Abstract
Growth of lateral organs is a complex mechanism that starts with formation of lateral primordia.Basal developmental programs like polarity, organ identity and environmental cues influence the final organ size achieved via coordinated cell division and expansion. recent evidence shows that the precise balance between these two processes, known as compensation mechanisms, seems to be influenced by the identity of the organ. Furthermore, studies of mutants affected in floral organ size suggest the existence of developmental compartments within different floral whorls that show distinct compensation behaviors.
Key words: Antirrhinum majus, cell division, cell expansion, COMPACTA ÄHNLICH, compensation, floral size, FORMOSA, NITIDA, organ identity
Global Aspect of Lateral Organ Growth
Formation of lateral organs starts with the recruitment of meristematic cells that acquire a primordial fate. Newly formed primordia start extending as a result of an organ polarity program.1 A meristem signal is required to establish polarity within a developing organ.2 How much an organ grows and what shape it achieves implicates two additional basal modulators, the organ identity imposed on the growing primordium and the level of interactions with the environment in terms of size and/or differentiation.
Flowers of angiosperms like Arabidopsis thaliana, Antirrhinum majus or Petunia sp. have in common the formation of concentric whorls of organs that include sepals, petals, stamens and carpels. Its precise arrangement is due to combinatorial genetic functions that give raise to the four identities found in the flower.3–5 The formation of whorls requires the activation of the so-called organ identity genes that in Antirrhinum correspond mainly to DEFICIENS, GLOBOSA and PLENA.6 Direct protein-protein interactions between these MADS-box genes give rise to heterodimers7,8 and ternary complexes9–11 that drive target gene expression required for floral morphogenesis. There is evidence that genes involved in polarity interact with early genes of SAM maintenance1 and also with organ identity genes12,13 affecting both organ growth and development.
Besides genetic developmental control, plants are able to change their morphogenesis adapting to environmental condition and modifying growth, flowering and sometimes survival.14
Understanding Control of Floral Organ Size
Recent work has focused on understanding how floral size is controlled.15 Two issues have been addressed; one is the existence of genes that control floral size in a specific way. Indeed, QTL affecting floral size have been found in Arabidopsis,16 Petunia17 and tomato.18 The other issue is the genetic dissection of floral organ size control and its relation with floral organ identity genes.
Lateral organ growth can be divided in two phases: one, which includes cell mass increment coupled to cell proliferation and a second one, that takes place once cells exit the proliferative period and growth is mainly due to cell expansion.19,20 Final organ cell number depends on the number of cells founding primordia, active dividing cells and cell proliferation rate and period.19,21,22 Genetic analysis in Arabidopsis revealed several independent routes controlling plant organ size via changes in the cell division interval. For instance the auxin signaling through ARGOS and AINTEGUMENTA (ANT) extend the cell proliferation phase.22,23 KLUH, which promotes growth through a non-cell autonomous signaling, seems to maintain cell division until primordia reach a determinate size.24 JAGGED (JAG) and NUBBIN (NUB) act together to promote cell proliferation in marginal regions of lateral organs. Genes with opposite effects include BIG BROTHER (BB)25 and DA1,26 which restrict proliferation.
Cell expansion can induce organ size changes through various ways, including vacuolation, ploidy level, biosynthesis of cell wall components and cytoskeleton.27,28 For instance the bHLH transcription factor BIGPETALp (BPEp), activated by floral identity genes, was shown to regulate petal size via cell expansion.29
Organ Level Coordination between Cell Division and Expansion
In several higher organisms, normal organ size has been observed in mutants with aberrant or deficient cell division due to increased cell expansion.30 This phenomenon is known as compensation mechanism and it adds another level of control on organ size by monitoring and coordinating cell division and expansion.31 The current hypothesis suggests that it could be a common procedure to control the size of determinate organs.32 Recently published data suggest that reduction in cell expansion, by a decrease in endoreduplication, could also trigger compensation via increased cell proliferation to attain a normal leaf size.33
There are several studies in leaves showing reduction in cell number, which conduces to an increment in cell expansion. Mutation in TANGLED gene in corn shows aberrant cell division but no morphological defects,34 mutations in SHORT INTEGUMENT 2 gene also reduce cell number in integument but display almost normal morphology.35 In Arabidopsis, null mutations in ANGUSTIFOLIA3 restrict the proliferation period with a partial compensatory increment in cell expansion and narrower leaves.36 Furthermore, several other mutations that induce compensation have been described as struwelpeter, swell-map, G-protein α-subunit1 and deformed roots and leaves.37–40 A compensation mechanisms triggered by variations in cell division41 has been identified in floral size mutants of Antirrhinum. The mutant compacta ähnlich (coan) shows a reduction in floral organ size in all floral whorls and this change is mainly due to a reduction in cell division (see below).42 Although the mutants described in Arabidopsis and Antirrhinum show compensation mechanisms, lateral organs tend to be smaller than wild type.
The opposite compensation phenomenon has been documented in Arabidopsis integuments overexpressing KNAT1, with additional cell cycle rounds and reduced cell expansion, and also, in the mutant swellmap38;43 and in the Antirrhinum mutant formosa (fo) with increased floral organ size due to higher cell number but a reduction in cell area.41
Organ Identity Coming into Play
Several lines of evidence suggest that floral organ identity plays a key role as a regulator of organ size. Weak hypomorphic alleles of DEFICIENS (DEF)44 in Antirrhinum, or single mutants of PhDEF and PhGLO1 in petunia45 where a full loss of organ identity is only achieved in the double mutant,45 share a reduced petal growth as phenotype. These observations suggest that final organ size requires high expression of B function genes. Direct evidence has been obtained in temperature sensitive alleles of DEF that show decreased organ size under non-permissive temperatures.46
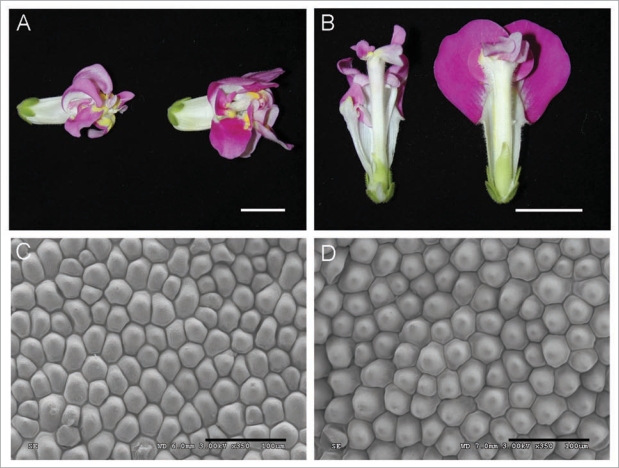
Several mutants affecting organ size do it in an organ-identity dependent fashion. Ectopic expression of ANT causes increased cell division in all floral whorls,22 and also shows enhanced cell expansion in petals, stamens and pistils but not in sepals.47 In tomato, the OVATE gene represses growth in fruits, but additional gene copy numbers expand the size of all lateral organs with a stronger influence in sepals and stamens than in petals and carpels.48 Transgenic tobacco expressing dominant negative constructs of Cdc2 kinase display reduction in cell number in all lateral organs, that triggers compensation in leaves but not in flowers.49 Mutations in the Antirrhinum FO gene increase cell division in the three inner floral whorls and displays compensation with a reduction in cell expansion in petals and pistils but not in stamens.41 In contrast the mutant coan has reduced cell division in petals, stamens and pistils but compensation is triggered in petals via increased cell expansion. These differences in organ size control depending on identity is also supported by a double mutant between fo, affecting floral organ size, an a flower identity mutant, plena (ple), in a C function gene. The ple fo double mutant only shows size differences in petals of second whorl which have normal identity (Fig. 1A and B). Inner whorls with altered identity don't show any significant size variation between ple single mutant and fo ple. However fo effect over petal size in ple genetic background is stronger than in single mutant with an increment in dorsal petal expansion that doubles the one observed in single fo mutant. Cell compensation is not observed in fo ple petals anymore, showing an increment in conical cell expansion in contrast to the reduction observed in single fo mutant (Fig. 1C and D). This would suggest an implication of PLE in organ size control in a non-autonomous way. Several previous works in C function (AGAMOUS) mutation in Arabidopsis suggest its non-autonomous role in specifying the pattern of cell division in different layers of the second whorl,50 intercellular communication between different whorls50 and in petal development.51 Moreover, the gene BPEp mainly expressed in petals, has been described to be regulated by identity genes including AGAMOUS.29
Figure 1.
Pictures showing flowers of ple single mutant and fo ple double mutant (left and right respectively) (A) lateral view and (B) longitudinal section. White bars represent 1 cm. SEM images of petal conical cells (C) ple mutant and (D) fo ple double mutant. Black bars represent 100 µm.
Developmental Compartments Show Distinct Compensation Behaviors
Petals are highly specialized organs comprised of distinct regions like the tube or the petal blade. The latter has two cell types distinguished by shape and function since conical cells are involved in light scattering and scent production.52,53 In coan cell behavior is complex as cell size is only affected in petal regions with epidermal conical cells while flat cells adjacent to the petal tube do not show this compensation process.42 The opposite was observed in another floral size mutant of Antirrhinum, Nitida (Ni), where the reduction in floral size in petals also induces decreased expansion of flat cells while no variation was observed in conical petal cells.42
The evidences described confirm compensation could be triggered both in leaves and flowers but seems to depend on organ identity and even on different developmental compartments inside organs. This suggests the existence of a global control of organ size integrating cell division, expansion and development, but there is no mechanism clarifying communication between those programs yet. Hopefully forthcoming studies would reveal the intricate network controlling final organ size.
Acknowledgements
This work was supported by Biocarm (BANANASAI), Fundación Séneca de la Región de Murcia and MEC-AGL2007-61384. Grants from AECI to L.D.-B. Thanks to Izaskun Mallona for comments on the manuscript.
Footnotes
Previously published online: http://www.landesbioscience.com/journals/psb/article/9394
References
- 1.Kidner CA, Timmermans MCP. Mixing and matching pathways in leaf polarity. Curr Opin Plant Biol. 2007;10:13–20. doi: 10.1016/j.pbi.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Reinhardt D, Frenz M, Mandel T, Kuhlemeier C. Microsurgical and laser ablation analysis of leaf positioning and dorsoventral patterning in tomato. Development. 2005;132:15–26. doi: 10.1242/dev.01544. [DOI] [PubMed] [Google Scholar]
- 3.Coen ES, Meyerowitz EM. The war of the whorls—genetic interactions controlling flower Development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 4.Egea Gutierrez-Cortines M, Davies B. Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends Plant Sci. 2000;5:473–478. doi: 10.1016/s1360-1385(00)01761-1. [DOI] [PubMed] [Google Scholar]
- 5.Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell. 2006;18:1819. doi: 10.1105/tpc.106.042937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science. 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- 7.Davies B, EgeaCortines M, Silva ED, Saedler H, Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996;15:4330–4343. [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, et al. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causier B, Cook H, Davies B. An Antirrhinum ternary complex factor specifically interacts with C-function and SEPALLATA-like MADS-box factors. Plant Mol Biol. 2003;52:1051–1062. doi: 10.1023/a:1025426016267. [DOI] [PubMed] [Google Scholar]
- 10.Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 12.Franks RG, Wang CX, Levin JZ, Liu ZC. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- 13.Meyerowitz EM. Genes and gene interactions that establish patterns in flower development. J Cell Biochem. 1995:329. [Google Scholar]
- 14.Hardtke CS, Torii KU. Plant growth and developmentùthe new wave. Curr Opin Plant Biol. 2008;11:1–3. [Google Scholar]
- 15.Weiss J, Delgado-Benarroch L, Egea-Cortines M. Genetic control of floral size and proportions. Int J Dev Biol. 2005;49:513–525. doi: 10.1387/ijdb.051998jw. [DOI] [PubMed] [Google Scholar]
- 16.Juenger T, Perez-Perez JM, Bernal S, Micol JL. Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: evidence for modular genetic architecture. Evol Dev. 2005;7:259–271. doi: 10.1111/j.1525-142X.2005.05028.x. [DOI] [PubMed] [Google Scholar]
- 17.Galliot C, Stuurman J, Kuhlemeier C. The genetic dissection of floral pollination syndromes. Curr Opin Plant Biol. 2006;9:78–82. doi: 10.1016/j.pbi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Frary A, Fritz LA, Tanksley SD. A comparative study of the genetic bases of natural variation in tomato leaf, sepal and petal morphology. Theor Appl Genet. 2004;109:523–533. doi: 10.1007/s00122-004-1669-x. [DOI] [PubMed] [Google Scholar]
- 19.Anastasiou E, Lenhard M. Growing up to one's standard. Curr Opin Plant Biol. 2007;10:63–69. doi: 10.1016/j.pbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Bogre L, Magyar Z, Lopez-Juez E. New clues to organ size control in plants. Genome Biol. 2008;9:226. doi: 10.1186/gb-2008-9-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizukami Y. A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol. 2001;4:533–539. doi: 10.1016/s1369-5266(00)00212-0. [DOI] [PubMed] [Google Scholar]
- 22.Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell. 2003;15:1951–1961. doi: 10.1105/tpc.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell. 2007;13:843–856. doi: 10.1016/j.devcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol. 2006;16:272–279. doi: 10.1016/j.cub.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Zheng L, Corke F, Smith C, Bevan MW. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Gene Dev. 2008;22:1331. doi: 10.1101/gad.463608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joubes J, Chevalier C. Endoreduplication in higher plants. Plant Mol Biol. 2000;43:735–745. doi: 10.1023/a:1006446417196. [DOI] [PubMed] [Google Scholar]
- 28.Smith LG, Oppenheimer DG. Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol. 2005;21:271–295. doi: 10.1146/annurev.cellbio.21.122303.114901. [DOI] [PubMed] [Google Scholar]
- 29.Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, et al. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J Plant Res. 2006;119:37–42. doi: 10.1007/s10265-005-0232-4. [DOI] [PubMed] [Google Scholar]
- 31.Tsukaya H. Interpretation of mutants in leaf morphology: Genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between cell and organismal theories. Int Rev Cytol. 2002;217:1–39. doi: 10.1016/s0074-7696(02)17011-2. [DOI] [PubMed] [Google Scholar]
- 32.Tsukaya H. Controlling size in multicellular organs: focus on the leaf. PLoS Biol. 2008;6:174. doi: 10.1371/journal.pbio.0060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larson-Rabin Z, Li Z, Masson PH, Day CD. FZR2/CCS52A1 Expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 2009;149:874–884. doi: 10.1104/pp.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith LG, Hake S, Sylvester AW. The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development. 1996;122:481–489. doi: 10.1242/dev.122.2.481. [DOI] [PubMed] [Google Scholar]
- 35.Broadhvest J, Baker SC, Gasser CS. SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics. 2000;155:899–907. doi: 10.1093/genetics/155.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 37.Autran D, Jonak C, Belcram K, Beemster GTS, Kronenberger J, Grandjean O, et al. Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 2002;21:6036–6049. doi: 10.1093/emboj/cdf614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clay NK, Nelson T. The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell. 2005;17:1994–2008. doi: 10.1105/tpc.105.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- 40.Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, et al. DRL1, a homolog of the yeast TOT4/KT112 protein, has a function in meristem activity and organ growth in plants. Plant Cell. 2003;15:639–654. doi: 10.1105/tpc.007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delgado-Benarroch L, Causier B, Weiss J, Egea-Cortines M. FORMOSA controls cell division and expansion during floral development in Antirrhinum majus. Planta. 2009;229:1219–1229. doi: 10.1007/s00425-009-0910-x. [DOI] [PubMed] [Google Scholar]
- 42.Delgado-Benarroch L, Weiss J, Egea-Cortines M. The mutants compacta ähnlich, Nitida and Grandiflora define developmental compartments and a compensation mechanism in floral development in Antirrhinum majus. J Plant Res. 2009 doi: 10.1007/s10265-009-0236-6. [DOI] [PubMed] [Google Scholar]
- 43.Truernit E, Haseloff J. Arabidopsis thaliana outer ovule integument morphogenesis: Ectopic expression of KNAT 1 reveals a compensation mechanism. BMC Plant Biol. 2008;8:35. doi: 10.1186/1471-2229-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, et al. Deficiens, A homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus—the protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. The duplicated B-class heterodimer model: Whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell. 2004;16:741–754. doi: 10.1105/tpc.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zachgo S, Silva E, Motte P, Trobner W, Saedler H, Schwarz-Sommer Z. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development. 1995;121:2861–2875. doi: 10.1242/dev.121.9.2861. [DOI] [PubMed] [Google Scholar]
- 47.Krizek BA. Ectopic expression AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev Genet. 1999;25:224–236. doi: 10.1002/(SICI)1520-6408(1999)25:3<224::AID-DVG5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 48.Liu JP, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemerly A, Engler JD, Bergounioux C, VanMontagu M, Engler G, Inze D, et al. Dominant-negative mutants of the Cdc2 kinase uncouple cell-dvision from iterative plant development. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenik PD, Irish VF. Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development. 2000;127:1267–1276. doi: 10.1242/dev.127.6.1267. [DOI] [PubMed] [Google Scholar]
- 51.Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, et al. The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:8571–8576. doi: 10.1073/pnas.1033043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dudareva N, Murfitt LM, Mann CJ, Gorenstein N, Kolosova N, Kish CM, et al. Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell. 2000;12:949–961. doi: 10.1105/tpc.12.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin C, Bhatt K, Baumann K, Jin H, Zachgo S, Roberts K, et al. The mechanics of cell fate determination in petals. Philos Trans R Soc Lond B Biol Sci. 2002;357:809–813. doi: 10.1098/rstb.2002.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]