Abstract
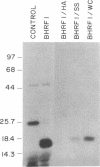
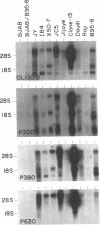
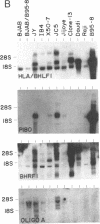
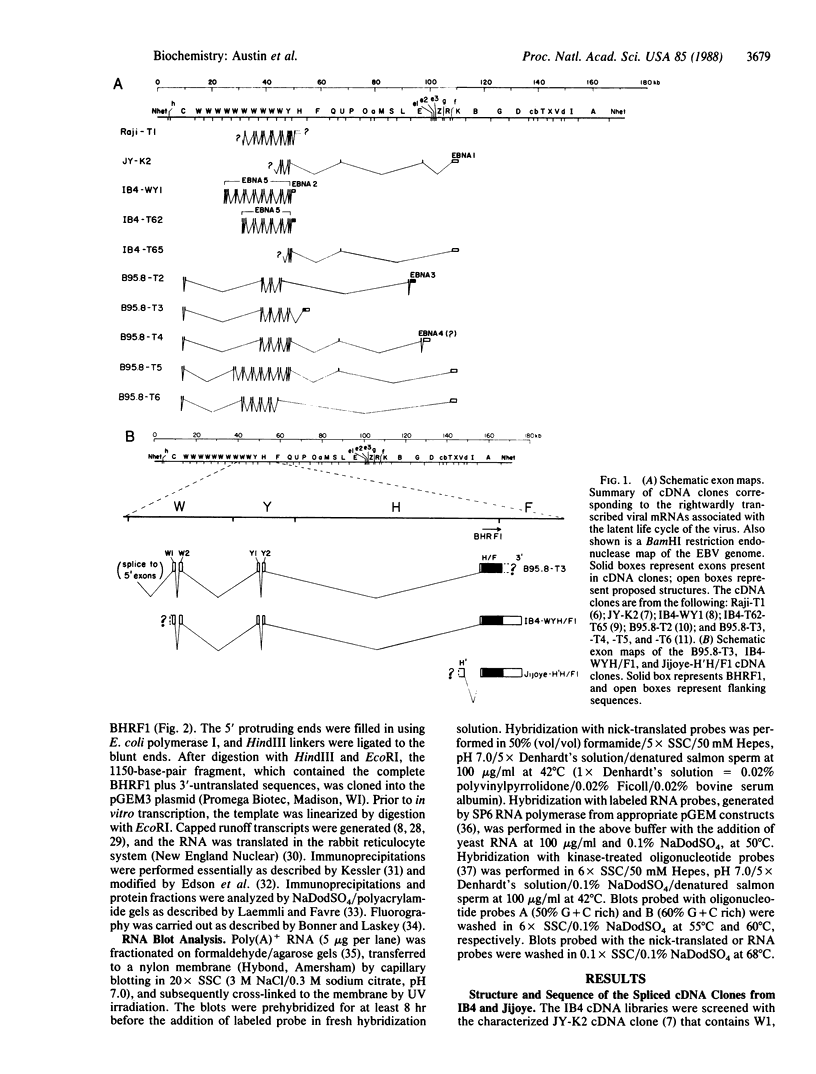
Several cDNA clones containing the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) have been recovered from the tightly latent lymphoblastoid cell line IB4. These clones contain the 5' leader exons encoded in the major internal repeat 1 and the viral BamHI fragment Y, identified in the rightwardly transcribed viral mRNAs associated with the latent viral life cycle. In addition, a cDNA clone containing BHRF1 from the Burkitt lymphoma cell line Jijoye was also recovered and exhibits a distinctive splicing pattern. In vitro transcription and translation of BHRF1, followed by immunoprecipitation with Epstein-Barr virus-positive human sera, indicates that this viral antigen is expressed during infection. RNA blot analyses with a wide panel of lymphoblastoid and Burkitt lymphoma cell lines revealed a complex pattern of transcription. Hybridization data obtained with several probes is presented.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bodescot M., Brison O., Perricaudet M. An Epstein-Barr virus transcription unit is at least 84 kilobases long. Nucleic Acids Res. 1986 Mar 25;14(6):2611–2620. doi: 10.1093/nar/14.6.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodescot M., Chambraud B., Farrell P., Perricaudet M. Spliced RNA from the IR1-U2 region of Epstein-Barr virus: presence of an open reading frame for a repetitive polypeptide. EMBO J. 1984 Aug;3(8):1913–1917. doi: 10.1002/j.1460-2075.1984.tb02067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodescot M., Perricaudet M. Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 1986 Sep 11;14(17):7103–7114. doi: 10.1093/nar/14.17.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodescot M., Perricaudet M., Farrell P. J. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J Virol. 1987 Nov;61(11):3424–3430. doi: 10.1128/jvi.61.11.3424-3430.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brandsma J., Miller G. Nucleic acid spot hybridization: rapid quantitative screening of lymphoid cell lines for Epstein-Barr viral DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6851–6855. doi: 10.1073/pnas.77.11.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier-Greco A., Manet E., Chavrier P., Mosnier C., Daillie J., Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986 Dec 1;5(12):3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M. L., Smith S. D., Sklar J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell. 1986 Oct 10;47(1):19–28. doi: 10.1016/0092-8674(86)90362-4. [DOI] [PubMed] [Google Scholar]
- Edson C. M., Cohen L. K., Henle W., Strominger J. L. An unusually high-titer human anti-Epstein Barr virus (EBV) serum and its use in the study of EBV-specific proteins synthesized in vitro and in vivo. J Immunol. 1983 Feb;130(2):919–924. [PubMed] [Google Scholar]
- Fennewald S., van Santen V., Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J Virol. 1984 Aug;51(2):411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hennessy K., Fennewald S., Hummel M., Cole T., Kieff E. A membrane protein encoded by Epstein-Barr virus in latent growth-transforming infection. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7207–7211. doi: 10.1073/pnas.81.22.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Jeang K. T., Hayward S. D. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J Virol. 1983 Oct;48(1):135–148. doi: 10.1128/jvi.48.1.135-148.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- King W., Thomas-Powell A. L., Raab-Traub N., Hawke M., Kieff E. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J Virol. 1980 Nov;36(2):506–518. doi: 10.1128/jvi.36.2.506-518.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E., Klein G., Nadkarni J. S., Nadkarni J. J., Wigzell H., Clifford P. Surface IgM-kappa specificity on a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 1968 Jul;28(7):1300–1310. [PubMed] [Google Scholar]
- Klein G., Lindahl T., Jondal M., Leibold W., Menézes J., Nilsson K., Sundström C. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3283–3286. doi: 10.1073/pnas.71.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A. 1973 Jan;70(1):190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PULVERTAFT J. V. A STUDY OF MALIGNANT TUMOURS IN NIGERIA BY SHORT-TERM TISSUE CULTURE. J Clin Pathol. 1965 May;18:261–273. doi: 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson B. M., Roberts B. E., Kuff E. L. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4370–4374. doi: 10.1073/pnas.74.10.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner A. J., Tsai E. C., Strominger J. L., Speck S. H. Isolation and characterization of cDNA clones corresponding to transcripts from the BamHI H and F regions of the Epstein-Barr virus genome. J Virol. 1987 Sep;61(9):2902–2909. doi: 10.1128/jvi.61.9.2902-2909.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson M., Gradoville L., Heston L., Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982 Dec;44(3):834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe D. T., Farrell P. J., Miller G. Novel nuclear antigens recognized by human sera in lymphocytes latently infected by Epstein-Barr virus. Virology. 1987 Jan;156(1):153–162. doi: 10.1016/0042-6822(87)90446-6. [DOI] [PubMed] [Google Scholar]
- Sample J., Hummel M., Braun D., Birkenbach M., Kieff E. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5096–5100. doi: 10.1073/pnas.83.14.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S. H., Pfitzner A., Strominger J. L. An Epstein-Barr virus transcript from a latently infected, growth-transformed B-cell line encodes a highly repetitive polypeptide. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9298–9302. doi: 10.1073/pnas.83.24.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S. H., Strominger J. L. Analysis of the transcript encoding the latent Epstein-Barr virus nuclear antigen I: a potentially polycistronic message generated by long-range splicing of several exons. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8305–8309. doi: 10.1073/pnas.82.24.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S. H., Strominger J. L. Epstein-Barr virus transformation. Prog Nucleic Acid Res Mol Biol. 1987;34:189–207. [PubMed] [Google Scholar]
- Terhorst C., Parham P., Mann D. L., Strominger J. L. Structure of HLA antigens: amino-acid and carbohydrate compositions and NH2-terminal sequences of four antigen preparations. Proc Natl Acad Sci U S A. 1976 Mar;73(3):910–914. doi: 10.1073/pnas.73.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G., Miller G. Recovery of Epstein-Barr virus from nonproducer neonatal human lymphoid cell transformants. Virology. 1979 Jun;95(2):351–358. doi: 10.1016/0042-6822(79)90490-2. [DOI] [PubMed] [Google Scholar]