Abstract
Individuals are frequently observed to have long segments of uninterrupted sequences of homozygous markers. One of the major mechanisms that gives rise to such long homozygous segments is consanguineous marriages, where parents pass shared chromosomal segments to their child. Such chromosomal segments are also known as autozygous segments. The clinical evidence that progeny from inbred individuals may have reduced health and fitness because of homozygosity of recessive alleles is well-known. As the length of such homozygous segments depends on the degree of parental consanguinity, it would be logical to observe shorter homozygous segments in more outbred populations. However, a recent study identified long homozygous regions, thus likely to be autozygous segments in the HapMap populations. While an abundance of homozygous segments may significantly reduce the ability to fine map disease genes using association studies, detecting tracts of extended homozygosity related to disease status seems the natural next step in genome-wide association studies beyond allele, genotype and haplotype association analyses. In this study, we propose a new algorithm to map disease-related segments based on autozygosity using case-control data. The underlying rationale for the proposed method is that shared autozygosity regions that differ between diseased and healthy individuals may harbor mutations underlying diseases. Specifically, our algorithm uses a sliding-window framework and employs a LOD score measure of autozygosity coupled with permutation-based methods to identify disease related regions. We illustrate the advantage of the algorithm with its application to a genome-wide association study on Parkinson's disease.
Keywords: Homozygosity, IBD
Introduction
Individuals are frequently observed to have long segments of uninterrupted sequences of homozygous markers. These long homozygous segments arise through numerous mechanisms, including consanguineous marriages, in which parents pass shared chromosomal segments to their child. Such chromosomal segments are also known as autozygous segments, i.e., the two alleles in a homozygous genotype are identical by descent (IBD). Statistically, the length of such homozygous segments depends on the degree of parental consanguinity, because it is on average reduced via recombination and related processes that break up chromosomal segments over generations. Therefore, we expect to see shorter homozygous segments in more outbred populations [Gibson et al., 2006]. However, based on HapMap SNP data, Gibson et al. [2006] identified 1393 homozygous regions exceeding 1 Mb in length, and thus likely to be autozygous segments among 209 unrelated HapMap individuals, with the longest spanning 17.9 Mb in a Japanese individual. Li et al. [2006] observed frequent occurrence of long contiguous stretches of homozygosity ranging from 2.94 to 26.27 Mb in Han Chinese, Taiwanese aborigines, Caucasians, and African Americans. In another study, using 276 unrelated controls recruited for an association study of Parkinson's disease, Simon-Sanchez et al. [2007] identified 26 samples with extended homozygosity spanning 5 Mb or longer. Long homozygous segments have also been observed in CEPH individuals [Broman and weber, 1999]. The origin of these segments is unclear. An alternative mechanism other than consanguinity that may increase homozygosity is linkage disequilibrium (LD) in a population. But homozygous segments created this way, although identical by state, would not be autozygous and would likely be very short. Heterozygous deletions can sometimes be detected by apparent homozygosity as well, but we presume these segments would also likely be very short. Other explanations of long homozygous segments include deletions or chromosomal abnormalities such as uniparental disomy, when a person receives two copies of a chromosome or part of a chromosome from one parent and no copies from the other parent. However, a recent study using 276 unrelated controls recruited for an association study of Parkinson's disease [Simon-Sanchez et al., 2007] rules out the possibility that the observed homozygosity was due to deletion through the examination of SNP hybridization intensity to estimate copy numbers. Moreover, the authors re-assayed DNA extracted directly from blood and showed that the effects of lymphoblast cell lines (LCL) creation and passage on genotypes and genetic architecture are minimal. Another recent study using trios from CEPH families found no excess of apparent transmission errors in the regions of extended homozygosity [Curtis, 2007], thus rules out the possibility of uniparental disomy. In another work, Curtis et al. [2008] also identified regions of extended homozygosity over 1Mb in CEPH populations and argued that the alternative explanation of the presence of the tracts of long homozygous segments is the presence of extended haplotypes with high frequencies in the populations so that these extended haplotypes were inherited by chance from both parents of a subject.
The clinical implication of long homozygous segments are well known. Progeny from inbred individuals may have reduced health and fitness because of homozygosity of recessive alleles [Morton 1978; Stoll et al., 1994; Ober et al., 1999; Rudan et al., 2003]. Wright [1922] first introduced the inbreeding coefficient to measure how close two people are genetically related to each other, Lander and Botstein [1987] later proposed a homozygosity mapping method that uses the inbreeding coefficient and searches for an autozygous genome region in inbred individuals afflicted by the disease of interest. As their method specifically looked at offspring from consanguineous matings and affected with a recessive trait, it is rather restrictive. Broman and Weber [1999] pointed out that it might be possible to extend the homozygosity (autozygosity) mapping to genetically more complex disorders from rare recessive disorders. This is because there often will be greater disease risk with the presence of two copies of a relatively common allele than the presence of a single copy. Most of the current studies of inbreeding in humans have relied on genealogical structure. Recently, Leutenegger et al. [2006] developed a homozygosity mapping method for rare recessive traits in a population of individuals with unknown pedigree structure.
The rapid technological advance in genotyping SNP assays has made genome-wide association studies realistic and manageable. Recently, several genome-wide association studies have shown great power in identifying SNPs associated with complex diseases [Scott et al., 2007; Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research, 2007; The Wellcome Trust Case Control Consortium, 2007]. All those studies had large numbers of cases and controls. In our collaborative work on genome-wide molecular profiling of colon cancer patients, Bacolod et al. [Accepted] applied autozygosity mapping method as an alternative approach to identify disease associated SNPs, which used a smaller number of cases and controls. In a recent review paper on genome-wide association studies, the authors pointed out that the most obvious application of genome-wide association studies beyond simple association is the detection of tracts of extended homozygosity [Gibbs and Singleton 2006]. This has become possible when a SNP genotyping platform provides high resolution and essentially complete genomic coverage. The observation that the abundance of homozygous segments will significantly reduce the ability to fine map disease genes using association studies also supports the idea of autozygosity mapping with genome-wide association studies [Gibson et al., 2006]. However, those recent reports with tracts of homozygosity identified in unrelated individuals only concentrated on identifying tracts on the individual level, but did not try to link the identified tracts to disease status.
We developed an algorithm that examines strength of shared autozygous segments measured by a LOD score between cases and controls under a genome-wide association study framework for disease mapping. The underlying rationale for the proposed method is that shared autozygous regions, which are distinguished from the chance homozygosity that differ between diseased and healthy individuals may harbor mutations underlying diseases. The proposed method is expected to be powerful when applied to isolated populations or subgroups in which consanguinity is high, for example, with selected case and control individuals from Ashkenazi Jewish populations. The proposed method is also expected to be powerful in non-isolated or non-subgroup populations because ascertainment procedures which select case individuals from the general population tend to make them distantly related, especially for rare recessive traits [Terwilliger 2000]. This observation has been previously cited as a disadvantage of case-control studies [Terwilliger 2000] but we turn this into a search tool in our proposed method. We illustrate the advantage of the proposed algorithm in an application of a genome-wide association study with Parkinson's disease [Fung et al., 2006].
Method
We develop a new algorithm to map disease related autozygous segments with case-control data from genome-wide association studies. Assume there are n1 unrelated cases, n2 unrelated controls, and M biallelic (denoted by allele A and allele a) single nuclear polymorphism (SNP) markers distributed throughout the genome. The autozygosity mapping algorithm is divided into four steps.
Step 1: Estimate Allele Frequency
Allele frequency of each SNP marker is estimated for case and control groups separately simply using the allele counting method.
Step 2: Quantify Strength of Autozygosity
For each individual, we quantify the strength of autozygosity for a given chromosomal segment i, where we form segments by moving a window of size w (Mb) from one end of a chromosome to the other with b (Mb) step size. Note that two adjacent segments overlap with each other (with length w-b Mb) if the step size b is smaller than the window size w. A smaller segment is defined at the end of a chromosome. Within each segment i, there are in total Ki SNP markers, where we observe a genotype Gk at the kth SNP. Define Xk =1 if SNP k is autozygous and 0 otherwise. The probability of observing a genotype Gk is determined by the autozygosity status Xk and is a function of the allele frequency at marker k under the assumption of Hardy-Weinberg equilibrium (Table 1). For SNP markers with missing genotypes, we have Pr(Gk|Xk=x) =1 for all possible autozygous status. We also include an error and mutation model similar to that in Broman and Weber [1999]. Assuming linkage equilibrium between all markers, the strength of autozygosity for a given chromosomal segment i of individual j is quantified by a LOD score comparing the hypothesis that the segment is autozygous versus it is not autozygous with the following formula [Broman et al., 1999],
Table 1.
Probabilities of the observed genotype Gk given the autozygosity status and the error and mutation model.
| Probability of Gk when | ||||
|---|---|---|---|---|
| Observed Genotype Gk | Autozygous at k | Not Autozygous at k | ||
| AA |
|
|
||
| AB | 2εpApB | 2pApB | ||
| BB |
|
|
||
| Missing | 1 | 1 | ||
ε refers to the rate of genotyping errors and mutations.
pA refers to the allele frequency of allele A.
With a fixed window size w and a step size b, a total of N possible segments will be formed across the autosomal genome for each individual case and each individual control.
Step 3: Compare Strength of Autozygosity of Each Segment Between Cases and Controls
After Step 2, for each chromosomal segment formed by a sliding window, we will have n1 LOD scores for n1 cases, with each LOD score measuring the strength of autozygosity for the specific segment for one case; and n2 LOD scores for n2 controls, with each LOD score measuring the strength of autozygosity for the specific segment for one control. To determine whether any of the N chromosomal segments contributes to the disease status because of autozygosity, we compare the strength of autozygosity between n1 cases and n2 controls using the two-sample t-test, one sided, for each segment. Large value of the t statistic for one segment might indicate the role of chromosomal segment in predicting the disease status because of autozygosity.
Step 4: Significance Test
When the step size b is smaller than the window size w, each segment overlaps with several of its neighboring segments, making the test statistics for these overlapping segments positively correlated (such correlations may also be present because the SNPs are not in linkage equilibrium as assumed under our model). Thus, standard tests for the significance of test statistics are not valid. Therefore, we propose to assess the significance level associated with the t statistic Ti for segment i with permutation procedures while maintaining the autocorrelation between t statistics of the neighboring segments. We permute the disease status among n1 cases and n2 controls, which generates a new data set in which the null hypothesis holds that no segment is associated with the disease status because of autozygosity. With the jth permuted data set pj, we obtain the t statistic for segment i and the genome-wide maximum statistic among all . We repeat the permutation procedures 1,000 times to generate the distribution of the test statistic under the null hypothesis. Thus, the genome-wide adjusted p-value for the t statistic Ti comparing autozygosity strength of the ith segment between case and control groups is the proportion of the genome-wide maximum statistics from the permuted data that are equal or greater than the observed statistic Ti [Westfall and Young, 1993].
We also record the 100(1–α)% percentile of the maximum as the 100(1–α)% genome-wide threshold for the statistic Ti, which can be used to detect the presence of an autozygous segment in the autosomal genome that is associated with the disease status so as to control the overall Type I error rate to be α or less [Churchill and Doerge, 1994].
Results
We applied the algorithm to a genome-wide association study of Parkinson's disease with 500K SNP chip data [Fung et al., 2006]. The raw data were made publicly available by Coriell Institute for Medical Research (NJ, USA). There are 270 Parkinson's disease patients and 271 neurologically normal controls, and 408,787 SNP markers genotyped for each individual. All subjects were Caucasians. We considered 396,591 autosomal SNP markers excluding 12,196 SNP markers on the sex chromosomes. For quality control of the genotyping data, we removed SNPs that were monomorphic, thus uninformative; SNPs for which no homozygotes were observed in the entire sample or which had genotypic frequencies departing from Hardy-Weinberg equilibrium after Bonferroni correction. Of the 396,591 SNPs, 395,245 were informative and passed the quality-control checks and were therefore carried over into the autozygosity analysis.
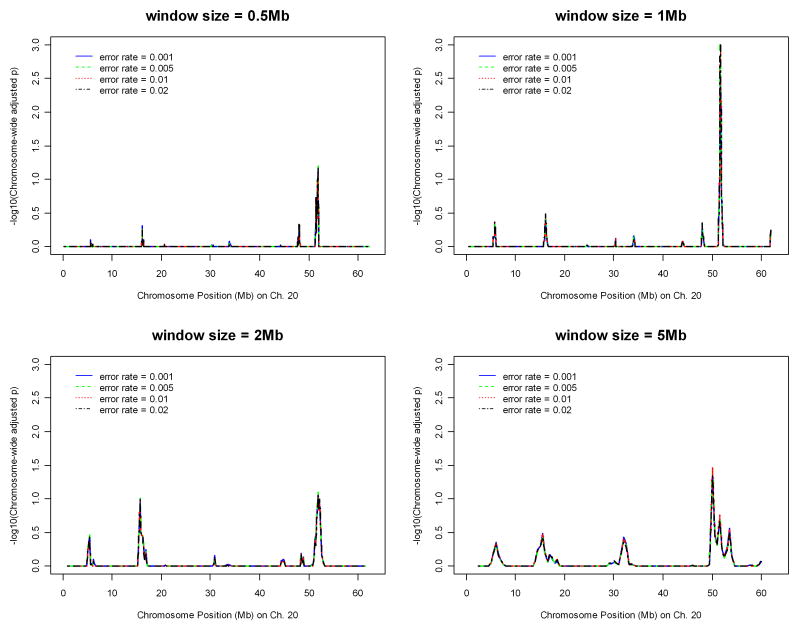
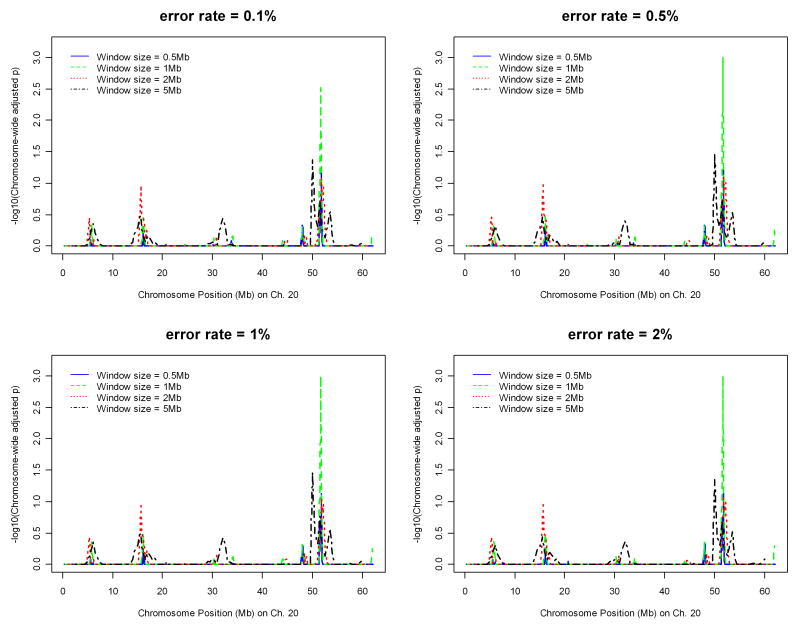
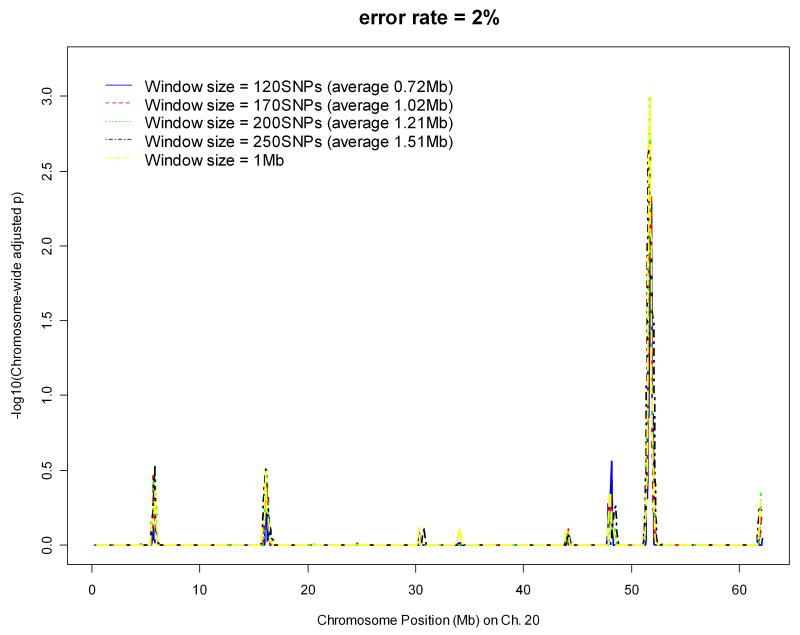
With significance suggested on chromosome 20 from preliminary runs on Parkinson's disease, we applied the algorithm on the data of chromosome 20 only with different parameter values to determine the best set of the parameter values ad hoc. Figures 1 and 2 display the effects of window sizes, and error rates on IBD mapping, respectively, where the step sizes are fixed as one tenth of the window sizes. Figure 1 suggests that window size 1 Mb might give the most powerful results among four window sizes ranging from 0.5 Mb to 5 Mb. Figure 2 shows that error rates 0.5%, 1%, and 2% all give similar results. Therefore, in the whole-genome analysis of the Parkinson's disease data, we chose the window size to be 1Mb, the step size to be 0.1Mb, and the error rate to be 2%. Instead of fixing the window size based on number of base pairs, we also investigated the effect of window size by fixing the number of SNPs falling into one window. The error rate was fixed at 2% and step size was fixed as one tenth of the number of SNPs falling in one window. Figure 3 suggests that the results will not be affected by how window size is fixed. The same pattern was observed and the same peak was detected.
Figure 1.
Effect of window size (with base pair length) on IBD mapping with chromosome 20 data of the Parkinson's disease.
Figure 2.
Effect of error rate on IBD mapping with chromosome 20 data of the Parkinson's disease.
Figure 3.
Effect of window size (with number of SNPs falling into one window) on IBD mapping with chromosome 20 data of the Parkinson's disease.
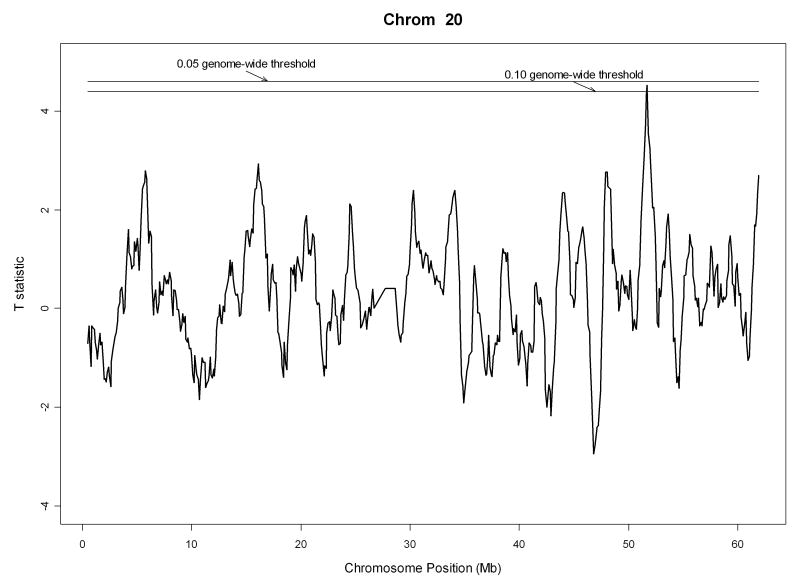
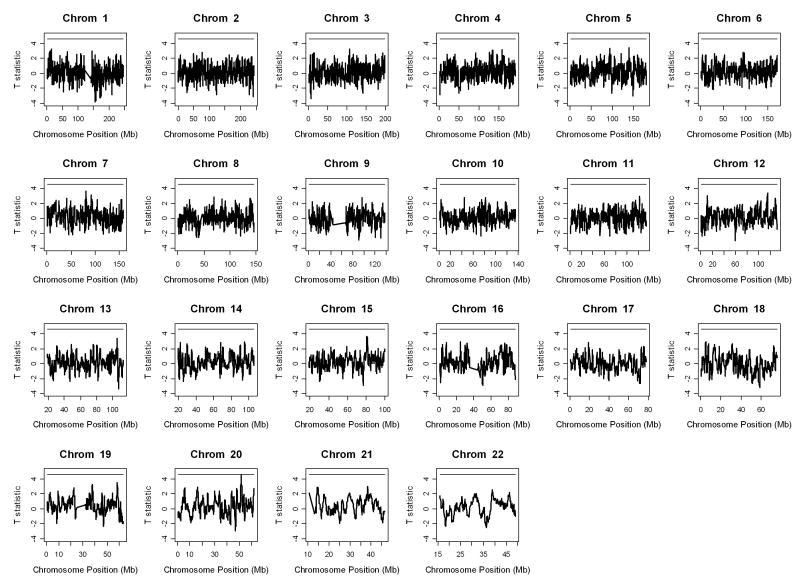
We moved a window of size 1 Mb from one end of a chromosome to the other with a step size of 0.1 Mb, making adjacent segments overlap by 0.9 Mb. This formed a total of 26,668 segments genome-wide. We also assumed an error and mutation rate of 2% in the autozygosity analysis. The pattern of genome-wide IBD comparisons between Parkinson's cases and normal controls is displayed in Figure 3 together with the 0.05 genome-wide significance threshold of t statistics. There is one region on chromosome 20 (from 51211799 bp to 52211798 bp) that has t statistic 4.51 comparing strength of autozygosity between Parkinson's cases and normal controls with genome-wide adjusted p-value 0.053. The IBD pattern for chromosome 20 only is individually displayed in Figure 5. Within the identified region, on average there were 225 SNPs (ranging from 218 to 226) genotyped for cases and 226 SNPs (ranging from 215 to 226) genotyped for normal controls, making the average distance between two SNPs 4.4 kb. We then examined the identified region for Parkinson's disease. Table 2 displays all genes in the region from 51211799 bp to 52211798 bp on chromosome 20 together with their descriptions.
Figure 5.
Pattern of the IBD comparisons on chromosome 20 between Parkinson's cases and normal controls. The top horizontal bar is the 0.05 genome-wide significance threshold of t statistics. The bottom horizontal bar is the 0.1 genome-wide significance threshold of t statistics.
Table 2.
Genes within the identified region on chromosome 20 for the Parkinson's disease.
| Genes Symbols | Descriptions |
|---|---|
| LOC728805 | hypothetical protein LOC728805 |
| TSHZ2 | teashirt family zinc finger 2 |
| PPIAP10 | peptidylprolyl isomerase A (cyclophilin A) pseudogene 10 |
| ZNF217 | zinc finger protein 217 |
| LOC391257 | SUMO1 pseudogene 1 |
| BCAS1 | breast carcinoma amplified sequence 1 |
| CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 |
| PFDN4 | prefoldin subunit 4 |
It is known that the etiology of many neurodegenerative diseases such as Parkinson's and Alzheimer's disease is related to aggregation of inappropriately folded proteins in the brain. For Parkinson's disease, these protein clumps, known as Lewy bodies, contain α-synuclein, ubiquitin, α/β-tubulin and other misfolded proteins. We notice that gene LOC391257 in the identified region is a SUMO1 (small ubiquitin-like modifier) pseudogene. If gene LOC391257 is transcribed, it may interfere with SUMO1, which was found to be able to counteract ubiquitin and stabilize proteins against degradation by the 26S proteasome. The fact that parkin is a ubiquitin ligase suggests that disturbance of protein degradation by the ubiquitin-proteasome system might have a critical role in neurodegeneration [Hattori and Mizuno, 2004; Oria et al., 2005]. Another interesting gene in the identified region is CYP24A1. Cytochrome P450 members have been implicated in detoxifying a number of risk factors of Parkinson's disease. More specifically, studies have shown that smoking activates P450 genes and has protective effect on development of Parkinson's disease [Miksys and Tyndale, 2006; Elbaz et al., 2007; Duric et al., 2007]. Lastly, gene PFDN4, which encodes prefoldin subunit 4, is a member of the chaperonins that assist in proper folding of its target proteins such as actin and tubulins [Simons et al., 2004].
Consistent with what has been observed previously [Gibson et al., 2006], homozygous tracts are extremely common in unrelated Parkinson's cases and normal controls. All 270 Parkinson's cases and 271 normal controls had at least one segment with LOD score of autozygosity greater than 25 (an arbitrary high value showing very strong autozygosity). Within segments of LOD > 25, the percentage of homozygous genotypes within a specific segment ranges from 86.2% to 100% for cases, and 83.6% to 100% for controls. More specifically, among segments of LOD > 25, 75.8% segments of cases have 100% homozygous genotypes and 70.2% segments of controls have 100% homozygous genotypes. Within the identified segment on chromosome 20, the percentage of homozygous genotypes is 66.3% and 64.2% for cases and controls, respectively with mean LOD scores of -105.3 for cases and -114.8 for controls. The longest segment with complete homozygosity within the identified region among all 270 Parkinson's disease patients is 0.552 Mb in length and that among 271 normal controls is 0.325 Mb. The average length of the longest segment with complete homozygosity of each patient within the identified region is 0.114 Mb and that of each normal control is 0.102 Mb.
To check the false positive rate of the algorithm, we applied the algorithm to the chromosome 20 data of the Parkinson's study, where the two groups were formed by randomly splitting 271 normal controls (135 vs. 136). The same parameter settings were applied, i.e., 1 Mb window size, 0.1 Mb step size and 2% error rate. Among the 606 segments formed along chromosome 20, 20 were significant at the 0.05 significance level, making the false positive rate 0.033.
Discussion
Genome-wide association studies have been proposed as a method to identify common genetic variability that underlies complex diseases. Gibbs and Singleton [2006] suggested that the application of genome-wide association studies beyond allele, haplotype, and genotype associations consists of detecting tracts of extended homozygosity, which might contain a genetic susceptibility variant that is responsible for the underlying disease. The most likely explanation for such long tracts of homozygosity is autozygosity, wherein the same chromosomal segments inherited from a common ancestor were passed from (distantly related) parents to a child. The length of autozygosity tracts may be influenced by the degree of consanguinity in the marriage, mutation rate, population structure, uniparental disomy (UPD), natural selection, recombination process, and linkage disequilibrium (LD) pattern [Gibson et al., 2006]. Broman and Weber [1999] observed long tracts of homozygosity in the CEPH families, Gibson et al. [2006] observed tracts exceeding 1Mb in length among the unrelated HapMap individuals, and Li et al. [2006] observed frequent occurrence of long contiguous stretches of homozygosity in Han Chinese, Taiwanese aborigines, Caucasians, and African Americans.
In this paper, we proposed an algorithm to predict disease status under a genome-wide association study framework with case-control data through examining the strength of autozygosity for apparently recessive disorders. Given the current SNP density, unlike that in Broman and Weber [1999], where all possible subsets of contiguous markers were considered, we propose a sliding-window method with a fixed window size. A permutation procedure that takes into account the dependence among adjacent segments is applied to obtain empirical significance. The sliding window idea was also applied in the work of Lin et al. [2004] to identify shared loss-of-heterozygosity (LOH) regions based on paired normal and tumor samples from the same prostate cancer patient. Huang et al. [2004] used a probabilistic method to calculate the probability of a stretch of SNPs with fixed length all being homozygous. The algorithm was incorporated in the Affymetrix's CNAT copy number and LOH calculation. The feasibility of the proposed algorithm has been demonstrated through an application of a genome-wide association study with Parkinson's disease [Fung et al., 2006]. Note that we applied the parameter checking procedures ad hoc only on chromosome 20 in order to be computationally more efficient. With a new data set on a different disease with a different population, similar procedures can be applied. That is, run the algorithm on the whole genome with one set of parameters and narrow down the parameter checking procedures to chromosomes that show signals. The proposed algorithm has a disadvantage that it does not take into account the fact that the tracts of homozygosity might not be homozygous for the same allele, or the patterns of homozygosity within one window might not be the same for different individuals. From several recent reports, we observed that different tracts of homozygosity are distributed across the genome differently in different unrelated individuals. Therefore, instead of mapping certain specific tracts, our algorithm maps the disease based on the strength of autozygosity with a LOD score measuring how likely for a segment to be an autozygous segment. We consider our proposed algorithm a screening tool to identify segments possibly harboring disease variants/haplotypes that warrants further studies within the identified region to identify true disease associations. One region on chromosome 20 was identified to have different autozygosity strength between Parkinson's cases and normal controls after genome-wide multiple comparisons adjustment. We do not expect to replicate previous findings with the proposed method as previous studies of gene mapping with Parkinson's disease were either association studies looking at the difference in allele frequency or genotype frequency between cases and controls or linkage studies looking at recombinations. Although several genes in the identified region are consistent with influencing Parkinson's disease pathology [Hattori et al., 2004; Oria et al., 2005; Miksys et al., 2006; Elbaz et al., 2007; Duric et al., 2007; Simons et al., 2004; Sun et al., 2007], further study on this suggestive region is needed to narrow down the disease related segment and to understand the functions of the candidate genes that may possibly be associated with Parkinson's disease.
With frequent observations of long tracts of homozygosity in outbred populations, and with current high-density SNP maps, the natural next step for genome-wide association studies is detecting tracts of extended autozygosity in individuals with recessive diseases. In this study, we have demonstrated the power and feasibility of an autozygosity association mapping algorithm with genome-wide association studies. Although the results from the application on the 500K genome-wide association study with Parkinson's disease are promising, future studies are needed to improve the current algorithm for autozygosity association mapping. To define a new LOD score of autozygosity that takes LD between SNPs into account, and to propose a new algorithm that defines segments with different lengths according to the percentage of homozygosity will be our future research.
Figure 4.
Pattern of the genome-wide IBD comparisons between Parkinson's cases and normal controls. The horizontal bar is the 0.05 genome-wide significance threshold of t statistics.
Acknowledgments
Support through China NSFC grant no. 30730057 (J.O) and grant P01 CA065930 (J.O. & F.B) from the National Cancer Institute is gratefully acknowledged.
References
- Bacolod MD, Schemmann GS, Wang S, Shattock R, Giardina SF, Zeng ZS, Shia J, Stengel RF, Gerry N, Hoh J, Kirchoff T, Gold B, Christman MF, Offit K, Gerald WL, Notterman DA, Ott J, Paty PB, Barany F. The Signatures of Autozygosity Among Patients Afflicted with Colorectal Cancer. Cancer Res. 2008;68:2610–2621. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K, Weber J. Long Homozygous Chromosomal Segments in Reference Families from the Centre d'Etude du Polymorphisme Humain. Am J Hum Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D. Extended homozygosity is not usually due to cytogenetic abnormality. BMC Genetics. 2007;8:67. doi: 10.1186/1471-2156-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Vine AE, Knight J. Study of Regions of Extended Homozygosity Provides a Powerful Method to Explore Haplotype Structure of Human Populations. Ann Human Genet. 2008;72:261–278. doi: 10.1111/j.1469-1809.2007.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Duric G, Svetel M, Nikolaevic SI, Dragadevic N, Gavrilovic J, Kostic VS. Polymorphisms in the genes of cytochrome oxidase P450 2D6 (CYP2D6), paraoxonase 1 (PON1) and apolipoprotein E (APOE) as risk factors for Parkinson's disease. Vojnosanit Pregl. 2007;64(1):25–30. doi: 10.2298/vsp0701025d. [DOI] [PubMed] [Google Scholar]
- Elbaz A, Dufouil C, Alperovitch A. Interaction between genes and environment in neurodegenerative diseases. C R Biol. 2007;330(4):318–328. doi: 10.1016/j.crvi.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Fung HC, Scholz S, Matarin M, Simon-Sanchez J, Hernandez D, Britton A, Gibbs JR, Langefeld C, Stiegert ML, Schymick J, Okun MS, Mandel RJ, Fernandez HH, Foote KD, Rodriguez RL, Peckham E, De Vrieze FW, Gwinn-Hardy K, Hardy JA, Singleton A. Genome-wide genotyping in Parkingson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- Gibson J, Morton N, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;5(5):789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Gibbs JR, Singleton A. Application of Genome-Wide Single Nucleotide Polymorphism Typing: Simple Association and Beyond. PLoS Gene. 2006;2(10):1511–1517. doi: 10.1371/journal.pgen.0020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Mizuno Y. Pathogenetic mechanisms of parkin in Parkinson's disease. Lancet. 2004;364(9435):722–724. doi: 10.1016/S0140-6736(04)16901-8. [DOI] [PubMed] [Google Scholar]
- Huang J, Wei W, Zhang J, Liu GY, Bignell GR, Stratton MR, Futreal PA, Wooster R, Jones KW, Shapero MH. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genome. 2004;4:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- Leutenegger AL, Labalme A, Genin E, Toutain A, Steichen E, Clerger-Darpoux F, Edery P. Using Genomic Inbreeding Coefficient Estimates for Homozygosity Mapping of Rare Recessive Traits: Application to Taybi-Linder Syndrome. Am J Hum Genet. 2006;79:62–66. doi: 10.1086/504640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LH, Ho SF, Chen CH, Wei CY, Wong WC, Li LY, Hung SI, Chung WH, Pan WH, Lee MT, Tsai FJ, Chang CF, Wu JY, Chen YT. Long Contiguous Stretches of Homozygosity in the Human Genome. Hum Mutat. 2006;27(11):1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-vased loss-of-heterozygosity data. Bioinfomatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- Miksys S, Tyndale RF. Nicotine induces brain CYP enzymes: relevance to Parkinson's disease. J Neural Transm Suppl. 2006;70:177–80. doi: 10.1007/978-3-211-45295-0_28. [DOI] [PubMed] [Google Scholar]
- Morton N. Effect of inbreeding on IQ and mental retardation. Proc Natl Acad Sci USA. 1978;75:3906–3908. doi: 10.1073/pnas.75.8.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Hyslop T, Hauck WW. Inbreeding effects on fertility in humans: evidence for reproductive compensation. Am J Hum Genet. 1999;64:225–231. doi: 10.1086/302198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oria RB, Patrick PD, Zhang H, Lorntz Breyette, Castro Costa CM, Brito GAC, Barrett LJ, Lima AAM, Guerrant RL. APOE4 Protects the Cognitive Development in Children with Heavy Diarrhea Burdens in Northeast Brazil. Pediatr Res. 2005;57:310–316. doi: 10.1203/01.PDR.0000148719.82468.CA. [DOI] [PubMed] [Google Scholar]
- Rudan I, Rudan D, Campbell H, Carothers A, Wright A, Smolej-Narancic N, Janicijevic B, Jin L, Chakraborty R, Deka R, Rudan P. Inbreeding and risk of late onset complex disease. J Med Genet. 2003;40:925–932. doi: 10.1136/jmg.40.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons CT, Staes A, Rommelaere H, Ampe C, Lewis SA, Cowan NJ. Selective contribution of eukaryotic prefoldin subunits to actin and tubulin binding. J Biol Chem. 2004;279(6):4196–203. doi: 10.1074/jbc.M306053200. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, Wavrant de Vrieze F, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16(1):1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Stoll C, Alembik Y, Dott B, Feingold J. Parental consanguinity as a cause of increased incidence of birth defects in a study of 131,760 consecutive births. Am J Med Genet. 1994;49:114–117. doi: 10.1002/ajmg.1320490123. [DOI] [PubMed] [Google Scholar]
- Sun F, Kanthasamy A, Anantharam V, Kanthasamy AG. Environmental neurotoxic chemicals-induced ubiquitin proteasome system dysfunction in the pathogenesis and progression of Parkinson's disease. Pharmacol Ther. 2007;114:327–344. doi: 10.1016/j.pharmthera.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Terwilliger G. Inflated False-Positive Rates in Hardy-Weinberg and Linkage-Equilibrium Tests are Due to Sampling on the Basis of Rare Familial Phenotypes in Finite Populations. Am J Hum Genet. 2000;67:258–259. doi: 10.1086/302964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing: examples and methods for p-value adjustment. New York: Wiley; 1993. [Google Scholar]
- Wright S. Coefficients of inbreeding and relationship. Am Nat. 1922;56:330–339. [Google Scholar]