Abstract
The COP9 signalosome (CSN) is a conserved eukaryotic protein complex implicated in the regulation of cullin-RING type E3 ubiquitin ligases by cleaving the small peptide RUB/Nedd8 from cullins. However, detailed analysis of CSN physiological functions in Arabidopsis has been hampered by the early seedling-lethality of csn null mutants. We and others have now identified a number of viable hypomorphic csn mutants which start to reveal novel CSN-dependent activities in adult Arabidopsis plants.1 Here, we present a detailed comparative analysis of the csn5a-1 and csn2-5 mutants as a mean to improve understanding of CSN functions in plant cells. Our observations point to CSN-independent activities of CSN5 and suggest a role of the CSN in cytoskeleton assembly/organization.
Key words: Arabidopsis, root skewing, CSN, COP9 signalosome, SCF, ubiquitin, TIR1, auxin
Introduction
The control of protein turnover is crucial for virtually all physiological processes. The small peptide ubiquitin can be conjugated to target proteins through the sequential actions of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin protein ligases (E3) usually to induce their degradation by the 26S proteasome.2 E3s determine substrate specificity of the ubiquitination machinery, and the largest class of E3s is represented by the cullin-RING ligases (CRLs). According to sequence-based predictions, Arabidopsis expresses approximately 800 different CRLs potentially regulating the abundance of an even larger array of substrate proteins. CRLs all share a common architecture: The catalytically active RING protein RBX1 (Hrt1/Roc1) and a substrate-recognition subunit are brought together on a cullin protein (reviewed in ref. 3). However, cullins not only act as a passive scaffold since modification of cullins by the ubiquitin-like peptide RUB/Nedd8 is needed for CRL activities and is essential for viability in many different organisms. Recent structural work showed that major conformational changes occur upon cullin neddylation, leading to an open and catalytically active conformation.4 Reciprocally, mutants lacking the COP9 signalosome (CSN), which cleaves RUB/Nedd8 from cullins, exhibit pleiotropic phenotypes that culminate in Arabidopsis in early seedling lethality. CSN is an eight subunit complex conserved among eukaryotes and deletion of one of its subunit leads to complete loss of CSN activity in Arabidopsis. This has largely hampered analyses on CSN functions. The identification of viable csn reduced function (hypomorphic) mutant lines has allowed analysis of developmental and physiological responses in more mature plants and, for example, revealed the importance of CSN in responses to the plant hormone auxin.5–7
We identified an additional hypomorphic csn mutant in a screen for enhancers of the auxin resistance phenotype of sgt1b-3 that carries a mutation in one of the two functional Arabidopsis SGT1 (suppressor of G2 allele of skp1) genes.1 SGT1 was initially identified in yeast as a dose-dependent suppressor of skp1-4, a mutation resulting in defects in kinetochore function, and was also shown to associate with cullin1-based SCF-type (Skp, Cullin, F-Box) CRLs in different organisms.8–10 In Arabidopsis, the response to auxin is mediated by the well-characterized CRLs SCFTIR1/AFB,11,12 but the precise role of SGT1 in auxin signaling remains unclear. Our newly identified csn2-5 mutant carries a point mutation in the gene encoding the CSN2 subunit, one of the six PCI (proteasome, COP9 signalosome, eIF3) domain-containing subunits of CSN. Although the resulting mutant protein accumulates at reduced levels some intact CSN can still form in the csn2-5 mutant, seen as an intermediate accumulation of neddylated cullins compared to wild type and csn null mutants. We used this mild mutant to analyze further the regulation of SCFTIR1 as an archetypal CRL. We show that CUL1 and the F box protein TIR1, the substrate receptor of SCFTIR1 complex, are destabilized in csn2-5 mutant tissues, thereby providing a possible explanation for the hitherto poorly understood auxin resistance phenotype of Arabidopsis csn mutants. Our results further point to posttranslational modification of TIR1 (most likely by ubiquitination) and the proteasome-mediated degradation of TIR1, ASK1 and CUL1. Similar observations had previously been reported for other organisms.13–15 However, while the ubiquitination and subsequent degradation of substrate receptor proteins is well established in various systems, the reduced accumulation or increased turnover of other CRL components has remained a subject of debate that could not be rationalized until now. In this addendum, we discuss some additional observations made during the characterization of csn2-5 mutant plants that provide insight to CSN functions and implicate the CSN in novel developmental processes.
Different Phenotypes ofHypomorphic csn2 and csn5 Mutants: Evidence for CSN-Independent Functionsof CSN5
The archetypal CSN present in Arabidopsis is composed of six PCI and two MPN (MPR1, PAD1 N terminal) domain subunits. Among these subunits, CSN5 has some unique features: not only does it harbor the metallopeptidase activity needed for cullin deneddylation16 but it is also the only subunit that is fully stable and detectable as a monomer or in smaller subcomplexes in wild type extracts (reviewed in ref. 17). Also, CSN5 was the only CSN subunit initially identified by sequence comparisons in budding yeast which possesses a more divergent CSN-like complex.18,19 These points suggest that CSN5 might fulfill additional CSN-independent functions inside the cell as supported by studies performed in animal cell culture systems. For example, ectopic expression of HA-Jab1 (CSN5) led to downregulation of the cell cycle regulator p27, although it did not change cullin neddylation and was not detectably incorporated into CSN, but rather present as a monomer and part of a smaller subcomplex.20 In plants, however, no indications for CSN-independent roles of CSN5 have been provided so far and no differences between csn3, csn4 and csn5 mutants could be detected by transcriptional profiling.21 The csn2-5 mutant and the csn5a-1 mutant (which carries a mutation in one of two Arabidopsis CSN5 loci) both accumulate reduced amounts of CSN, but while the CSN5 monomer is unaffected by csn2-5, it is barely detectable in csn5a mutant lines.1,5,17 We compared these two mutants in different biochemical and physiological assays. Very similar results were obtained for defects in cullin deneddylation assessed by western blotting, and physiological assays for presumed CRL-mediated responses to the phytohormones auxin, jasmonate and ethylene. While csn5a-1 mutants displayed a stronger constitutive photomorphogenesis (cop) phenotype and were strongly dwarfed, csn2-5 plants were virtually indistinguishable from wild type (see ref. 1). Thus, physiological defects of csn mutants may not correlate strictly with defects in cullin deneddylation. Also, relatively strong changes in cullin neddylation patterns can be tolerated without major phenotypic consequences. As the presence of the CSN5 monomer seems to be a major difference between the two mutants, CSN-independent functions of this protein, which are retained in csn2-5 mutant but lost in csn5a-1, might account for the above phenotypic differences. Monomeric CSN5 could have important functions in general development but would act independently of CSN's deneddylation activity and CRLs. However, while this explanation for the stronger phenotype of the csn5a-1 mutant is plausible, it should be noted that different results have been reported for Schizosaccharomyces pombe: csn1 and csn2 mutants exhibit more severe phenotypes than csn5 mutant strains.22 These differences highlight the importance of studying multiple model organisms to reach a more fundamental understanding of CSN functions.
Hypomorphic csn Mutants Exhibit Phenotypes Suggesting Defectsin Cytoskeleton Assembly
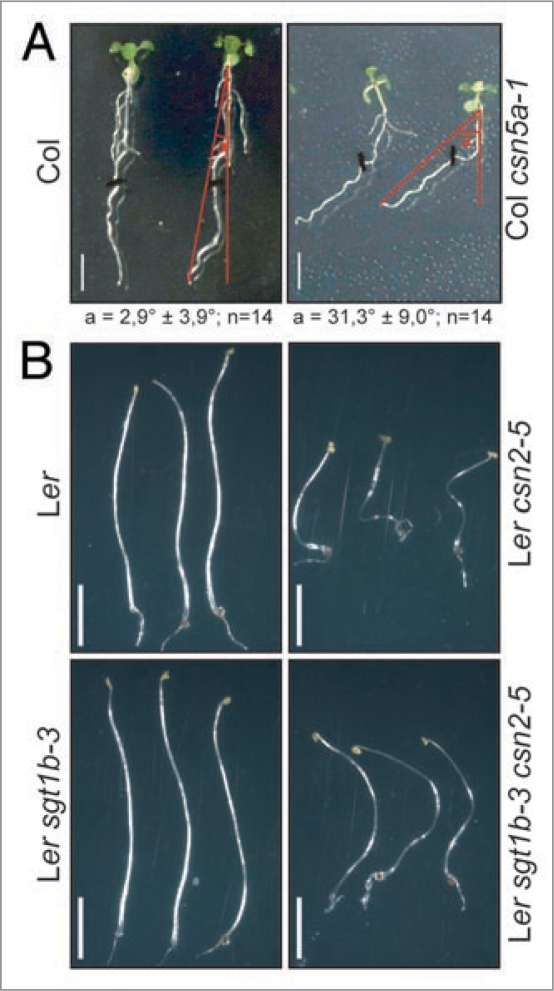
Auxin is a key player controlling lateral root formation (reviewed in ref. 23). Consistent with CSN's role as an indirect regulator of auxin signaling, defects in lateral root formation were previously described for CSN5 hypomorphic mutants.5,6 When we performed similar analyses with csn2-5 and csn5a-1, we detected differences in lateral root numbers, but these appeared to be mainly due to delayed and reduced growth of mutant primary roots, since no significant differences in lateral root density were found (data not shown). However, under these experimental conditions, we detected a skewed root growth phenotype for the csn5a-1 mutant: the average skewing angle of Col roots was approximately 3° compared to 31° of the csn5a-1 mutant (Fig. 1A; n = 14; p < 0.001). A similar phenotype was not observed for the Ler csn2-5 mutant, possibly due to more pronounced initial root skewing of wild type Ler plants (33.5° ± 7°). The basis for root skewing is currently not well understood, but auxin transport, gravity and root-gel contacts are considered to be of major importance (reviewed in ref. 24). Interestingly, exacerbated root skewing can be induced by cytoskeleton-affecting drugs, and several root skewing mutants are affected in genes regulating cytoskeleton stability. In parallel, we observed a ‘curly hypocotyl’ phenotype reminiscent of defects in cytoskeleton formation when seedlings were grown in darkness (Fig. 1B). Under these conditions, control plants grew straight, but hypocotyls of csn2-5 and csn2-5 sgt1b-3 double mutant seedlings grew in a helix-like shape performing typically 1–2 right-handed turns. In this case, a similar phenotype could not be detected for csn5a-1, but hypocotyls of csn5a-1 seedlings were extremely short because of its dwarfism and strong constitutive photo-morphogenesis (data not shown), which might mask such curly phenotype.
Figure 1.
Hypomorphic csn mutants exhibit phenotypes regularly associated with defects in microtubule assembly. (A) Plants were grown on inclined 0.1x MS plates for eight days and photographed from above. The skewing angle was measured from the root tip to a vertical vector as indicated in the pictures. (B) Plants were grown on horizontal 0.1x MS plates without sucrose for six days in darkness and transferred on a new plate for documentation. Scale bar = 0.5 cm.
Both the observed root skewing and ‘curly hypocotyls’ could result indirectly from defects in responses to hormones such as auxin and ethylene (reviewed in ref. 24). But although csn mutations confer auxin resistance, we did not detect altered gravitropism response or altered ethylene response (data not shown) in csn5a-1 and csn2-5. Besides, tir1 mutants did not present any agravitropic or modified skewing phenotypes. Therefore, it would be interesting to analyze cytoskeleton assembly and organization in hypomorphic csn mutants further and assess, for example, orientation of cellulose fibrils along the cell wall, which follows the progression of cortical microtubules (reviewed in ref. 25). Such observations would be of particular interest since CRLs and the CSN-dependent Nedd8 deconjugation pathways have been directly implicated in cytoskel eton formation in C. elegans.26,27
Acknowledgements
J.S. and J.E.P. are grateful to the Deutsche Forschungsgemeinschaft for SFB grant 635.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9526
References
- 1.Stuttmann J, Lechner E, Guérois R, Parker JE, Nussaume L, Genschik P, et al. COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J Biol Chem. 2009;284:7920–7930. doi: 10.1074/jbc.M809069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohmann EM, Kuhnle C, Schwechheimer C. Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell. 2005;17:1967–1978. doi: 10.1105/tpc.105.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Ito H, Quint M, Huang H, Noël LD, Gray WM. Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc Natl Acad Sci USA. 2008;105:8470–8475. doi: 10.1073/pnas.0804144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Heiter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 9.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell. 2002;14:1483–1496. doi: 10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 12.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 14.Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Q, Cheng P, He Q, Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 17.Gusmaroli G, Figueroa P, Serino G, Deng XW. Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell. 2007;19:564–581. doi: 10.1105/tpc.106.047571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maytal-Kivity V, Pick E, Piran R, Hofmann K, Glickman MH. The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int J Biochem Cell Biol. 2003;35:706–715. doi: 10.1016/s1357-2725(02)00378-3. [DOI] [PubMed] [Google Scholar]
- 19.Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002;3:15. doi: 10.1186/1471-2156-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomoda K, Kato JY, Tatsumi E, Takahashi T, Matsuo Y, Yoneda-Kato N. The Jab1/COP9 signalosome sub-complex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell cycle progression. Blood. 2005;105:775–783. doi: 10.1182/blood-2004-04-1242. [DOI] [PubMed] [Google Scholar]
- 21.Dohmann EM, Levesque MP, Isono E, Schmid M, Schwechheimer C. Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Physiol. 2008;147:1369–1379. doi: 10.1104/pp.108.121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mundt KE, Liu C, Carr AM. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Smet I, Vanneste S, Inze D, Beeckman T. Lateral root initiation or the birth of a new meristem. Plant Mol Biol. 2006;60:871–887. doi: 10.1007/s11103-005-4547-2. [DOI] [PubMed] [Google Scholar]
- 24.Oliva M, Dunand C. Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol. 2007;176:37–43. doi: 10.1111/j.1469-8137.2007.02184.x. [DOI] [PubMed] [Google Scholar]
- 25.Thitamadee S, Tuchihara K, Hashimoto T. Microtubule basis for left-handed helical growth in Arabidopsis. Nature. 2002;417:193–196. doi: 10.1038/417193a. [DOI] [PubMed] [Google Scholar]
- 26.Kurz T, Pintard L, Willis JH, Hamill DR, Gonczy P, Peter M, et al. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science. 2002;295:1294–1298. doi: 10.1126/science.1067765. [DOI] [PubMed] [Google Scholar]
- 27.Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitinligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]