Abstract
At least theoretically, plants may synthesize nitric oxide (NO) either by reduction of N in higher oxidations states, or by oxidation of more reduced N-compounds. The well established reductive pathway uses nitrite as a substrate, produced by cytosolic nitrate reductase. The only oxidative pathway described so far comprises nitric oxide synthase (NOS)-like activity, where guanidino-N from L-arginine is oxidized to NO. In our previous paper we have demonstrated yet another form of oxidative NO formation, whereby hydroxylamine (HA), but also the AOX-inhibitor salicylhydroxamate (SHAM) is oxidized to NO by tobacco suspension cells. Oxidation of HA to NO was also demonstrated in vitro by using ROS producing enzymes. Apparently superoxide radicals and/or hydrogen peroxide served as oxidants. Another unexpected observation pointed to a special role for superoxide dismutase in oxidative NO formation.
Key words: hydroxylamine, nitric oxide, oxidative NO formation, reactive oxygen species, salicyl hydroxamate, superoxide dismutase
Introduction
In animals the free radical NO is generated in a well characterized oxygen- and NADPH-dependent reaction from the amino acid L-arginine by the enzyme family of nitric oxide synthases (reviewed in ref. 1). Because plants appear basically able to grow and to complete their life cycle in the absence of nitrate and nitrite, e.g., with ammonium as the only N-source, they must possess nitrite-independent, oxidative pathways for NO production. Indeed, numerous publications have given indirect hints that a “plant NOS” might exist, but the potential NOS gene in Arabidopsis (AtNOS1) has been renamed recently into AtNOA1 (NO-associated 1), in order to make clear that AtNOS 1 has no NOS activity but is somehow involved in regulating NO and NO dependent reactions, possibly via formation of cGMP as secondary messenger.2
In animals, besides L-arginine, hydroxylamine was considered another substrate for oxidative NO formation in animals.3 HA was shown to cause vasodilatation just like NO-releasing compounds.4 Hydroxylamine is also an important intermediate in the process of nitrification, the conversion of ammonia to nitrite. Bacteria are able to oxidize HA to nitrite or even directly to NO5 via the enzyme hydroxylamine- oxidoreductase. All these facts led us to the question if similar reactions would exist in plants.
Plant Cells Oxidize Hydroxylamines to NO
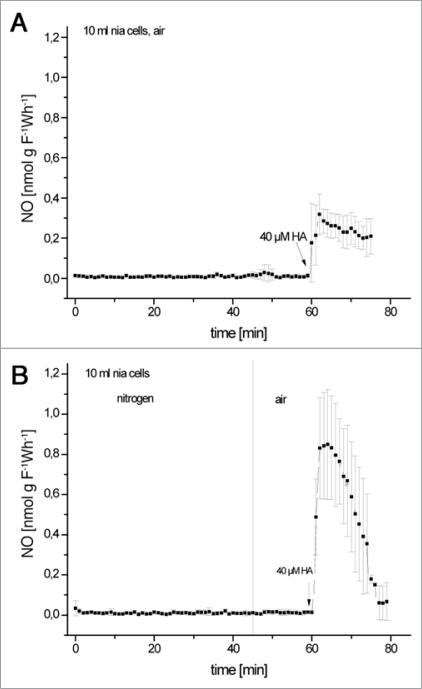
We followed NO-emission by gas-phase chemiluminescence from wildtype tobacco suspension cells and from cell cultures of a nia double mutant (nia 30) which is deficient in nitrate reductase, has negligible nitrite-concentrations and which should therefore be unable to produce NO in a reductive way. When HA was applied to cells in concentrations of 4 or 40 µM, NO was emitted. This was not the case when the cells were kept in nitrogen, suggesting that the process is oxygen-dependent. Salicylhydroxamat (SHAM), which is very often used as an inhibitor of alternative oxidase (AOX), was also oxidized to NO.
Suspecting reactive oxygen species (ROS) as oxidants for hydroxylamines, we provoked formation of ROS by tobacco suspension cells e.g., by treatment with heavy metals or elicitors, which increased NO emission after HA or SHAM addition. Here (Fig. 1) we show in addition, that NO-emission from hydroxylamine was also increased when cells were first exposed to anoxia and then re-oxygenated, a condition also known to provoke ROS formation.6
Figure 1.
Rates of NO-emission from 40 µm Ha by tobacco suspension cells exposed to NO-free air for 1 hour (A) or anoxia for 45 minutes and re-oxygenation for 15 minutes (B). Bars give Sd (n = 4).
Cell-Free System
Oxidation of hydroxylamines to NO was also observed in cell-free enzyme solutions containing a superoxide-generating system, xanthine oxidase (XOD) plus xanthine. NO may be rapidly oxidized, even more in the presence of ROS, or may react with organic cell constituents, thereby escaping detection with chemiluminescence. We therefore also analyzed the oxidation products of NO, nitrite and (here) nitrate. Vanadium(III)-chloride (100 mM in 1 M HCl) was used to reduce nitrate to nitrite at 70°C, the latter then reacts with added Griess reagents. With this assay we indeed observed nitrite and nitrate formation in addition to NO (Table 1). In the in vitro-system with XOD as a source for superoxide radicals, we also added superoxide dismutase (SOD) to decompose O2.− to H2O2 and O2. Unexpectedly, SOD did not abolish, but strongly increased NO emission and nitrite/nitrate formation from HA + XOD (Table 1). However, addition of hydrogen peroxide (100 µM, not shown) or of a hydrogen peroxide-producing enzyme system (GOD + glucose) to a solution of HA or SHAM produced only a minor NO-emission. Thus it appears that SOD interacts with NO production in an unknown way, probably independent of hydrogen peroxide formation.
Table 1.
Formation of NO, nitrite and nitrate from 40 µm HA and 50 µm SHAM, in solutions containing ROS-forming and/or scavenging enzymes
| Reaction mixture | NO-emission [ppb] | Nitrite formation [nmoles/10 ml] | Nitrite + nitrate formation |
| + HA (400 nmoles/10 ml) | |||
| no addition | 0.08 ± 0.02 | 1.57 ± 0.25 | 25.4 ± 4.4 |
| + XOD + xanthine | 1.16 ± 0.20 | 24.89 ± 0.59 | 97.6 ± 37.5 |
| + XOD + xanthine + SOD | 2.67 ± 0.52 | 142.68 ± 3.07 | 267.2 ± 5.62 |
| + SOD, air | 0.78 ± 0.23 | 2.82 ± 0.49 | 22.02 ± 11.02 |
| + GOD + glucose | 0.19 ± 0.07 | 7.70 ± 5.20 | Not measured |
| + SHAM (500 nmoles/10 ml) | |||
| no addition | 0.00 ± 0.02 | 1.86 ± 1.08 | 5.98 ± 1.74 |
| + XOD + xanthine | 0.19 ± 0.08 | 29.46 ± 0.29 | 50.80 ± 1.37 |
| + XOD + xanthine + SOD | 0.33 ± 0.07 | 20.66 ± 0.04 | 90.34 ± 19.03 |
| + SOD, air | 0.68 ± 0.10 | 3.12 ± 1.24 | 23.45 ± 5.89 |
| + GOD + glucose | 0.02 ± 0.01 | 7.10 ± 4.08 | Not measured |
While our work demonstrates that plant cells are basically able to oxidize hydroxylamines to nitric oxide, future experiments are required to decide whether HA's occur naturally in plants to serve as substrates for oxidative NO formation. The specific role of SOD for NO formation also deserves further attention.
Abbreviations
- cGMP
cyclic guanos-inmonophosphat
- HA
hydroxylamine
- NO
nitric oxide
- NOS
nitric oxide synthase
- SHAM
salicylhydroxamic acid
- SOD
superoxide dismutase
- XOD
xanthine oxidase
- LS
Linsmaier and Skoog
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9378
References
- 1.Marletta AM. Nitric oxide synthase: aspects concerning structure and catalysis. Cell. 1994;78:927–930. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 2.Moreau M, Lee GI, Wang Y, Crane BR, Klessig DF. AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J Biol Chem. 2008;283:32957–32967. doi: 10.1074/jbc.M804838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markert M, Carnal B, Mauël J. Nitric oxide production by activated human neutrophils exposed to sodium azide and hydroxylamine: The role of oxygen radicals. Biochem Biophys Res Commun. 1994;189:1245–1249. doi: 10.1006/bbrc.1994.1364. [DOI] [PubMed] [Google Scholar]
- 4.DeMaster EG, Raij L, Archer SL, Weir EK. Hydroxylamine is a vasorelaxant and a possible intermediate in the oxidative conversion of L-Arginine to nitric oxide. Biochem Biophys Res Commun. 1989;163:527–533. doi: 10.1016/0006-291x(89)92169-4. [DOI] [PubMed] [Google Scholar]
- 5.Hooper AB, Terry KR. Hydroxylamine oxidoreductase of nitrosomonas: production of nitric oxide from hydroxylamine. Biochim Biophys Acta. 1979;571:12–20. doi: 10.1016/0005-2744(79)90220-1. [DOI] [PubMed] [Google Scholar]
- 6.Blokhina O, Virolainen E, Fagerstedt KV. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot. 2003;91:179–194. doi: 10.1093/aob/mcf118. [DOI] [PMC free article] [PubMed] [Google Scholar]