Abstract
Vascular smooth muscle cells (VSMCs) switch from a contractile to a synthetic phenotype in human cardiovascular disease such as atherosclerosis and restenosis after angioplasty. VSMCs show reduced expression of contractile proteins and are capable of responding to mitogens by increasing expression of growth factor receptors. Fibroblast growth factor receptor-1 (FGFR1) signaling is one of several signaling pathways involved in this VSMC phenotypic switching. The aim of the present study was to examine the signaling pathway downstream of FGFR1 in the regulation of SM marker gene expression. We found that FGFR1 activated Akt/mTOR pathway and that the mTOR inhibitor rapamycin partially reversed FGFR1-mediated downregulated SM marker gene expression. Furthermore, we showed that mTOR forms a multi-protein complex with FGFR1 in VSMCs. These findings provide novel information for future development of therapeutic strategies for the treatment of human cardiovascular disease.
Introduction
Vascular smooth muscle cells (VSMCs) in the human arterial intima exhibit phenotypic plasticity that involved dedifferentiation from a contractile to a synthetic phenotype during atherosclerosis and restenosis after vascular injury. Synthetic VSMCs have reduced expression of contractile proteins such as smooth muscle α-actin (SM α-actin) that are required for cell contraction, and increased proliferation, migration and growth factor receptor expression [1, 2].
The fibroblast growth factor system is a key signaling pathway for the proliferation and migration of VSMCs in vivo [3]. FGFR1 is the predominant form of FGFRs expressed in VSMCs and its activation by ligand leads to different cellular responses e.g. proliferation, migration, and survival depending upon the activation of specific downstream pathways by the receptor [4]. Fibroblast growth factor receptor substrate 2 (FRS2), an adaptor protein, constitutively associates with FGFR1 and is a major downstream mediator of FGFR signaling [5, 6]. FRS2 is tyrosine phosphorylated on six sites, four of which bind the adaptor protein Grb2 leading to activation of the PI3 kinase pathway, and two which bind SHP2 which activates Ras and the ERK pathway [7, 8].
Previous studies in our lab indicated that FRS2 was required for FGFR1-mediated downregulation of VSMC marker genes (submitted). Here we show FGFR1/FRS2 complex activates Akt/mTOR signaling in VSMCs to regulate smooth muscle marker gene expression. Furthermore, we showed for the first time that FGFR1 forms a multi-protein complex with mTOR that is dependent upon FGFR1 kinase activity and FRS2. Our results provide novel information on FGFR1 signaling in VSMC phenotypic modulation and also set the stage for future design and development of new therapeutic agents in the treatment of human cardiovascular disease.
Materials and methods
Cell lines and reagents
293T cells (human embryonic kidney cells, ATCC CRL-11268), PAC1 cells [9], and primary bovine aortic vascular smooth muscle cells (BVSMC) [10] were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin-streptomycin (cellgro®). Recombinant human EGF, FGF-2, and PDGF-BB were purchased from PeproTech. DMEM and FBS were obtained from HyClone Laboratories Inc. Rapamycin was purchased from LC Laboratories. Akt1 siRNA was provided by Dr. Alex Toker [11]. EGFRvIII was provided by Dr. William Gullick [12]. EGFR K721A was provided by Dr. Sarah Parsons [13]. GST-Rheb was provided by Dr. Kun-Liang Guan [14]. SM α-actin promoter luciferase plasmid was provided by Dr. Gary K. Owens [15]. PDGFRβ D850N and PDGFRβ K634A plasmids were provided by Dr. Carl-Henrik Heldin [16]. mTOR full-length was purchased from Addgene. mTOR deletion constructs (1-1482, 1348-2549) were provided by Dr. David Sabatini [17].
Akt1 knockdown by RNA interference
To stably repress Akt1 expression in PAC1 VSMCs, we used pLKO.1 lentiviral shRNA constructs. The sequences of the oligonucleotides were described previously [11]. The lentivirus was packed by co-transfection of 293T cells with the shRNA expression vector, V-SVG and delta-VPR plasmid, using GeneJuice reagent (Novagen). After transfection for 48 h, the supernatants containing lentiviral particles were harvested. Monolayer PAC1 cells grown to about 60% confluence were transducted with above lentivirus-containing supernatant in the presence of 5 μg/ml polybrene and exposed to 1 μg/ml puromycin after 12 h of transduction. After 48 h of selection, cells were trypsinized, counted, and replated on 6-cm dishes for further Akt1 protein knockdown analysis.
Cell lysis, immunoprecipitation, and Western blot analysis
Cell lysis, immunoprecipitation, and Western blot analysis procedures were described previously [17, 18]. The following antibodies were used for immunoprecipitation and immunoblot analysis: Antibodies against Akt, HA-Tag, mTOR, mTOR Ser2448, p70 S6 kinase, p-p70 S6 kinase, PDGFRα, and PDGFRβ were purchased from Cell Signaling. Antibodies against pERK, SM α-actin, and β-tubulin were purchased from Sigma. Antibody against SM22α antibody was purchased from Abcam. Antibodies against EGFR and myc-Tag were purchased from Santa Cruz. Mouse monoclonal antibody against Xenopus FGFR1 (5G11) has been described previously [19]. For statistical analysis, the intensity of individual band in a given Western blot analysis was quantified using Image Quant analyzing software (Molecular Dynamics) and normalization to β-tubulin.
Creation of FGFR1 K562E mutant constructs and generation of PAC1 cells stably expressing FGFR1 K562E mutant constructs
We used previously described Xenopus constitutively active FGFR1 K562E construct [20] as a template to create FGFR1 K562E mutants (FGFR1 K562E: -FRS2 and FGFR1 K562E: -FRS2/-Crk/-PLCγ) using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s recommendation. Production of high-titer retroviruses was described previously [21]. Parental PAC1 cells were transducted with the viruses and then the cells were hygromycin selected. Single colonies were pooled for analysis of FGFR1 stable expression.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde and permeabilized in 0.1% Tween-20 and 0.1 % Triton X-100. Cells were incubated in mouse monoclonal SM α-actin (Sigma) at 4°C overnight followed by detecting using Alexa Fluor® 546-congugated (Invitrogen) secondary antibody for 1 h at room temperature. Control experiments were performed by substitution of the primary antibody with normal mouse serum IgG (Santa Cruz).
35S-labeling and pulse-chase experiments
PAC1 stable cell lines were first starved in serum-free DMEM lacking methionine and cysteine (Invitrogen) supplemented with 2 mM L-glutamine and 1 mM sodium pyruvate for 30 min at 37°C. Cells were pulsed-labeled for 2 h with 250 μCi ProMix L[35S]-methionine/cysteine (Amersham Biosciences), washed with PBS and chased with complete culture medium. At the indicated times, cells were lysed in HNTG buffer and protein content of the cell lysates was determined as described above. SM α-actin immunoprecipitates were prepared, samples were resolved by SDS-PAGE, and radiolabeled proteins were visualized by autoradiography. Band intensities corresponding to SM α-actin were quantified using Image Quant software (Molecular Dynamics).
GST pull-down assays
GST fusion protein preparation and GST pull-down assays were described previously [22].
Statistical analysis
Statistical analyses were performed using Student’s t-test, with a significant difference determined as p < 0.05. Data are presented as means ± SD.
Results and Discussion
Akt1 plays an important role in modulating VSMC phenotype
FRS2 is an adaptor protein that constitutive associates with FGFR1. Upon FGFR1 activation, FRS2 mediates signals to the ERK and Akt pathways [7, 8]. Martin and colleagues show that Akt1 and Akt2 have different functions in VSMCs, because Akt1 inhibits and Akt2 promotes SM marker gene expression [23]. In addition, they show that the mTOR inhibitor rapamycin increases VSMC contractile function by upregulating SM marker gene expression at both the transcriptional and translational levels [24]. Because Akt1 is the predominant form in VSMCs we examined the Akt1 signaling pathway.
To determine whether Akt1 signaling activity is required for FGFR1-mediated downregulation of SM α-actin gene expression, we used the shRNA approach. Endogenous Akt1 levels were reduced by ~80% as determined by Western blot analysis with anti-Akt antibody (Figure 1B). Akt1 knockdown cells showed increased cell size a sign of hypertrophy (Figure 1A). In addition, there was a ~4 fold increase in SM α-actin expression in Akt1 knockdowned cells compared to non-targeting control cells (Figure 1B). These results indicate that Akt1 is important in modulating VSMC phenotype in part by regulating SM α-actin expression.
Figure 1. Effects of Akt1 knockdown on PAC1 VSMCs.
A. Phase contrast images of PAC1 VSMC morphology after 48 h transducted with lentivirus expressing vector control or siRNA against Akt1. B. (Left panel) Western blot analysis of extracts from PAC1 VSMC transducted with control or Akt1 siRNA. (Middle and Right panels) Bar graphs showed quantitative analysis of three independent experiments on the expression of Akt1 and SM α-actin.
However, PAC1 cells did not survive beyond 72 h when endogenous Akt1 was reduced. Similar results were obtained using a dnAkt1 mutant (Akt1 K179M) approach. These results point out that Akt1 plays a key role in PAC1 VSMC survival. Therefore, we focused our studies on the Akt1 downstream target mTOR.
mTOR play an important role in mediating FGFR1/FRS2 downstream signaling
FGFs bind and activate FGFRs [25] and heparin sulfate proteoglycan (HSPG) family such as syndecans [26]. One major challenge in studying FGFR signaling is that most cells express FGFs, FGFRs, and HSPG. To overcome this problem we developed a constitutively activated FGFR1 (FGFR1 K562E) with mutations that abolish signaling through specific pathways. This approach abrogates the need for ligand stimulation, thus prevents ligand-mediated, FGFR1-independent signaling outputs. Importantly, our laboratory has an antibody that uniquely recognizes these Xenopus FGFR1 mutants but not FGFR1 in human, mouse and rat. In addition, the Xenopus FGFR1 functions are indistinguishable from human, mouse, and rat FGFR1 [27].
mTOR is a downstream target of Akt1, and to examine the importance of mTOR in mediating FGFR1-induced VSMC phenotypic modulation, we used a specific mTOR inhibitor, rapamycin [28]. Western blot analysis showed that FGFR1 K562E increased mTOR phosphorylated at Ser2448, a residue phosphorylated by Akt (Figure 2A, lane 3), compared to the control (lane 1) and the FRS2 deletion mutant, FGFR1 K562E: -FRS2 (lane 5). We next sought to determine whether specific inhibit mTOR pathway affected FGFR1-mediated VSMC phenotypic modulation. Using immunofluorescence staining for SM α-actin, we found that rapamycin treatment partially reversed FGFR1 K562E-induced PAC1 morphological changes (Figure 2B, panel 4) compared to DMSO-treated FGFR1 K562E cells (panel 3). Control cells treated with rapamycin (panel 2) showed increased cell size and SM α-actin stress fibers compare to DMSO-treated control cells (panel 1). In the presence of rapamycin, Western blot analysis and SM α-actin promoter luciferase assays confirmed FGFR1 K562E inhibition of SM α-actin gene expression was markedly attenuated (Figure 2C–D), compare to the controls and FGFR1 K562E with DMSO treatment. We also notice that rapamycin had no effect on SM-MHC protein expression levels in FGFR1 K562E overexpressing cells (Figure 2C). It is possible that longer exposure to rapamycin is required to synthesize the SM-MHC protein.
Figure 2. Effects of rapamycin on FGFR1-mediated downregulation of SM α-actin.
A. PAC1 stable cell lines were treated with DMSO or 10 nM rapamycin for 48 h and analyzed for mTOR kinase activity (Ser2448) by Western blot. The level of total mTOR served as loading control. B. Immunofluorescence staining of SM α-actin in PAC1 stable cell lines after treatment with DMSO or 10 nM rapamycin for 48 h. Images magnified 200× were acquired at similar exposure levels. C. PAC1 stable cell lines were treated with DMSO or 10 nM rapamycin for 48 h and analyzed for SM marker gene expression. β-tubulin served as loading control. D. PAC1 stable cells were transiently transfected with SM α-actin luciferase reporter and pRL-TK Renilla luciferase. After 24 h, cells were switched to 0.5% FBS with DMSO (black bars) or 10 nM rapamycin (gray bars) and incubated for an additional 48 h. Results were representative of three experiments and displayed as mean ± SD. * p < 0.05, as compared with the control. RLU, relative luciferase units. E. Quantification of the amount of newly synthesized SM α-actin in PAC1 stable cell lines treated with DMSO or 10 nM rapamycin for 48 h.
To confirm these findings, we labeled PAC1 stable cell lines with 35S-methionine/cysteine for 2 h to examine SM α-actin protein synthesis. This brief labeling protocol allows detection of alterations in SM α-actin synthesis that are independent of protein half-life. We found that FGFR1 K562E inhibited de novo SM α-actin protein synthesis at each time point compared to the control (Figure 2E). Rapamycin treatment increased SM α-actin protein synthesis in control cells and it also partially reversed the effects of FGFR1 K562E on SM α-actin protein synthesis. Together, these results demonstrate that FRS2 contributed to FGFR1 K562E-mediated mTOR phosphorylation, and the inhibition of mTOR signaling by rapamycin was concomitant with the partially reversed VSMC morphology and gain of SM α-actin protein expression and synthesis. These results suggest that FGFR1-mediated VSMC phenotypic modulation is driven, at least in part, by increased FRS2-dependent mTOR signaling. These observations also suggest that post-transcriptional regulation plays an important role in the control of FGFR1-mediated SM α-actin expression. FGFR1 may regulate SM α-actin expression by multiple mechanisms, including the regulation of SM α-actin gene expression, SM α-actin mRNA stability, and SM α-actin protein translation. It would be interesting to test these possibilities in the future.
FGFR1/FRS2 and mTOR form a complex
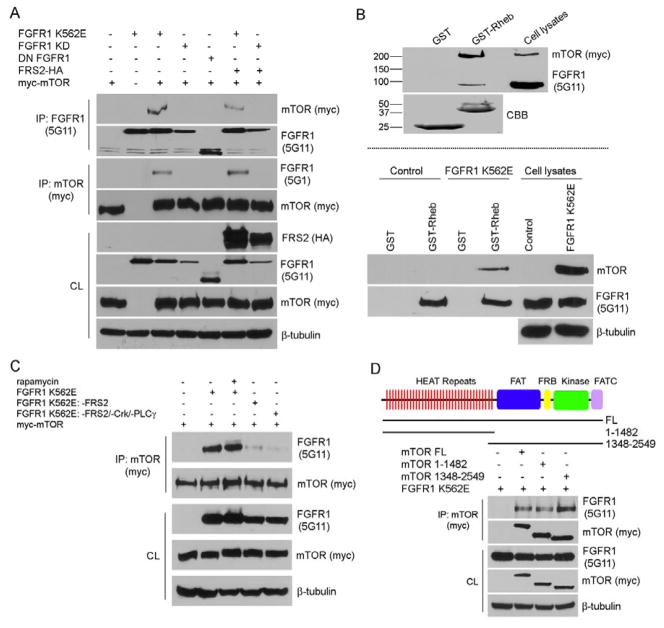
Because FRS2 deletion mutant of FGFR1 affects FGFR1-mediated mTOR activation at Ser2448 site (Figure 2A) and because rapamycin partially reversed FGFR1 K562E-mediated downregulation of SM marker gene expression, we tested whether FGFR1/FRS2 and mTOR form multi-protein complexes. In transfected 293T cells, mTOR co-immunoprecipitated with activated FGFR1/FRS2, but do not co-immunoprecipitate with kinase dead FGFR1 (FGFR1 KD) (Figure 3A). To determine if FGFR1-mTOR form a complex in VSMCs we used GST-Rheb (ras homologue enriched in brain) pull-down method as an alternative approach, because commercially available mTOR antibodies are not effective for immunoprecipitation of endogenous mTOR protein. Rheb positively regulates mTOR signaling and co-immunoprecipitate with mTOR [29]. In addition, elegant work from Sabatini and colleagues showed that the association of mTOR and its binding partner Raptor is very sensitive to non-ionic detergents, such as Triton-X 100 or NP-40, which are in general used in the preparation of cell lysates. We adapted their protein extraction protocol using CHAPS lysis buffer in this study [17]. Our results showed that GST-Rheb fusion proteins were able to pull-down both mTOR and FGFR1 in transiently transfected 293T cells and in PAC1 stable cells overexpressing FGFR1 K562E (Figure 3B).
Figure 3. FGFR1, FRS2, and mTOR form multiple complexes.
A and C–D. 293T cells were transiently transfected with different constructs as indicated. After serum starvation overnight, FGFR1 or mTOR (myc) was immunoprecipitated (IP) and subjected to Western blot analysis. The amount of transfected proteins in the cell lysates (CL) were also analyzed by Western blotting. 10 nM of rapamycin was used in this experiment in panel C. β-tubulin served as loading control. B. Cells were lysed, and the cell lysates were pre-cleaned with glutathione Sepharose alone before incubation with GST or GST-Rheb fusion proteins bound to glutathione Sepharose. Precipitates were subjected to immunoblot analysis. The input of GST and GST-Rheb construct levels shown in Upper panel were the same as in Lower panel. All results are representative of three separate experiments. CBB: coomassie brilliant blue.
Figure 3A showed that exogenous HA-tagged FRS2 can not rescue FGFR1 KD-mTOR complex formation, however, FGFR1 FRS2 deletion mutant (FGFR1 K562E: -FRS2) almost completely abrogated the FGFR1-mTOR complex formation (Figure 3C lane 4). Similar results were seen by using a FGFR1 kinase deficient mutant (FGFR1 K562E: -FRS2/Crk/-PLCγ) (lane 5). Rapamycin was found to destabilize the interaction of raptor with mTOR; however, it has no effect on FGFR1-mTOR complex formation (lane 3).
To further define the interaction domains required for FGFR1/mTOR complex formation, we used mTOR deletion constructs. Our results showed that both N-terminal and C-terminal mTOR deletion mutants bind to a constitutively active form of FGFR1 (Figure 3D). This interaction is likley mediated through the mTOR N-terminal HEAT repeats and the C-terminal FAT domain. Both of which have been previously reported as protein-protein interacting domains [30]. Our results demonstrate that mTOR binds to FGFR1 and that FRS2 is essential to formation of this complex. These results also suggest that formation of this complex regulates FGFR1-mediated downstream signaling.
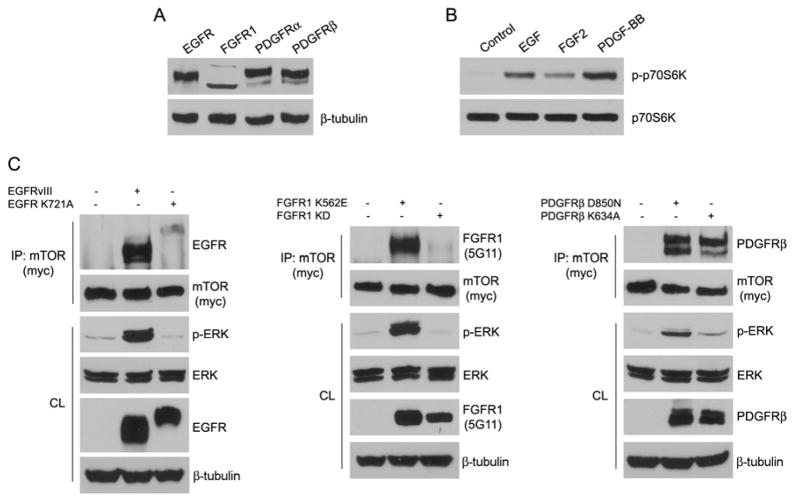
Besides FGF2, other growth factors may activate the mTOR signaling pathway [31]. Primary BVSMCs express EGFR, FGFR1, PDGFRα, and PDGFRβ (Figure 4A). EGF, FGF2, and PDGF-BB stimulation activated mTOR signaling using p70S6K1 phosphorylation as the readout (Figure 4B). Because mTOR co-immunoprecipitated with FGFR1, we tested whether EGFR or PDGFRβ formed complexes with mTOR. We co-transfected 293T cells with active and kinase dead forms of EGFR, FGFR1, PDGFRβ, and mTOR, followed by immunoprecipitation with mTOR (myc) antibodies. Our results showed that mTOR co-immunoprecipitated with constitutively active forms of EGFR and FGFR1, but not the kinase dead mutants. Interestingly, mTOR co-immunoprecipitated with a constitutively active form of PDGFRβ, but unlike with EGFR and FGFR1, mTOR co-immunoprecipitated with a kinase dead PDGFRβ (Figure 4C). These observations provide the first evidence that mTOR is capable of forming complexes with receptor tyrosine kinases (RTK). The interaction of FGFR1 and other RTKs with mTOR may be important to initiating this signaling pathway. Additional studies are underway to confirm this hypothesis and determine additional functional consequences of the formation of RTK/mTOR complexes.
Figure 4. Effects of RTKs on mTOR signaling pathway.
A. VSMCs were analyzed for expression of RTK family by immunoblotting. B. BVSMCs were serum starved overnight, stimulated with growth factors (20 ng/ml EGF, 20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 20 min, and analyzed for p70S6K phosphorylation by immunoblotting. Total p70S6K served as loading control. C. 293T cells were transiently transfected with different constructs as indicated. After serum starvation overnight, mTOR was immunoprecipitated (IP) and subjected to immunoblot analysis. The amount of transfected proteins in the cell lysate (CL) were also analyzed by immunoblotting. β-tubulin served as loading control.
In summary, we provide a novel mechanism for mTOR signaling in relaying FGFR1-mediated signals for VSMC phenotypic modulation. The key findings of this study are: (i) FGFR1 activates mTOR signaling in an FRS2 dependent manner; (ii) rapamycin treatment partially reversed FGFR1-mediated downregulation of SM marker gene expression; (iii) FGFR1/FRS2 and mTOR form a multi-protein complex in VSMCs. Work is ongoing to further characterize the physiological function of FGFR1/mTOR complex.
Acknowledgments
This work was supported by National Institutes of Health Grant R01-DK073871 (to R.E.F.), National Institutes of Health/National Center for Research Resources P20 RR1555 (to R.E.F.) and by an American Heart Association Founders Affiliate predoctoral fellowship 0715788T (to P.Y.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK. Molecular control of vascular smooth muscle cell differentiation and phenotypic plasticity. Novartis Found Symp. 2007;283:174–191. doi: 10.1002/9780470319413.ch14. discussion 191–173, 238–141. [DOI] [PubMed] [Google Scholar]
- 3.Lindner V, Reidy MA. Proliferation of smooth muscle cells after vascular injury is inhibited by an antibody against basic fibroblast growth factor. Proc Natl Acad Sci U S A. 1991;88:3739–3743. doi: 10.1073/pnas.88.9.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoine M, Wirz W, Tag CG, Mavituna M, Emans N, Korff T, Stoldt V, Gressner AM, Kiefer P. Expression pattern of fibroblast growth factors (FGFs), their receptors and antagonists in primary endothelial cells and vascular smooth muscle cells. Growth Factors. 2005;23:87–95. doi: 10.1080/08977190500096004. [DOI] [PubMed] [Google Scholar]
- 5.Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc Natl Acad Sci U S A. 2001;98:6074–6079. doi: 10.1073/pnas.111114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 8.Lamothe B, Yamada M, Schaeper U, Birchmeier W, Lax I, Schlessinger J. The docking protein Gab1 is an essential component of an indirect mechanism for fibroblast growth factor stimulation of the phosphatidylinositol 3-kinase/Akt antiapoptotic pathway. Mol Cell Biol. 2004;24:5657–5666. doi: 10.1128/MCB.24.13.5657-5666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firulli AB, Han D, Kelly-Roloff L, Koteliansky VE, Schwartz SM, Olson EN, Miano JM. A comparative molecular analysis of four rat smooth muscle cell lines. In Vitro Cell Dev Biol Anim. 1998;34:217–226. doi: 10.1007/s11626-998-0127-5. [DOI] [PubMed] [Google Scholar]
- 10.Silverman ES, Khachigian LM, Santiago FS, Williams AJ, Lindner V, Collins T. Vascular smooth muscle cells express the transcriptional corepressor NAB2 in response to injury. Am J Pathol. 1999;155:1311–1317. doi: 10.1016/S0002-9440(10)65233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, Reiser P, Hills D, Gullick WJ, Wels W. Expression of an oncogenic mutant EGF receptor markedly increases the sensitivity of cells to an EGF-receptor-specific antibody-toxin. Int J Cancer. 1998;75:878–884. doi: 10.1002/(sici)1097-0215(19980316)75:6<878::aid-ijc10>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM. STAT5b, a Mediator of Synergism between c-Src and the Epidermal Growth Factor Receptor. J Biol Chem. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 14.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN, Owens GK. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 16.Chiara F, Goumans MJ, Forsberg H, Ahgren A, Rasola A, Aspenstrom P, Wernstedt C, Hellberg C, Heldin CH, Heuchel R. A gain of function mutation in the activation loop of platelet-derived growth factor beta-receptor deregulates its kinase activity. J Biol Chem. 2004;279:42516–42527. doi: 10.1074/jbc.M406051200. [DOI] [PubMed] [Google Scholar]
- 17.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 18.Kovalenko D, Yang X, Nadeau RJ, Harkins LK, Friesel R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J Biol Chem. 2003;278:14087–14091. doi: 10.1074/jbc.C200606200. [DOI] [PubMed] [Google Scholar]
- 19.Mood K, Friesel R, Daar IO. SNT1/FRS2 mediates germinal vesicle breakdown induced by an activated FGF receptor1 in Xenopus oocytes. J Biol Chem. 2002;277:33196–33204. doi: 10.1074/jbc.M203894200. [DOI] [PubMed] [Google Scholar]
- 20.Neilson KM, Friesel R. Ligand-independent activation of fibroblast growth factor receptors by point mutations in the extracellular, transmembrane, and kinase domains. J Biol Chem. 1996;271:25049–25057. doi: 10.1074/jbc.271.40.25049. [DOI] [PubMed] [Google Scholar]
- 21.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Webster JB, Kovalenko D, Nadeau RJ, Zubanova O, Chen PY, Friesel R. Sprouty genes are expressed in osteoblasts and inhibit fibroblast growth factor-mediated osteoblast responses. Calcif Tissue Int. 2006;78:233–240. doi: 10.1007/s00223-005-0231-4. [DOI] [PubMed] [Google Scholar]
- 23.Martin KA, Merenick BL, Ding M, Fetalvero KM, Rzucidlo EM, Kozul CD, Brown DJ, Chiu HY, Shyu M, Drapeau BL, Wagner RJ, Powell RJ. Rapamycin promotes vascular smooth muscle cell differentiation through insulin receptor substrate-1/phosphatidylinositol 3-kinase/Akt2 feedback signaling. J Biol Chem. 2007;282:36112–36120. doi: 10.1074/jbc.M703914200. [DOI] [PubMed] [Google Scholar]
- 24.Martin KA, Rzucidlo EM, Merenick BL, Fingar DC, Brown DJ, Wagner RJ, Powell RJ. The mTOR/p70 S6K1 pathway regulates vascular smooth muscle cell differentiation. Am J Physiol Cell Physiol. 2004;286:C507–517. doi: 10.1152/ajpcell.00201.2003. [DOI] [PubMed] [Google Scholar]
- 25.Friesel R, Maciag T. Fibroblast growth factor prototype release and fibroblast growth factor receptor signaling. Thromb Haemost. 1999;82:748–754. [PubMed] [Google Scholar]
- 26.Horowitz A, Tkachenko E, Simons M. Fibroblast growth factor-specific modulation of cellular response by syndecan-4. J Cell Biol. 2002;157:715–725. doi: 10.1083/jcb.200112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friesel R, Dawid IB. cDNA cloning and developmental expression of fibroblast growth factor receptors from Xenopus laevis. Mol Cell Biol. 1991;11:2481–2488. doi: 10.1128/mcb.11.5.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edinger AL, Linardic CM, Chiang GG, Thompson CB, Abraham RT. Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res. 2003;63:8451–8460. [PubMed] [Google Scholar]
- 29.Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 30.Alarcon CM, Heitman J, Cardenas ME. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol Biol Cell. 1999;10:2531–2546. doi: 10.1091/mbc.10.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung J, Grammer TC, Lemon KP, Kazlauskas A, Blenis J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature. 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]