Abstract
Expression of the three SBE genes, encoding starch branching enzymes, in the sorghum endosperm exhibited a diurnal rhythm during a 24-h cycle. Remarkably, the oscillation in SBE expression was maintained in cultured spikes after a 48-h dark treatment, also when fed a continuous solution of sucrose or abscisic acid. Our findings suggest that the rhythmicity in SBE expression in the endosperm is independent of cues from the photosynthetic source and that the oscillator resides within the endosperm itself.
Key words: barley, circadian rhythms, diurnal regulation, endosperm, oscillation, sbe, sorghum, starch
Starch, is the predominant storage carbohydrate in plants and the second most abundant biopolymer on earth, after cellulose. The path of starch synthesis rests on the conversion of sucrose to ADP-glucose (ADPG) and subsequent formation of the α-1,4 polyglucans amylose and amylopectin. Amylose is an essentially linear α-1,4 glucan with few α-1,6 glucosidic branches while amylopectin is highly branched. Granule-bound starch synthase (GBSS) catalyses the formation of new glucosidic linkages in amylose by transferring the glucose residue of ADPG to the non-reducing end of an existing α-1,4-linked glucan chain, thereby elongating it. Soluble starch synthase (SS) performs the same action for amylopectin. Branch points are introduced by starch branching enzyme (SBE), and debranching enzyme (DBE) may be involved in tailoring the branched glucans into a form capable of crystallization. All enzymes, including AGPase, exist in several distinct isoforms with specific roles in different organs and during different developmental stages. Other enzymes, such as glucan water dikinase (GWDK), starch phosphorylase (SP) and the D-enzyme, also participate in starch synthesis although their functions remain to be resolved. In the plant, starch is deposited as granules in chloroplasts of source organs such as leaves (transitory starch) or in amyloplasts of sink organs such as seeds, tubers and roots (storage starch). Typically, starch granules consist of 70% amylopectin and 30% amylose. Pakaging of amylopectin and amylose in starch granules is an intricate and complex process that is not fully understood. (reviewed in ref. 1).
Photocycles, thermocycles, and the circadian clock together phase cellular activities in plants to generate diurnal oscillations in gene expression, metabolism and physiology.2,3 In Arabidopsis leaves, genes encoding enzymes involved in starch metabolism, particularly degradation, showed diurnal expression patterns.4 Circadian regulation of the GBSSI gene in leaves has been reported for Arabidopsis,5 sweet potato6 and snapdragon.7 Diurnal oscillations of starch synthesis gene expression has been observed also in sink organs; for the growth ring formation starch granules in potato tubers,8 for the gene encoding the catalytic subunit of AGPase in potato tubers,9 and the SBEI and SBEII genes in cassava storage roots.10,11
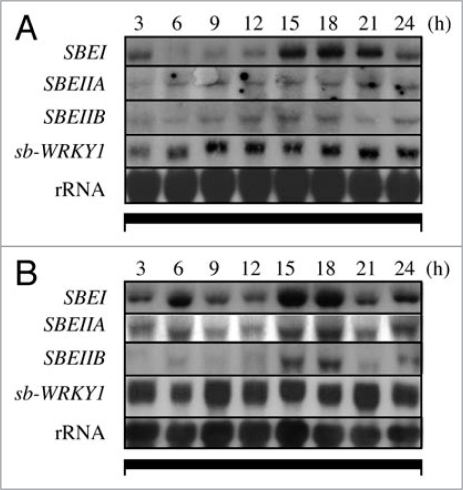
We have previously reported on the temporal and spatial expression profiles for the sorghum SBEI and SBEIIB genes.12,13 We have also shown that SBE expression levels in sorghum endosperm showed a diurnal fluctuation with an induction in the light and decline in the dark.14 To test whether the oscillations in endosperm transcript levels were acutely dependent on light or other cues from the source, we followed SBE expression in isolated seeds fed with a continuous supply of sucrose or ABA, two chemicals that have been shown to induce starch synthesis genes.11,15–17 For these experiments, plants were first transferred to darkness for 24 h to remove endogenous sucrose form the sink tissues. Spikes from the pre-conditioned plants were detached and cultured in water, sucrose or ABA in darkness. We found that even in sucrose or ABA-fed seeds, SBE expression in the endosperm oscillated with a period of approximately 24 h (Fig. 1). The oscillatory pattern was observed also in the absence of exogenous inducers and in seeds isolated from plants grown under continuous dark (DD) conditions, although the rhythm in SBE expression for DD plants was less overt as compared to light-dark grown (LD) plants or seeds supplied with sucrose or ABA (data not shown). These results indicate that, (1) the oscillation in SBE expression in the endosperm is not dependent on information from the source, and, (2) the oscillator resides in the endosperm tissues. The nature of the oscillator is unknown.
Figure 1.
Oscillation of SBe expression in endosperms from cultured spikes detached from sorghum plants pre-conditioned to darkness for 24 h. Spikes were supplied with a continuous solution of 200 mm sucrose (a) or 10 µm ABA (B) and cultured for 24 h in darkness prior to sampling.
Acknowledgements
This study was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas), Sida/SAREC under the auspices of the East Africa Regional Program and Research Network for Biotechnology, Biosafety and Biotechnology Policy Development program (BIO-EARN), and in part by U.S. Department of Energy Contract DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory.
Abbreviations
- ABA
abscisic acid
- GBSSI
granule-bound starch synthase I
- LD
light-dark cycle
- DD
continuous dark
- SBE
starch branching enzyme
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9424
References
- 1.Tetlow IJ, Morell MK, Emes MJ. Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot. 2004;55:2131–2145. doi: 10.1093/jxb/erh248. [DOI] [PubMed] [Google Scholar]
- 2.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. Plos Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Ann Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 4.Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenorio G, Orea A, Romero JM, Merida A. Oscillation of mRNA level and activity of granule-bound starch synthase I in Arabidopsis leaves during the day/night cycle. Plant Mol Biol. 2003;51:949–958. doi: 10.1023/a:1023053420632. [DOI] [PubMed] [Google Scholar]
- 6.Wang SJ, Yeh KW, Tsai CY. Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 2001;161:635–644. [Google Scholar]
- 7.Merida A, Rodriguez-Galan JM, Vincent C, Romero JM. Expression of the granule-bound starch synthase I (Waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant Physiol. 1999;120:401–409. doi: 10.1104/pp.120.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilling E, Smith AM. Growth ring formation in the starch granules of potato tubers. Plant Physiol. 2003;132:365–371. doi: 10.1104/pp.102.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geigenberger P, Stitt M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000;23:795–806. doi: 10.1046/j.1365-313x.2000.00848.x. [DOI] [PubMed] [Google Scholar]
- 10.Baguma Y, Sun C, Ahlandsberg S, Mutisya J, Palmqvist S, Rubaihayo PR, et al. Expression patterns of the gene encoding starch branching enzyme II in the storage roots of cassava (Manihot esculenta Crantz) Plant Sci. 2003;164:833–839. [Google Scholar]
- 11.Baguma Y, Sun C, Borén M, Olsson H, Rosenquist S, Mutisya J, et al. Sugar-meiated semidian oscillation of gene expression in the cassava storage root regulates starch synthesis. Plant Signal Behav. 2008;3:439–445. doi: 10.4161/psb.3.7.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutisya J, Sathish P, Sun CX, Andersson L, Ahlandsberg S, Baguma Y, et al. Starch branching enzymes in sorghum (Sorghum bicolor) and barley (Hordeum vulgare): Comparative analyses of enzyme structure and gene expression. J Plant Physiol. 2003;160:921–930. doi: 10.1078/0176-1617-00960. [DOI] [PubMed] [Google Scholar]
- 13.Mutisya J, Sun CX, Palmqvist S, Baguma Y, Odhiambo B, Jansson C. Transcriptional regulation of the sbellb genes in sorghum (Sorghum bicolor) and barley (Hordeum vulgare): Importance of the barley sbellb second intron. J Plant Physiol. 2006;163:770–780. doi: 10.1016/j.jplph.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 14.Mutisya J, Sun C, Rosenquist S, Baguma Y, Jansson C. Diurnal oscillation of SBE expression in sorghum endosperm. J Plant Physiol. 2009;166:428–434. doi: 10.1016/j.jplph.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Finkelstein RR, Gibson SI. ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol. 2002;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- 16.Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–433. doi: 10.1046/j.1365-313x.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- 17.Sun CX, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]