Abstract
The bacteriophage T4 encodes proteins that are responsible for tightly regulating mRNA synthesis throughout phage development in Escherichia coli. The three classes of T4 promoters (early, middle, and late) are utilized sequentially by the host RNA polymerase as a result of phage-induced modifications. One such modification is the tight binding of the T4 AsiA protein to the σ70 subunit of the RNA polymerase. This interaction is pivotal for the transition between T4 early and middle transcription, since it both inhibits recognition of host and T4 early promoters and stimulates T4 middle mode synthesis. The activation of T4 middle transcription also requires the T4 MotA protein, bound specifically to its recognition sequence, the “Mot box,” which is centered at position −30 of these promoters. Accordingly, the two T4 proteins working in concert are sufficient to effectively switch the transcription specificity of the RNA polymerase holoenzyme. Herein, we investigate the mechanism of transcription activation and report that, while the presence of MotA and AsiA increases the initial recruitment of RNA polymerase to a T4 middle promoter, it does not alter the intrinsic stability of the discrete complexes formed. In addition, we have characterized the RNA polymerase-promoter species by UV laser footprinting and followed their evolution from open into initiating complexes. These data, combined with in vitro transcription assays, indicate that AsiA and MotA facilitate promoter escape, thereby stimulating the production of full-length transcripts.
Gene expression is highly regulated during bacteriophage T4 development, resulting in the sequential transcription of genes from the T4 early, middle, and late promoters (1). The T4 early promoters contain bacterial-like DNA consensus sequences located at −10 and −35 with respect to the start site of transcription. These promoter regions are recognized by the σ70 subunit of Escherichia coli RNA polymerase (RNAP), leading to transcription by the unmodified host enzyme (2). Shortly after infection, T4-encoded proteins begin to modify the E. coli RNAP. The α subunits of the RNAP are ADP-ribosylated by the phage proteins Alt and Mod, and the RNAP holoenzyme becomes tightly associated with the T4 proteins RpbA and AsiA.
The interaction between the T4 AsiA protein and σ70 is known to be crucial for the switch in transcription specificity (reviewed in ref. 3). The presence of AsiA decreases transcription from T4 early and bacterial promoters (4–7), whose efficient recognition depends on specific interactions between the σ70 subunit and promoter sequences at both −10 and −35. Those regions within bacterial σ factors responsible for promoter recognition have been determined; the conserved σ region 2.4 contacts the −10 element, whereas the conserved domain 4.2 interacts with the −35 hexamer (reviewed in ref. 8). Evidence that AsiA binds within region 4 of σ (9–11) has led to the proposal that interaction with AsiA alters the conformation of σ70 in a way that blocks the formation of specific σ70–DNA contacts near −35. This idea is substantiated by our recent work, which indicates that AsiA diminishes the initial promoter binding of the RNAP to a T4 early promoter (12).
In contrast, the σ70–AsiA interaction is necessary for the enhanced recognition of T4 middle promoters (7, 12–14). These promoters lack a conserved −35 hexamer and are characterized by a “Mot box” sequence, (t/a)(t/a)TGCTT(t/c)A, centered at −30, that is recognized by the phage-encoded DNA-binding protein MotA (15–17). T4 middle promoters also possess a strong −10 consensus element, which is often directly preceded by the “extended −10” motif, 5′-TGN-3′ (17). A number of promoters that display such extended −10 character are transcribed by RNAP in the absence of a −35 hexamer (18, 19), thus potentially explaining the low level of T4 middle synthesis by unmodified RNAP (14, 20). Although it has been demonstrated that AsiA confers MotA dependence on synthesis from these promoters (7), the mechanism by which the two T4 proteins work together to stimulate transcription has yet to be elucidated.
MotA and AsiA together function to increase the level of RNAP–DNA complexes formed, as visualized by electrophoretic mobility-shift assays at both 4°C and 37°C (12). Since DNA strand opening and thus open complex formation is unlikely at 4°C, these data suggest that transcription stimulation promoted by the T4 factors occurs at least in part in the formation of initial closed complexes. However, the increase in gel-retarded complexes observed in the presence of MotA and AsiA was less pronounced than the measured effect on transcription, leading to the hypothesis that AsiA and MotA may function at more than one specific step in the transcription pathway (12).
In this work, we investigate this possibility by following each of the discrete species formed stepwise through the following simplified scheme for transcription (21, 22). ![]()
where R, P, and RP represent RNAP, the promoter DNA, and the various RNAP–promoter complexes, respectively. RPC includes all complexes wherein the DNA in the promoter region is closed, while RPO signifies open promoter complexes. RPinit denotes those species undergoing the reiterative synthesis of short abortive products (AP), which is followed by promoter escape and the formation of the stable elongation complex, RPE.
MATERIALS AND METHODS
Materials.
MotA and AsiA proteins were purified as described previously (7, 12). Pure E. coli RNAP was purchased from Amersham-Pharmacia Biotech, or purified according to the method of Burgess and Jendrisak (23).
DNA Fragments and Oligonucleotides.
All PrIIB2 DNA fragments were obtained by performing PCR on T4 DNA. The 158-bp fragment used for UV laser crosslinking has been described (7); the 260-bp PrIIB2 fragment has an identical 5′ end on the nontemplate (NT) strand but carries an additional 102-bp downstream from the promoter.
BIAcore.
A 260-bp DNA fragment containing the PrIIB2 promoter was immobilized on a streptavidin surface, via a biotin at the 5′ end of the NT strand. Proteins were passed over this surface at room temperature (22°C) or 37°C (data not shown) for 1 min at a flow rate of 10 μl/min. The buffer was composed of 40 mM Hepes (pH 8.0), 100 mM KCl, 10 mM MgCl2, and 0.005% Surfactant p20. The quantity of RNAP bound to the DNA, expressed in resonance units (RU), was measured as a function of RNAP concentration (as indicated) in the presence of MotA and AsiA (each at 200 nM) or their absence. The nucleoprotein complexes remaining 1 min after the termination of injection were monitored for 5 min (t = 120–420 s) to assess the rate of dissociation (koff), which was determined by using the BIAcore Evaluation software provided. The proteins were also passed across a surface that lacked DNA, to control for bulk refractive index effects and nonspecific binding; quantities expressed have been corrected by subtraction of these values.
UV Laser Photofootprinting.
Complexes were formed in a 10 μl volume, in a buffer containing 10 mM Hepes (pH 8.0), 100 mM KCl, 10 mM MgCl2, 3% glycerol, and 5 mM DTT. A 158-bp DNA fragment (2.5 nM) was incubated for 15 min at 37°C with the following components as indicated: RNAP (10 nM), AsiA (200 nM), and MotA (200 nM). Identical experiments performed at 22°C (data not shown) showed that the static complexes formed at the two temperatures are the same. Where mentioned, the samples were challenged by heparin (100 μg/ml) for 1 min, before the addition of GTP and ATP, or CTP (1 mM each, final concentration) and continued incubation for 2 min. The samples were irradiated by a single pulse of high-intensity UV light (266 nm), which induces the rapid formation of covalent crosslinks (both DNA-DNA and DNA-protein) (24). The resulting modifications were detected by primer extension analysis as described by Buckle et al. (24).
In Vitro Transcription Assays.
Both abortive and run-off in vitro transcription assays were performed with a 260-bp fragment bearing the PrIIB2 promoter in standard transcription buffer (50 mM Tris⋅HCl, pH 7.9/100 mM KCl/10 mM MgCl2/0.1 mM EDTA/1 mM DTT). Abortive transcription was assayed after incubating the DNA (5 nM) with RNAP (100 nM) in the presence (200 nM) or absence of AsiA and/or MotA for 15 min in a 10-μl volume. Nucleotides (2 μl) were added at the following final concentrations: GTP (100 μM), UTP and ATP (200 μM), and [γ-32P]GTP (5 μCi at 4500 Ci/mmol; 1 Ci = 37 GBq); or UTP (25 μM), GTP and ATP (200 μM), and [α-32P]UTP (5 μCi at 3000 Ci/mmol); with heparin (200 μg/ml). Run-off transcription was performed in the same manner, with the exception that AsiA was added at increasing concentrations (as indicated) and that the NTP solution contained GTP (100 μM), UTP, ATP and CTP (200 μM), and [γ-32P]GTP (5 μCi at 4500 Ci/mmol). Synthesis was allowed for 10 min before quenching the reaction with 8 μl of stop solution (7 M urea). A small portion of each reaction mixture was resolved on a 25% polyacrylamide sequencing gel, and quantitation was carried out by using a PhosphorImager (Molecular Dynamics) and the ImageQuant software.
Promoter mutations used as controls are as follows: the −10 hexamer mutation (including the extended −10 region) changed TGATAAAAT to TTGGGAAAT, and the start site mutation bears a single nucleotide change at the +1 position (from GAG to AAG).
RESULTS
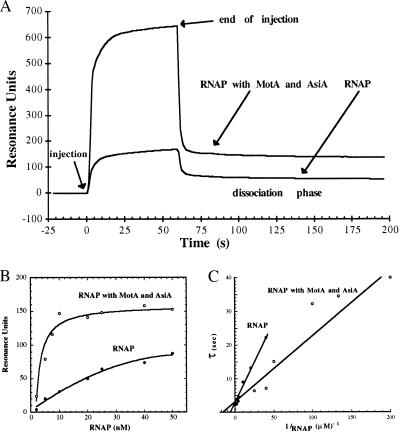
We employed surface plasmon resonance (SPR) techniques to assess the direct effects of the T4 proteins AsiA and MotA on binding of the E. coli RNAP to a T4 middle promoter. A DNA fragment carrying the T4 middle promoter PrIIB2 was immobilized on a streptavidin surface, by means of a 5′ biotin end-label. By passing the RNAP across this surface at increasing concentrations, both in the presence and in the absence of the T4 factors, we were able to measure the amount of RNAP bound to the promoter DNA at steady state, and the subsequent dissociation of the complexes formed. As shown in Fig. 1A, SPR analysis is consistent with previous data which demonstrate that AsiA and MotA function to recruit RNAP to the PrIIB2 promoter. A significant increase in the quantity of bound RNAP is observed only in the presence of both T4 proteins; the addition of MotA alone has no distinguishable effect on RNAP binding, whereas AsiA moderately decreases the quantity of RNAP bound to the promoter DNA (data not shown). The dissociation rate constant (koff) of RNAP complexes was determined with a single-exponential decay model. The value obtained for koff is the same, within experimental error, in the presence or absence of the T4 proteins: 4.18 (±0.67) × 10−4 s−1 for the RNAP; and 4.35 (±0.66) × 10−4 s−1 for the quaternary complexes in the presence of RNAP, MotA, and AsiA. Thus, while AsiA and MotA stimulate recognition of the PrIIB2 promoter, the stability of the RNAP–DNA complexes formed is not altered by the T4 proteins. In addition, these stable RNAP–DNA complexes (at either 22°C or 37°C) were not dissociated by challenge with heparin (data not shown), indicating that they are open complexes (12).
Figure 1.
Surface plasmon resonance analysis of RNAP binding to an immobilized T4 middle promoter. (A) The relative change in resonance angle, expressed as RU vs. time, is shown as RNAP (here 25 nM) is passed across the immobilized PrIIB2 DNA fragments, either alone or in the presence of an excess of both the AsiA and MotA proteins. The decrease in RU after injection reflects the real-time dissociation (koff) of the nucleoprotein complexes that remain. The values cited represent the average of five experiments each, performed over a range of RNAP concentrations between 10 and 50 nM. A semilogarithmic plot of the decrease in RU during this dissociation phase was linear with respect to time, and thus koff was determined by a direct fit of this phase to (A)t = (A)max⋅e−(koff⋅t), where (A)t is the concentration of nucleoprotein complex at time t; (A)max is the steady-state concentration of complex prior to dissociation (taken at t = 120 s); and koff is the dissociation rate constant. The relative contribution of MotA and AsiA, when present in stoichiometric amounts, is only 7% of the total mass represented by RNAP, and is therefore indiscernible, within experimental error. The (A)max values are expressed as a function of RNAP concentration, giving the binding isotherms shown in B. During the injection phase, the immediate initial increase in RU is due to the refractive index of the glycerol and salts present in the protein samples (t = 0–3 s). The ensuing increase in signal is due to the formation of RNAP–DNA complexes, which are in equilibrium with free RNAP in solution. Within a specific association period, the signal at time t, (A)t, may be expressed as (A)t = (A)max⋅(1 − e−(t/τ)), where τ = 1/k2(1 + 1/KB[RNAP]), where KB is the binding constant. This allows representation of the change in τ as a function of the change in RNAP concentration (22), as shown in C. The y intercept is thus 1/k2 and the x intercept is −KB, giving the following values of each: for RNAP k2 = 0.33 s−1, KB = 7.4 μM−1; RNAP plus the T4 proteins k2 = 0.3 s−1, KB = 17 μM−1.
Fig. 1B shows the steady-state response (RU) as a function of RNAP concentration both without (•) and with (○) the T4 proteins. This simple binding analysis shows differential degrees of promoter occupancy, characterized by distinct saturation curves. Taking into account the contribution to RU made by either protein or DNA (25), the steady-state signal observed at saturation (≈150 RU) correlates with a 1:1 stoichiometry for RNAP–promoter complexes. Since MotA and AsiA do not alter the dissociation rate constant (koff), the difference in apparent Kd values obtained (35.5 ± 8.7 nM for the RNAP and 4.7 ± 2 nM for the quaternary complexes) must denote effects on the association phase. Therefore, in spite of the inherent difficulties involved in measuring association events, we have determined the apparent association rates and expressed these as the inverse τ values (see legend to Fig. 1) as a function of RNAP concentration (Fig. 1C). As the final species is an open complex, this phase can be separated into two steps, the initial promoter binding (KB) and the isomerization to a more stable open complex (k2). Whereas the intercept of these plots with the ordinate (1/k2) does not appear to be altered by MotA and AsiA, that with the abscissa (−KB) is affected. Thus, these data indicate that stimulation of recognition occurs directly at KB.
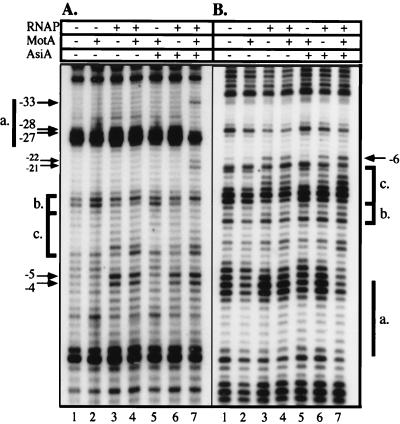
To characterize the species formed at the PrIIB2 promoter, we examined the discrete protein–DNA complexes by using UV laser photocrosslinking. PrIIB2 DNA was incubated for 15 min with RNAP, AsiA, and/or MotA as indicated, before irradiation by a short powerful UV pulse. Primer extension analysis was then performed on these samples, using labeled primers which were complementary to either the nontemplate (NT) or template (T) strand (Fig. 2 A and B, respectively). Assuming that primer extension will terminate before bases that have been modified by the UV irradiation (e.g., crosslinked), the resulting pattern will thus be specific to both the sequence and local structure of the DNA (24). If a given protein directly contacts the DNA, or the formation of nucleoprotein complexes leads to changes in the DNA conformation, this will be reflected by alterations in the photoreactivity, and thus the frequency of termination at that position (see Materials and Methods; ref. 24).
Figure 2.
UV laser photofootprinting of protein–DNA complexes at the PrIIB2 promoter. Complexes formed between the DNA and the indicated components were irradiated by a high-intensity UV pulse. Primer extension analysis was performed with primers complementary to the NT and T strands (A and B, respectively). For both panels, reaction mixtures contained DNA and the following: no proteins (lanes 1), MotA (lanes 2), RNAP (lanes 3), RNAP and MotA (lanes 4), MotA and AsiA (lanes 5), RNAP and AsiA (lanes 6), or RNAP plus MotA and AsiA (lanes 7). The location of the Mot box (a.) is indicated by a thick line. The extended −10 (b.) and −10 consensus regions (c.) are identified on each strand. Arrows denote the positions of selected characteristic changes in band intensity.
The irradiated PrIIB2 DNA alone shows a distinct termination pattern (Fig. 2, lanes 1), which is significantly altered neither by the presence of MotA (lanes 2) nor by the combination of MotA and AsiA (lanes 5). Upon the addition of RNAP, specific changes are seen in the footprinting pattern, most notably within the region between −20 and the transcription start site (lanes 3). The bands corresponding to positions −4 and −5 on the NT strand are significantly enhanced, as is the signal located at −6 on the T strand (marked by arrows). This pattern is not discernibly affected by the presence of MotA (lanes 4), while the RNAP plus AsiA gives a qualitatively similar photofootprint, but with a slightly weaker signal (lanes 6). Furthermore, the simultaneous presence of MotA and AsiA does not alter the pattern between positions −20 and +1. Hence, the structural characteristics of the RNAP–promoter complex appear to be identical within this region even in the presence of both of the T4 proteins. Challenge with heparin, which removes closed promoter complexes (12), does not alter the footprinting pattern of any of the RNAP–DNA species (see Fig. 3), confirming that these photofootprints represent open promoter complexes.
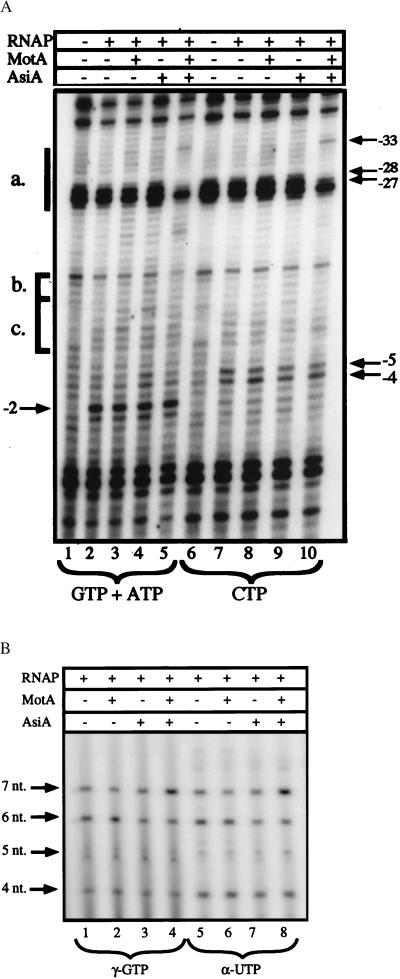
Figure 3.
Analysis of the RPO to RPinit transition by UV laser photofootprinting and in vitro abortive initiation assays. A shows the photofootprint of RNAP–PrIIB2 promoter complexes in the presence of either the initiating nucleotides GTP and ATP (lanes 1–5), which allow for the reiterative synthesis of a 3-nt product, or the nucleotide CTP (lanes 6–10), which should not be incorporated. The DNA was incubated for 15 min, either alone (lanes 1 and 6) or with the RNAP (lanes 2 and 7), in the presence of MotA (lanes 3 and 8), AsiA (lanes 4 and 9), and the two T4 proteins together (lanes 5 and 10). These complexes were challenged by heparin before the addition of the specified nucleotide(s), and the incubation was continued for 2 min. The samples were then irradiated and analyzed by primer extension on the NT strand. The Mot box (a.), extended −10 (b.), and −10 region (c.) are indicated as in Fig. 2. The positions of selected bands are marked by arrows. The abortive initiation products formed by the RPinit species are shown in B. Preformed complexes between PrIIB2 DNA (5 nM), RNAP (100 nM), and the T4 proteins (as indicated) were challenged by heparin, and supplied with selected NTPs. Lanes 1–4 received ATP and UTP at 200 μM, GTP at 100 μM, and 5 μCi of [γ-32P]GTP; lanes 5–8 received GTP and ATP at 200 μM, UTP at 25 μM, and 5 μCi of [α-32P]UTP. In both cases, this should allow for the synthesis of transcripts up to 7 nt in length, with the sequence 5′-GAGUUUA-3′. The position of each abortive product is shown by arrows.
The presence of the MotA and AsiA proteins leads to an alteration in the RNAP photofootprint upstream of −20 on the NT strand (Fig. 2A, lane 7). The most marked changes occur within the Mot box sequence, including a significant decrease in the termination frequency at positions −27 and −28, coupled with an increase in the signal at −33 (shown by arrows). Thus, specific interactions within the Mot box require the presence of all three components: RNAP, MotA, and AsiA. In addition, the bands at −21 and −22 are more intense, suggesting that the quaternary protein–promoter complex takes on a novel conformation in this region that requires both T4 proteins.
To investigate these species further, selected NTPs were added to heparin-resistant PrIIB2–RNAP complexes. The transcribed sequence of PrIIB2 is 5′-GAGUUUACC-3′, hence polymerization can be halted at either the 3-nt product (GAG) or the 7-nt product (GAGUUUA) by adding specific combinations of nucleotides. Fig. 3A shows the photofootprints of RNAP–DNA complexes after incubation with GTP and ATP (lanes 1–5), or CTP (lanes 6–10). Primer extension on the NT strand shows that neither nucleotide solution affects the termination pattern of the DNA alone (lanes 1 and 6). Nor does the addition of heparin and CTP significantly alter the footprint from that of open promoter complexes seen previously (compare Fig. 3A, lanes 6–10 to the corresponding lanes in Fig. 2A). The characteristic signals seen at −5 and −4 persist, as do the specific interactions observed within the Mot box in the presence of both T4 proteins (see arrows). In contrast, although the upstream events are not altered upon the addition of GTP and ATP, the region near the transcription start site shows significant changes in all RNAP-containing samples (Fig. 3A, lanes 2–5). The intense bands seen at positions −4 and −5 in the RPO are less prominent, whereas there is a salient increase in the frequency of termination at position −2. We note that although the sample containing RNAP and AsiA gives a slightly weaker footprint, AsiA neither alters nor prevents the formation of the RNAP–DNA contacts characteristic of RPO at this promoter. Moreover, in the presence of the initiating nucleotides GTP and ATP, each of the open complexes undergoes the same transition, which we interpret as the formation of the initiating complex (RPinit).
To assess the validity of this assumption, we directly assayed abortive transcription from these RNAP complexes in vitro. An excess of RNAP was used to minimize the initial binding effects of the two T4 proteins. Preformed RNAP–DNA complexes were challenged by heparin and supplied with the limiting substrates GTP, ATP, UTP, and either radiolabeled GTP or UTP to visualize the reiterative products (Fig. 3B). The use of [γ-32P]GTP (which will uniquely 5′ end-label the transcripts) allowed us to ensure that the abortive products arose solely from the PrIIB2 promoter. We observed only the expected transcripts, ranging up to 7 nt (labeled by arrows). Additionally, we have performed several control experiments to verify that the transcripts arose solely from the promoter (data not shown). Two promoter mutants were assayed for transcription activity, one in which the −10 hexamer was altered, and a second which bears a single nucleotide change at the +1 position (described in Materials and Methods). Abortive transcription was not detected from either of these mutant promoters whether labeling with [γ-32P]GTP, [γ-32P]ATP, or [α-32P]UTP. This finding demonstrates that the transcripts observed have the predicted 5′ end, and it suggests that the identity of the initiating nucleotide is important for transcription from the PrIIB2 promoter.
At the wild-type PrIIB2 promoter, significant levels of abortive transcripts are synthesized by all RNAP–DNA species, confirming their identification as initiating complexes. Transcription by the RNAP (Fig. 3B, lanes 1 and 5) is not significantly affected by MotA (lanes 2 and 6) or AsiA (lanes 3 and 7). The presence of both T4 factors yields an interesting effect: the synthesis of transcripts up to 6 nt long is slightly lower than that observed by RNAP, yet the production of 7-nt transcript is nearly doubled (compare lanes 1 and 5 to lanes 4 and 8). Thus, while the ratio of 6-nt to 7-nt product is 1.4 with the RNAP alone, it becomes 0.45 in the presence of MotA and AsiA. Taken together, these results suggest that whereas the RNAP is competent to initiate abortive synthesis it is deficient in the production of transcripts beyond a certain length.
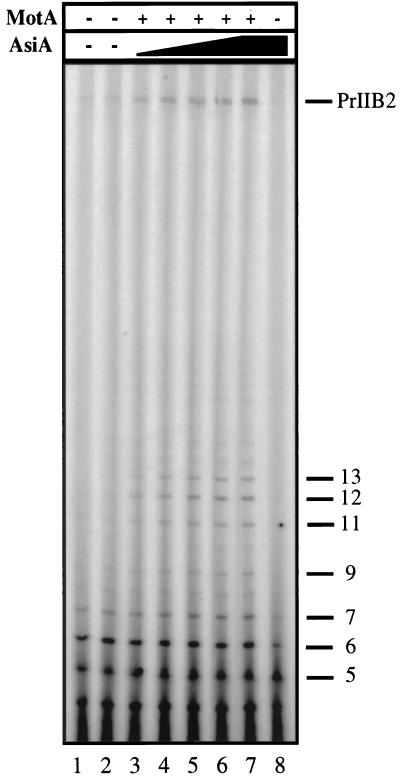
To investigate this phenomenon, we performed single- and multiple-round in vitro run-off transcription experiments. The multiple-round results are presented in Fig. 4. Transcripts are uniquely end-labeled with [γ-32P]GTP, so that comparative band intensities represent molar ratios. The RNAP alone, or with either T4 protein individually, produces significant levels of short reiterative transcripts and very little full-length product (lanes 1, 2, and 8). In the presence of an excess of MotA, increasing concentrations of AsiA stimulate the formation of full-length transcript and promote the appearance of intermediate-length products (lanes 3–7). As in the abortive transcription assay, the levels of transcript up to 6 nt in length are not enhanced by the T4 proteins, whereas longer transcripts are affected. Notably, the increase in intermediate-length (7–13 nt) products correlates directly with the appearance of full-length transcript. Thus, these data indicate that the T4 proteins aid in the transition from initiating to elongating complexes. Hence, in addition to affecting the initial recruitment of the RNAP to the promoter sequences, MotA and AsiA work together to facilitate promoter escape.
Figure 4.
In vitro run-off transcription assays at the PrIIB2 promoter. Transcripts formed by the RNAP at 100 nM (lane 1) in the presence of an excess (200 nM) of MotA (lane 2) or AsiA (lane 8) individually are compared with those produced in the presence of MotA (200 nM), when AsiA is added at increasing concentrations: 10 nM (lane 3), 25 nM (lane 4), 50 nM (lane 5), 100 nM (lane 6), or 200 nM (lane 7). Each transcript observed is uniquely end-labeled, by the use of [γ-32P]GTP, thereby permitting accurate comparison of various transcript production. For example, the molar ratios of transcripts in lane 7, as compared with the full-length product, are the following: 7 nt, 1.10; 9 nt, 0.35; and 12 nt, 0.78. The positions of full-length (PrIIB2) transcript as well as shorter products are indicated.
DISCUSSION
The AsiA protein of bacteriophage T4 binds tightly to the σ70 subunit of E. coli RNAP, thereby decreasing its affinity for E. coli and T4 early promoters and stimulating MotA-dependent T4 middle-mode synthesis. Our previous work has demonstrated that the MotA and AsiA proteins together stimulate the recruitment of RNAP to a T4 middle promoter (12). Using a BIAcore surface plasmon resonance unit, we confirm these data and investigate the complexes formed. The dissociation rate constant (koff) calculated for the RNAP–DNA complexes is identical, within experimental error, to that obtained from the quaternary complexes. The fact that overall complex formation is increased without an alteration in complex stability indicates that recognition is stimulated at the initial binding step. A growing number of transcription activators have been described that work by recruitment of the RNAP to the DNA (reviewed in ref. 26); these activators are thought to provide additional sites for protein–protein interactions in the vicinity of the promoter sequences, thereby increasing the affinity of RNAP for this region. It is likely that the T4 proteins function in this way, enabling interactions between the polymerase and a MotA-bound promoter that compensate for the loss of canonical DNA–RNAP contacts.
The UV laser photofootprints reveal intimate RNAP–promoter contacts occurring predominantly in the region between −20 and the transcription start site (Fig. 2, lanes 3, 4, and 6). The presence of the MotA protein alters neither the photofootprint of RNAP within this region nor the transcriptional capacity of the open promoter complexes. This result concurs with previous transcription and DNase I footprinting data, which assert that MotA has no effect in the absence of AsiA (7, 14). Although DNA binding by the AsiA–RNAP complex has been observed previously, the promoter complex formed by T4 modified polymerase (containing AsiA) proved unstable during analysis by DNase I (14). The nonperturbing nature of UV footprinting has allowed us to investigate this binding at PrIIB2 and to demonstrate that the contacts formed by the RNAP bound to AsiA are qualitatively the same as those formed by the RNAP alone. The concentration of AsiA used is sufficient to abolish open complex formation at an E. coli promoter, making it unlikely that this complex represents an RNAP lacking AsiA. Thus, the presence of the T4 proteins, either individually or together, does not alter the specific interactions observed between RNAP and the DNA from the −10 hexamer to +1. Since contacts within this region are implicated in both the isomerization to an open complex and transcription initiation, it is consistent that AsiA and/or MotA does not affect these steps in the transcription cycle.
The inability to footprint a specific complex between MotA alone and the Mot box sequence has been observed in prior work, suggesting that MotA binding may be stabilized by an interaction with the T4 modified RNAP (27). There is, however, a small but reproducible decrease in the intensity of the bands just downstream of the Mot box on the T strand (Fig. 2B, lanes 2 and 7). This may be indicative of an interaction between MotA and its target site, although we have not obtained any definitive evidence for such an interaction in the absence of RNAP and AsiA. On the contrary, in the presence of these proteins there is a pronounced decrease in the signals at −27 and −28, as a result of a structural change in the DNA in this region (Fig. 2A, lane 7). In addition, since the signal at position −33 appears only in the presence of both T4 proteins, it must be because of a specific interaction within the Mot box that is possible only in the quaternary complex. While the direct contact is most likely made by the MotA protein, which is known to bind to this sequence, the possibility remains that σ70 or AsiA also makes intimate contacts in this region. Likewise, in the presence of AsiA and MotA, new contacts are witnessed at −22 and −21, indicating that this complex possesses a modified conformation in the upstream segment. These data are in agreement with the unique DNase I footprint obtained from this species (14) and the distinctive mobility of the quaternary complex observed in native gel-shift assays (12).
The signals that characterize the open complexes are entirely conserved after the addition of heparin and the unincorporated nucleotide CTP (Fig. 3A, lanes 6–10). In contrast, the presence of the initiating nucleotides GTP and ATP leads to the loss of signal at −4 and −5, and a concomitant increase in signal at −2. We interpret this change as the formation of initiating complexes, a proposal that is confirmed by abortive transcription assays (Fig. 3B). The fact that each of the open promoter species is competent to initiate transcription demonstrates that the T4 proteins are not required for the formation of the first phosphodiester bond or at any other step between RPO and RPinit. Instead, their presence facilitates the transition from initiating complexes to stable elongating complexes (Fig. 4). This transition may occur by means of direct effects of AsiA and MotA on promoter escape, or alternatively, through indirect or transient effects on the structure of the RNAP–promoter complexes. In this way, the T4 proteins could induce the formation of a more productive open complex, thus alleviating the observed block in transcription experienced by RNAP at 6 nt (Fig. 3B, lane 4). Indeed, the RNAP alone does not synthesize intermediate-length transcripts (Fig. 4, lane 1), which suggests the formation of a functionally distinct RNAP–promoter complex in the presence of the T4 factors. Additionally, it is unlikely that the 7- to 13-nt products represent trapped kinetic transcription intermediates, because the addition of excess unlabeled NTPs did not “chase” them into full-length transcripts (data not shown). Hence, the synthesis of these intermediate-length transcripts results from premature termination, an effect whose in vivo relevance is as yet unknown. It is possible that the additional T4 modifications of the RNAP (α ADP-ribosylation, interaction with RpbA) play a role in diminishing RNAP–promoter interactions that affect transcript elongation.
In summary, our data indicate that AsiA and MotA function at two distinct steps in the transcription pathway. Initially, they increase recruitment of RNAP to the promoter sequences and stimulate the formation of a quaternary complex with unique structural characteristics. This species then possesses a greater capacity to make the transition between RPinit and RPE. Thus, T4 presents a remarkable system of transcription regulation, wherein the presence of AsiA effectively inhibits all nonenhanced transcription, thereby conferring complete MotA dependence on mRNA synthesis. In this way, T4 middle transcription is ensured to occur precisely within a given interval and under the specific conditions dictated by phage development.
Acknowledgments
We are grateful to Henri Buc for his helpful input and encouragement, and to Mohamed Ouhammouch for his continued support. We also thank Iain Pemberton for his critical reading of this manuscript. This work was supported by National Institutes of Health Grant GM50700 to E.N.B.; K.A. was supported by a National Science Foundation Graduate Fellowship.
ABBREVIATIONS
- RNAP
RNA polymerase
- T
template
- NT
nontemplate
- RU
resonance unit(s)
References
- 1.Mosig G, Hall D H. In: The Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 126–131. [Google Scholar]
- 2.Wilkens K, Rüger W. In: The Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 132–141. [Google Scholar]
- 3.Brody E N, Kassavetis G A, Ouhammouch M, Sanders G M, Tinker R L, Geiduschek E P. FEMS Microbiol Lett. 1995;128:1–8. doi: 10.1111/j.1574-6968.1995.tb07491.x. [DOI] [PubMed] [Google Scholar]
- 4.Stevens A. In: RNA Polymerase. Losick R, Chamberlin M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. pp. 617–627. [Google Scholar]
- 5.Stevens A. Biochim Biophys Acta. 1977;475:193–196. doi: 10.1016/0005-2787(77)90355-0. [DOI] [PubMed] [Google Scholar]
- 6.Orsini G, Ouhammouch M, Le Caer J, Brody E N. J Bacteriol. 1993;175:85–93. doi: 10.1128/jb.175.1.85-93.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouhammouch M, Adelman K, Harvey S R, Orsini G, Brody E N. Proc Natl Acad Sci USA. 1995;92:1451–1455. doi: 10.1073/pnas.92.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross C A, Lonetto M, Losick R. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 129–176. [Google Scholar]
- 9.Severinova E, Severinov K, Fenyo D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 10.Pahari S, Chatterji D. FEBS Lett. 1997;411:60–62. doi: 10.1016/s0014-5793(97)00668-6. [DOI] [PubMed] [Google Scholar]
- 11.Colland F, Orsini G, Brody E N, Buc H, Kolb A. Mol Microbiol. 1998;27:819–829. doi: 10.1046/j.1365-2958.1998.00729.x. [DOI] [PubMed] [Google Scholar]
- 12.Adelman K, Orsini G, Kolb A, Graziani L, Brody E N. J Biol Chem. 1997;272:27435–27443. doi: 10.1074/jbc.272.43.27435. [DOI] [PubMed] [Google Scholar]
- 13.Ouhammouch M, Orsini G, Brody E N. J Bacteriol. 1994;176:3956–3965. doi: 10.1128/jb.176.13.3956-3965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinton D M, March A R, Gerber J S, Sharma M. J Mol Biol. 1996;256:235–248. doi: 10.1006/jmbi.1996.0082. [DOI] [PubMed] [Google Scholar]
- 15.Guild N, Gayle M, Sweeney R, Hollingsworth T, Modeer T, Gold L. J Mol Biol. 1988;199:241–258. doi: 10.1016/0022-2836(88)90311-7. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt R P, Kreuzer K N. J Biol Chem. 1992;267:11399–11407. [PubMed] [Google Scholar]
- 17.Stitt B, Hinton D. In: The Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 142–160. [Google Scholar]
- 18.Keilty S, Rosenberg M. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 19.Minchin S, Busby S. Biochem J. 1993;289:771–775. doi: 10.1042/bj2890771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinton D M. J Biol Chem. 1991;266:18034–18044. [PubMed] [Google Scholar]
- 21.Record M T, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 792–821. [Google Scholar]
- 22.Buc H, McClure W R. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 23.Burgess R R, Jendrisak J J. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 24.Buckle M, Geiselmann J, Kolb A, Buc H. Nucleic Acids Res. 1991;19:833–840. doi: 10.1093/nar/19.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckle M, Williams R M, Negroni M, Buc H. Proc Natl Acad Sci USA. 1996;93:889–894. doi: 10.1073/pnas.93.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 27.March-Armegadzie R, Hinton D M. Mol Microbiol. 1995;15:649–660. doi: 10.1111/j.1365-2958.1995.tb02374.x. [DOI] [PubMed] [Google Scholar]