert whose sequence corresponds exactly to the previously determined DNA sequence of mule deer PrP codons 25-234 (GenBank Accession # AY228473) was selected for further experimental use.
MD rPrP was purified from cultures of pET-MD29-transformed BL21(DE3) cells that were induced with 1 mM IPTG during log phase growth at 37°C (OD600 0.6–1.0) and incubated for 6 hours. Cell pellets were collected by centrifugation and stored at −70°C. All purification steps including bacterial cell lysis, solubilization of inclusion body pellet, and protein purification by nickel affinity column liquid chromatography (AKTA Explorer system, GE Healthcare), dialysis and concentration were carried out using reagents, equipment and procedures as previously described.47 Final concentrations were calculated from measurements of UV absorbance at 280 nm, using an extinction coefficient of 2.59 based on a deduced peptide molecular weight of 23,000 and a peptide molar absorption coefficient of 59,610 M−1cm−1 approximated by the weighted sum of amino acid molar absorptivities at 280 nm.
Acknowledgements
The authors thank the North American Deer Farmers Association, the Colorado Section of the American Water Resources Association, the City of Fort Collins Water Board and the Colorado State University College Research Council and Infectious Disease Initiative. We thank Tom Holley and Les Nichols for help with water collections, Kathleen Ganzer, Judy Billica and Grant Jones for help with water quality analyses and Terry Spraker for CWD-infected brain samples, technical assistance and advice.
Abbreviations
- CWD
chronic wasting disease
- sPMCA
serial protein misfolding cyclic amplification
- PrPC
cellular prion protein
- PrPSc
disease-related, misfolded murine PrP
- PrPCWD
disease-related, misfolded cervid PrP
- PrPRES
protease-resistant PrP
- FCWTF
Fort Collins water treatment facility
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/9819
Supplementary Material
References
- 1.Johnson CJ, Phillips KE, Schramm PT, McKenzie D, Aiken JM, Pedersen JA. Prions adhere to soil minerals and remain infectious. PLoS Pathog. 2006;2:32. doi: 10.1371/journal.ppat.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller MW, Williams ES, Hobbs NT, Wolfe LL. Environmental sources of prion transmission in mule deer. Emerg Infect Dis. 2004;10:1003–1006. doi: 10.3201/eid1006.040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidel B, Thomzig A, Buschmann A, Groschup MH, Peters R, Beekes M, et al. Scrapie Agent (Strain 263K) can transmit disease via the oral route after persistence in soil over years. PLoS ONE. 2007;2:435. doi: 10.1371/journal.pone.0000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinckley GT, Johnson CJ, Jacobson KH, Bartholomay C, McMahon KD, McKenzie D, et al. Persistence of pathogenic prion protein during simulated wastewater treatment processes. Environ Sci Technol. 2008;42:5254–5259. doi: 10.1021/es703186e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathiason CK, Powers JG, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, et al. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science. 2006;314:133–136. doi: 10.1126/science.1132661. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Romero D, Barria MA, Leon P, Morales R, Soto C. Detection of infectious prions in urine. FEBS Lett. 2008;582:3161–3166. doi: 10.1016/j.febslet.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safar JG, Lessard P, Tamguney G, Freyman Y, Deering C, Letessier F, et al. Transmission and detection of prions in feces. J Infect Dis. 2008;198:81–89. doi: 10.1086/588193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams ES, Young S. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis. 1980;16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 9.Colorado Division of Wildlife, author. CWD Info and Testing. Fort Collins; 2009. http://wildlife.state.co.us/Hunting/BigGame/CWD/ [Google Scholar]
- 10.Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 11.Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 12.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 13.Angers RC, Browning SR, Seward TS, Sigurdson CJ, Miller MW, Hoover EA, et al. Prions in skeletal muscles of deer with chronic wasting disease. Science. 2006;311:1117. doi: 10.1126/science.1122864. [DOI] [PubMed] [Google Scholar]
- 14.Williams ES. Chronic wasting disease. Vet Pathol. 2005;42:530–549. doi: 10.1354/vp.42-5-530. [DOI] [PubMed] [Google Scholar]
- 15.Jewell JE, Brown J, Kreeger T, Williams ES. Prion protein in cardiac muscle of elk (Cervus elaphus nelsoni) and white-tailed deer (Odocoileus virginianus) infected with chronic wasting disease. J Gen Virol. 2006;87:3443–3450. doi: 10.1099/vir.0.81777-0. [DOI] [PubMed] [Google Scholar]
- 16.Spraker TR, Zink RR, Cummings BA, Wild MA, Miller MW, O'Rourke KI. Comparison of histological lesions and immunohistochemical staining of proteinase-resistant prion protein in a naturally occurring spongiform encephalopathy of free-ranging mule deer (Odocoileus hemionus) with those of chronic wasting disease of captive mule deer. Vet Pathol. 2002;39:110–119. doi: 10.1354/vp.39-1-110. [DOI] [PubMed] [Google Scholar]
- 17.Johnson CJ, Pedersen JA, Chappell RJ, McKenzie D, Aiken JM. Oral transmissibility of prion disease is enhanced by binding to soil particles. PLoS Pathog. 2007;3:93. doi: 10.1371/journal.ppat.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maluquer de Motes C, Cano MJ, Torres JM, Pumarola M, Girones R. Detection and survival of prion agents in aquatic environments. Water Res. 2008 doi: 10.1016/j.watres.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 19.Boyer EW, Hornberger GM, Bencala KE, McKnight DM. Effects of asynchronous snowmelt on flushing of dissolved organic carbon: a mixing model approach. Hydrological Processes. 2000;14:3291–3308. [Google Scholar]
- 20.Rand GM, Wells PG, McCarty LS. Introduction to Aquatic Toxicology. In: Rand GM, editor. Fundamentals of Aquatic Toxicology: Effects, Environmental Fate and Risk Assessment. Philadelphia: Taylor and Francis; 1995. pp. 11–12. [Google Scholar]
- 21.Schumacher BA National ESD, editor. Methods for the determination of total organic carbon (TOC) in soils and sediment. EPA; 2002. [Google Scholar]
- 22.Castilla J, Saa P, Soto C. Detection of prions in blood. Nat Med. 2005;11:982–985. doi: 10.1038/nm1286. [DOI] [PubMed] [Google Scholar]
- 23.Saa P, Castilla J, Soto C. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J Biol Chem. 2006;281:35245–35252. doi: 10.1074/jbc.M603964200. [DOI] [PubMed] [Google Scholar]
- 24.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 25.Soto C, Anderes L, Suardi S, Cardone F, Castilla J, Frossard MJ, et al. Pre-symptomatic detection of prions by cyclic amplification of protein misfolding. FEBS Lett. 2005;579:638–642. doi: 10.1016/j.febslet.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Kurt TD, Perrott MR, Wilusz CJ, Wilusz J, Supattapone S, Telling GC, et al. Efficient in vitro amplification of chronic wasting disease PrPRES. J Virol. 2007;81:9605–9608. doi: 10.1128/JVI.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerett C, Michel B, Pulford B, Spraker TR, Nichols TA, Johnson T, et al. In vitro strain adaptation of CWD prions by serial protein misfolding cyclic amplification. Virology. 2008;382:267–276. doi: 10.1016/j.virol.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browning SR, Mason GL, Seward T, Green M, Eliason GA, Mathiason C, et al. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J Virol. 2004;78:13345–13350. doi: 10.1128/JVI.78.23.13345-13350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angers RC, Seward TS, Napier D, Green M, Hoover E, Spraker T, et al. Chronic wasting disease prions in elk antler velvet. Emerg Infect Dis. 2009;15:696–703. doi: 10.3201/eid1505.081458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathiason CK, Hays SA, Powers J, Hayes-Klug J, Langenberg J, Dahmes SJ, et al. Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS One. 2009;4:5916. doi: 10.1371/journal.pone.0005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barria MA, Mukherjee A, Gonzalez-Romero D, Morales R, Soto C. De novo generation of infectious prions in vitro produces a new disease phenotype. PLoS Pathog. 2009;5:1000421. doi: 10.1371/journal.ppat.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders SE, Bartz JC, Bartelt-Hunt SL. Influence of Prion Strain on Prion Protein Adsorption to Soil in a Competitive Matrix. Environmental Science & Technology. 2009;43:5242–5248. doi: 10.1021/es900502f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurt TD, Telling GC, Zabel MD, Hoover EA. Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology. 2009;387:235–243. doi: 10.1016/j.virol.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 36.Haley NJ, Seelig DM, Zabel MD, Telling GC, Hoover EA. Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS One. 2009;4:4848. doi: 10.1371/journal.pone.0004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colorado Division of Water Resources, author. Fort Collins; 2009. http://www.dwr.state.co.us/SurfaceWater/data/detail_graph.aspx?ID=CLAFTCCO&MTYPE=DISCHRG. [Google Scholar]
- 38.Murayama Y, Yoshioka M, Okada H, Takata M, Yokoyama T, Mohri S. Urinary excretion and blood level of prions in scrapie-infected hamsters. J Gen Virol. 2007;88:2890–2898. doi: 10.1099/vir.0.82786-0. [DOI] [PubMed] [Google Scholar]
- 39.Eberhardt L, Van Etten RC. Evaluation of the pellet group count as a deer census method. J Wildl Manage. 1956;20:70–74. [Google Scholar]
- 40.McCullough DR. White-tailed deer pellet-group weights. J Wildl Manage. 1982;46:829–832. [Google Scholar]
- 41.Neff DJ, Wallmo OC, Morrison DC. A determination of defecation rate for elk. J Wildl Manage. 1965;29:406–407. [Google Scholar]
- 42.Swenson MJ. Dukes' Physiology of Domestic Animals. Ithaca: Comstock Publishing Associates; 1970. [Google Scholar]
- 43.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 44.Prusiner SB, Hadlow WJ, Eklund CM, Race RE, Cochran SP. Sedimentation characteristics of the scrapie agent from murine spleen and brain. Biochemistry. 1978;17:4987–4992. doi: 10.1021/bi00616a020. [DOI] [PubMed] [Google Scholar]
- 45.Peretz D, Supattapone S, Giles K, Vergara J, Freyman Y, Lessard P, et al. Inactivation of prions by acidic sodium dodecyl sulfate. J Virol. 2006;80:322–331. doi: 10.1128/JVI.80.1.322-331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.APHA, AWWA and WEF, author. Standard Methods for the Examination of Water and Wastewater. Washington: 1998. [Google Scholar]
- 47.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007;4:645–650. doi: 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- 48.Google Earth. 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.