Abstract
The finite proliferative potential of normal human cells leads to replicative cellular senescence, which is a critical barrier to tumour progression in vivo1–3. We show that human p53 isoforms (Δ133p53 and p53β)4 constitute an endogenous regulatory mechanism for p53-mediated replicative senescence. Induced p53β and diminished Δ133p53 were associated with replicative senescence, but not oncogene-induced senescence, in normal human fibroblasts. The replicatively senescent fibroblasts also expressed increased levels of miR-34a, a p53-induced microRNA5–9, the antisense inhibition of which delayed the onset of replicative senescence. The siRNA-mediated knockdown of endogenous Δ133p53 induced cellular senescence, which was attributed to the regulation of p21WAF1 and other p53 transcriptional target genes. In overexpression experiments, while p53β cooperated with full-length p53 to accelerate cellular senescence, Δ133p53 repressed miR-34a expression and extended cellular replicative lifespan, providing a functional connection of this microRNA to the p53 isoform-mediated regulation of senescence. The senescence-associated signature of p53 isoform expression (i.e., elevated p53β and reduced Δ133p53) was observed in vivo in colon adenomas with senescent phenotypes10, 11. The increased Δ133p53 and decreased p53β isoform expression found in colon carcinoma may signal an escape from the senescence barrier during the progression from adenoma to carcinoma.
Normal human somatic cells can undergo only a limited number of cell divisions, eventually reaching an irreversible proliferation arrest called replicative cellular senescence2, 12. Various cellular stresses (e.g., oncogene activation and DNA damage) can also induce cellular senescence1–3. Whether replicatively induced or prematurely stress-induced, cellular senescence constitutes a critical mechanism for tumour suppression in vivo and may contribute to organismal ageing1–3. The p53 signalling pathway plays a central role in the regulation of cellular senescence2, 3. Drosophila, zebrafish and humans are reported to have p53 isoforms4, 13; however, their regulation and function are poorly understood. Here, we examine the expression profiles of two human p53 isoforms, p53β (lacking the C-terminal oligomerisation domain due to an alternative mRNA splicing)4 and Δ133p53 (lacking the N-terminal transactivation and proline-rich domains due to the transcription from an alternative promoter in intron 4)4, during cellular senescence in vitro and in vivo, their biological activities in regulating cellular senescence, and the physiological role of miR-34a5–9 in p53-mediated senescence.
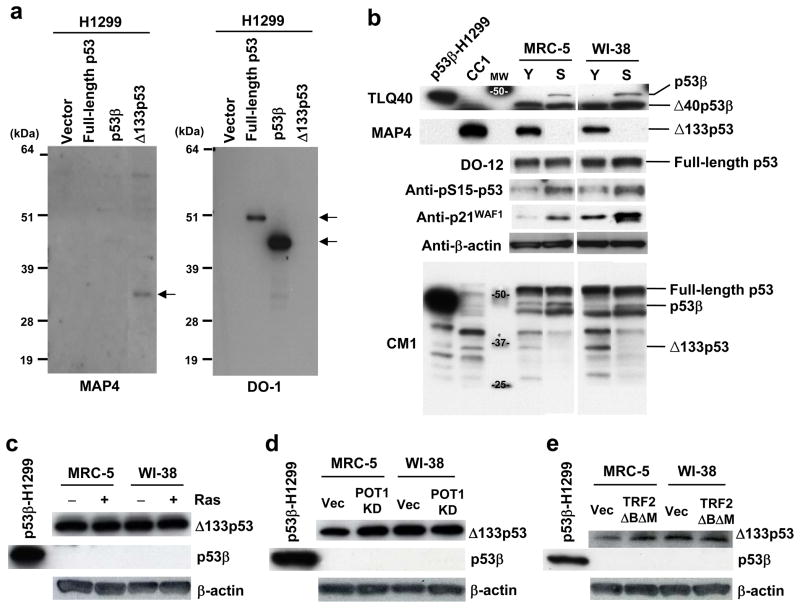
By using the p53β-specific antibody (TLQ40)4 and the Δ133p53-specific antibody (MAP4) (Fig. 1a), the endogenous expression levels of p53β and Δ133p53 were examined in normal human fibroblast strains (MRC-5 and WI-38) at early passage (both strains at passage number 30, Y in Fig. 1b) and at replicative senescence (MRC-5 at passage 65 and WI-38 at passage 58, S in Fig. 1b) (Supplementary Information, Fig. S1a). While the expression of full-length p53 (detected by DO-12) showed no changes during replicative senescence, p53β was specifically detected when the cells became senescent. In remarkable contrast, the expression of Δ133p53 was markedly diminished in the senescent cells. As reported14, increased amounts of p53 phosphorylated at serine 15 (pS15-p53) were associated with the replicative senescence (Fig. 1b). The result using CM1 antibody showed that p53β and Δ133p53 were expressed less abundantly than full-length p53 but still at readily detectable levels (Fig. 1b). Premature senescence induced by oncogenic Ras (Supplementary Information, Fig. S1b) or acute telomere dysfunction (by knockdown of POT115 or overexpression of a dominant-negative TRF2 mutant16) was not associated with either induced p53β or diminished Δ133p53 (Fig. 1c, d, e).
Figure 1.
Replicative senescence-associated changes in expression of endogenous p53 isoforms. (a) MAP4 antibody specifically recognizes Δ133p53. H1299 cells (p53-null) exogenously expressing full-length p53, p53β or Δ133p53 were analyzed in immunoblot using MAP4 (left) and DO-1 (right) antibodies. MAP4 detects Δ133p53, but not full-length p53 or p53β. (b) Induction of p53β and repression of Δ133p53 at replicative senescence. The immunoblot analyses were performed in early-passage (Y) and senescent (S) human fibroblast strains MRC-5 and WI-38. The examined passage numbers were 30 (Y) and 65 (S) for MRC-5; and 30 (Y) and 58 (S) for WI-38. TLQ40, an antibody detecting p53β isoforms; DO-12, an antibody used to detect full-length p53; CM1, an antibody used to simultaneously detect full-length p53, p53β and Δ133p53. Δ40p53β4 was a predominant form detected by TLQ40 and was constitutively expressed in both early-passage and senescent cells. p53 phosphorylated at serine 15 (pS15-p53) and p21WAF1 were also examined. β-actin was a loading control. H1299 cells overexpressing p53β and CC1 cells were used as the positive controls for p53β andΔ133p53, respectively. As indicated on full blots available in Supplementary Information, Fig. S10a, the data of TLQ40, MAP4, DO-12 and CM1 are composite images, but all the bands shown for each antibody were from the same blot. (c-e) No induction of p53β and no repression of Δ133p53 at oncogene-induced senescence (overexpression of H-RasV12)35 (c) and premature senescence with acute telomere dysfunction induced by shRNA knockdown of POT115 (d) or overexpression of a dominant-negative TRF2 mutant16 (e). Early-passage MRC-5 and WI-38 (at passage 32) were used. H1299 cells overexpressing p53β was the positive control for p53β. Vec, vector control. β-actin was a loading control.
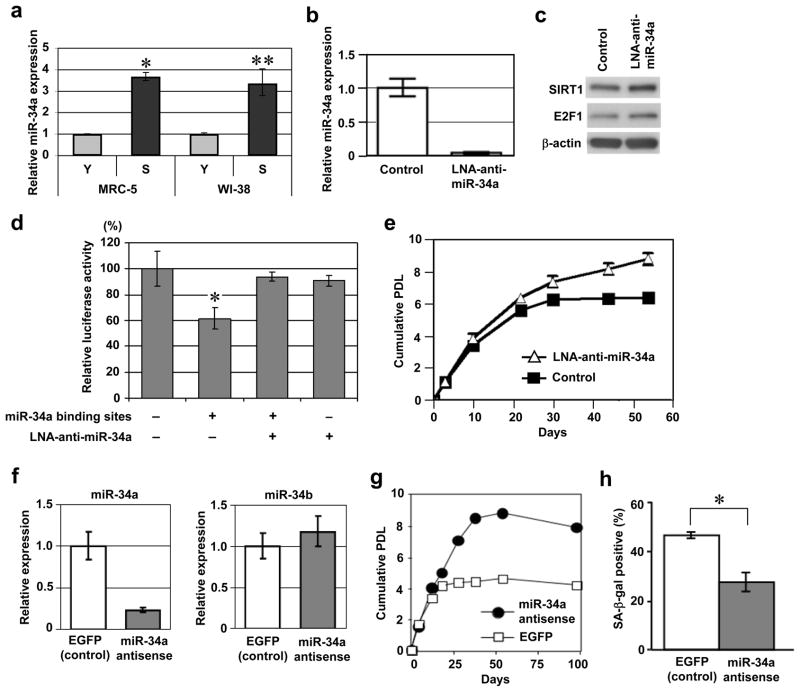
In addition to the upregulation of p21WAF1 (Fig. 1b)17, 18, the replicatively senescent MRC-5 and WI-38 fibroblasts expressed increased amounts of miR-34a (Fig. 2a), a microRNA that is transactivated by full-length p53 (Supplementary Information, Fig. S2a, b) and can induce cellular senescence when overexpressed5–9. A locked nucleic acids (LNA)-antisense oligonucleotide was used to specifically knockdown the endogenous expression of miR-34a. The specific effect of the LNA antisense was demonstrated by diminished levels of miR-34a in the qRT-PCR assay (Fig. 2b), increased protein amounts of SIRT1 and E2F1 (Fig. 2c), which were previously reported to be downregulated by miR-34a9, 19, and the abrogation of a miR-34a binding site-dependent decreased activity in the luciferase reporter assay20 (Fig. 2d). The antisense inhibition of miR-34a in late-passage MRC-5 fibroblasts resulted in an extension of the replicative lifespan by approximately three population doublings (PDLs) (Fig. 2e). The lifespan extension by miR-34a inhibition was reproducibly observed in an independent experiment using an antisense 2′-O-methyl oligonucleotide (Fig. 2f, g). The Nutlin-3a-induced senescence, which is dependent on the accumulation and activation of endogenous p5321, was also significantly, but partially (by approximately 50%), inhibited by the 2′-O-methyl oligonucleotide-mediated knockdown of endogenous miR-34a (Fig. 2h). These findings provide the first evidence that the endogenous levels of miR-34a, as one of the downstream effectors of the p53 signalling pathway, plays a physiological role in cellular senescence.
Figure 2.
Endogenous miR-34a is a regulator of replicative senescence. (a) Upregulation of miR-34a at replicative senescence. The same set of MRC-5 and WI-38 fibroblasts as in Fig. 1b were examined for miR-34a expression by real-time qRT-PCR. The data were normalized with control RNU66 expression and shown as the relative values (mean ± s.d. from triplicate sample). The reproducible results were obtained in three independent experiments.*, p < 0.001.**, p < 0.01. (b-e) LNA antisense knockdown of miR-34a extends cellular replicative lifespan. Late-passage MRC-5 fibroblasts (at passage 57) were transfected with antisense LNA oligonucleotide against miR-34a (LNA-anti-miR-34a) or control oligonucleotide. (b) Downregulation of miR-34a was confirmed by the real-time qRT-PCR, as in a. Error bars represent s.d. from triplicate sample. (c) Immunoblot analysis of the known targets of miR-34a (SIRT119 and E2F19). β-actin was a loading control. (d) Abrogation of the miR-34a binding site-dependent repression by LNA-anti-miR-34a in luciferase reporter assay. A luciferase reporter construct, pMIR-MYCN-WT containing two miR-34a binding sites (+) or pMIR-MYCN-MT1&2 with these sites mutated (−)20, and pRL-SV40 (control plasmid for normalization) were co-transfected with LNA-anti-miR-34a (+) or control oligonucleotide (−). Normalized luciferase activities (mean ± s.d. from triplicate samples) are shown relative to the value obtained with pMIR-MYCN-MT1&2 (−) and control oligonucleotide (−).*, p < 0.05. (e) The transfection of LNA-anti-miR-34a or control oligonucleotide was repeated every 4 days and the cumulative population doublings (PDL) were examined. (f-h) Antisense knockdown of miR-34a by 2′-O-methyl oligonucleotides. (f) Late-passage MRC-5 fibroblasts (at passage 58) were transfected with antisense 2′-O-methyl oligonucleotide against miR-34a or control oligonucleotide (EGFP) and analyzed by real-time qRT-PCR assays of miR-34a (left) and miR-34b (right). A non-specific effect on miR-34b was not observed. (g) The oligonucleotide transfection was repeated every 4 days and the cumulative PDL were examined, as in e. (h) hTERT-immortalized human fibroblasts (hTERT/NHF) were transfected with the 2′-O-methyl oligonucleotides (miR-34a antisense or control) and induced to senesce by treatment with 10 μM of Nutlin-3a for 72 h21. Summary of senescence-associated β-galactosidase (SA-β-gal) assay is shown. The data (mean ± s.d.) were from three independent experiments. *, p < 0.05.
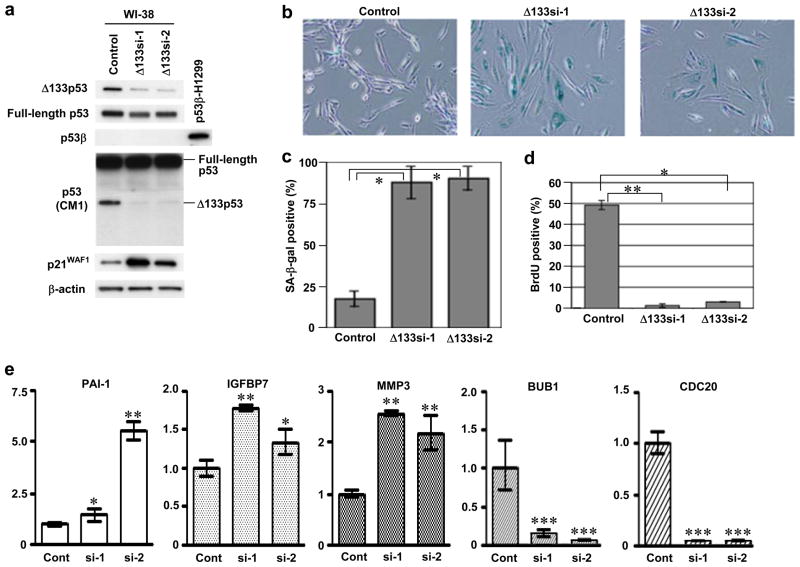
The endogenous expression of Δ133p53 was knocked down by RNA interference in early-passage WI-38 (Fig. 3) and MRC-5 (Supplementary Information, Fig. S3). Two small interfering RNA (siRNA) oligonucleotides (Δ133si-1 and Δ133si-2) efficiently downregulated the endogenous Δ133p53 with a minimal effect on full-length p53 and no induction of p53β (Fig. 3a; Supplementary Information, Fig. S3a). The cells transfected with Δ133si-1 and Δ133si-2 underwent a senescent growth arrest uniformly and rapidly (within 7 days), showing the flattened cell morphology (Fig. 3b; Supplementary Information, Fig. S3b), the induction of the senescence-associated β-galactosidase (SA-β-gal) activity (Fig. 3b, c; Supplementary Information, Fig. S3b), and the attenuation of BrdU incorporation (Fig. 3d; Supplementary Information, Fig. S3c). These results suggest that the endogenous expression of Δ133p53 is critical to the replicative potential of normal human fibroblasts. The Δ133p53 knockdown-induced senescence was accompanied by the upregulation of p21WAF1 (Fig. 3a; Supplementary Information, Fig. S3a). Also examined by qRT-PCR were PAI-1 (plasminogen activator inhibitor-1)22, IGFBP7 (insulin-like growth factor binding protein 7)23, MMP3 (matrix metallo-protease 3)24, BUB1 (budding uninhibited by benzimidazoles 1 homolog)25 and CDC20 (cell division cycle 20 homolog)26, which were reported to be upregulated (the former three) or downregulated (the latter two) at cellular senescence27 and predicted (IGFBP7: www2.ba.itb.cnr.it/p53FamTaG) or experimentally shown (the other four) to be transcriptionally regulated by p5322, 25–27. These genes were found to be changed in expression in the expected directions (Fig. 3e), consistent with the activation of the full-length p53 function upon a relief from the dominant-negative inhibition by Δ133p534. Unlike at replicative senescence, however, miR-34a was not upregulated at Δ133p53 knockdown-induced senescence (Supplementary Information, Fig. S2c). PUMA and BAX, two proapoptotic p53 target genes, were not upregulated either (data not shown), and consistently, immunoblot analyses of PARP [poly(ADP-ribose) polymerase] or caspase-3 did not show a sign of apoptosis in these siRNA-transfected fibroblasts (Supplementary Information, Fig. S3d).
Figure 3.
Knockdown of endogenous Δ133p53 induces cellular senescence. Two independent siRNAs (Δ133si-1 and Δ133si-2) were designed to target the sequences that are present in Δ133p53 mRNA as 5′ untranslated region but spliced out of full-length p53 mRNA as intron 4. Early-passage WI-38 fibroblasts (at passage 30) were transfected with Δ133si-1, Δ133si-2 or a control oligonucleotide twice (at day 1 and day 4), and at day 7 were used for immunoblot analyses (a) and examined for SA-β-gal activity (b, c), bromo-deoxyuridine (BrdU) incorporation (d) and p53 target gene expression (e). (a) siRNA-mediated repression of Δ133p53. Expressions of full-length p53 (DO-1 antibody), Δ133p53 (MAP4 antibody), p53β (TLQ40 antibody) and p21WAF1 were examined. The expression levels of full-length p53 and Δ133p53 were also confirmed by the CM1 antibody. β-actin was a loading control. H1299 expressing p53β was the positive control for TLQ40. Full blots are available in Supplementary Information, Fig. S10b. (b) Representative pictures of SA-β-gal staining. (c) Summary of SA-β-gal assay. The data (mean ± s.d.) were from three independent experiments.*, p < 0.01. (d) BrdU incorporation assay. The number of BrdU-positive cells/the total number of cells examined (at least 100 cells for each well) was recorded. Data are mean ± s.d. from triplicate wells.*, p < 0.05.**, p < 0.01. (e) Real-time qRT-PCR analysis of p53 target genes. The expression levels in the Δ133p53 siRNA-transfected cells (si-1 and si-2) are shown as the relative values to those in control cells (Cont). Data are mean ± s.d. from triplicate samples.*, p < 0.05.**, p < 0.01.***, p < 0.001.
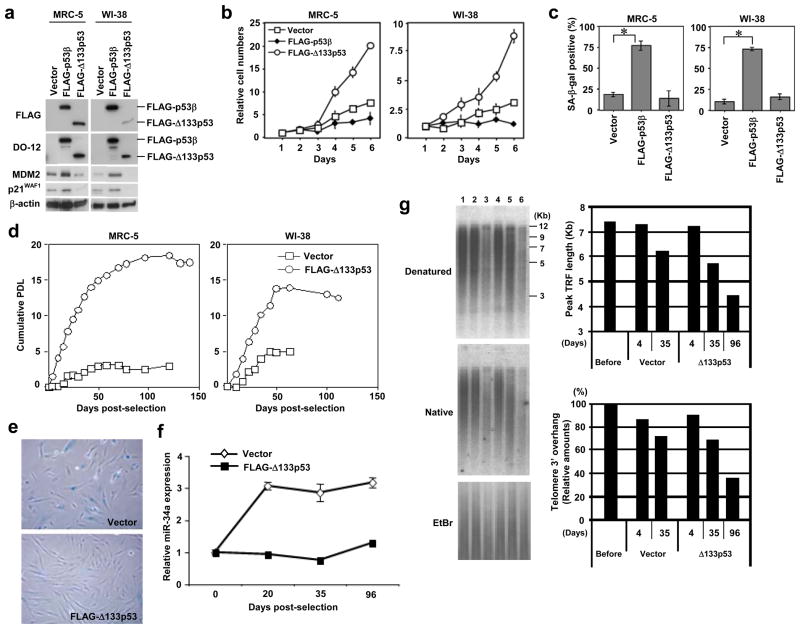
The FLAG-tagged p53β and Δ133p53 were retrovirally expressed in the early-passage human fibroblasts (Fig. S4a). p53β inhibited cell proliferation (Fig. 4b) and induced cellular senescence (Fig. 4c). p53β overexpression also upregulated p21WAF1 and MDM228 (Fig. 4a), as previously reported4, but not miR-34a (Supplementary Information, Fig. S2c). Among the p53 target genes that were regulated by Δ133p53 knockdown, MMP3 was similarly upregulated, BUB1 and CDC20 were downregulated to a lesser degree, and PAI-1 and IGFBP7 did not show a consistent change in WI-38 and MRC-5 fibroblasts overexpressing p53β (Supplementary Information, Fig. S4). p53β also inhibited cell proliferation and induced cellular senescence in a telomerase-immortalized fibroblast cell line (Supplementary Information, Fig. S5). However, the overexpression of p53β had no effect on cell proliferation, cellular senescence or the expression of p21WAF1 in p53-null MDAH041 fibroblasts (homozygous for a frameshift mutation at codon 184)29 (data not shown), indicating that p53β co-operates with full-length p53. In contrast to p53β, the overexpression of Δ133p53 in early-passage MRC-5 and WI-38 fibroblasts (Fig. 4a) accelerated cell proliferation (Fig. 4b) without inducing cellular senescence (Fig. 4c), and repressed the expression of p21WAF1 and MDM2 (Fig. 4a). Consistent with its dominant-negative activity against full-length p534, the overexpression of Δ133p53 did not affect cell proliferation in p53-null MDAH041 fibroblasts (data not shown).
Figure 4.
Overexpression of p53β induces senescence and overexpression of Δ133p53 extends replicative lifespan. (a-c) Early-passage MRC-5 and WI-38 fibroblasts (both at passage 32) were retrovirally transduced with vector alone, FLAG-tagged p53β or FLAG-tagged Δ133p53. (a) Immunoblot analyses of the overexpressed p53 isoforms, MDM2 and p21WAF1. Protein samples at 8 days after retroviral transduction were analyzed. The anti-FLAG and DO-12 antibodies detected FLAG-tagged p53β and FLAG-tagged Δ133p53. β-actin was a loading control. (b) Cell proliferation assay. The cells with vector alone (open squares), FLAG-tagged p53β (closed diamonds) and FLAG-tagged Δ133p53 (open circles) were used at 8 days after retroviral transduction. The data (mean ± s.d.) were from triplicate wells. (c) Summary of SA-β-gal assay. The same set of cells as in b were examined. The data (mean ± s.d.) were from three independent experiments. *, p < 0.01. (d-g) Extension of cellular replicative lifespan by Δ133p53. The FLAG- Δ133p53 retroviral vector or the control vector was transduced to human fibroblasts at late passage (MRC-5 at passage 53 and WI-38 at passage 51). (d) The cumulative PDL were calculated and plotted to days post-selection. Open squares, vector alone. Open circles, FLAG- Δ133p53. (e) SA-β-gal staining. The pictures at 36 days post-selection are shown. (f) Repression of miR-34a by Δ133p53. MRC-5 (at passage 53) before transduction (day 0), MRC-5 with control vector and MRC-5 overexpressing FLAG- Δ133p53 (at days 20, 35 and 96 post-selection) were examined for miR-34a expression by qRT-PCR, as in Fig. 2a. The value before transduction was defined as 1.0 and the expression levels in the other samples were expressed as the relative values (mean ± s.d. from triplicate sample). (g) Measurement of telomere length (denatured gel) and telomeric 3′ overhang (native gel) by in-gel hybridization. Lane 1, MRC-5 before transduction. Lanes 2 and 3, MRC-5 with vector control (days 4 and 35 post-selection). Lanes 4–6, MRC-5 overexpressing FLAG- Δ133p53 (days 4, 35 and 96 post-selection). The telomere lengths were measured as peak TRF (terminal restriction fragment) lengths. The amounts of telomeric 3′ overhang were normalized with loaded DNA amounts (EtBr) and shown as percent signals to the cells before transduction.
The biological effects of Δ133p53 were more evident when overexpressed in the late-passage human fibroblasts. In MRC-5 and WI-38, whereas the vector control cells underwent senescent growth arrest at only one to five PDLs after retroviral transduction, the Δ133p53-overexpressing cells reproducibly bypassed this normal senescence point and continued to proliferate for six to 15 more PDLs (Fig. 4d, e; Supplementary Information, Fig. S6). As shown in Fig. 4f, the expression of miR-34a in Δ133p53-overexpressing MRC-5 remained restricted to low levels throughout the replicative lifespan. Δ133p53 was also shown to inhibit the full-length p53-induced transcriptional activation of the miR-34a gene promoter (Supplementary Information, Fig. S2b). Taken together with the extension of replicative lifespan by miR-34a knockdown (Fig. 2), these findings suggest that the extension of replicative lifespan by Δ133p53 is attributed in part to its ability to dominant-negatively inhibit p53 induction of miR-34a, and provide a functional connection of this p53-induced microRNA to the p53 isoform-mediated regulation of replicative senescence. In the Δ133p53-overexpressing cells, both the overall length of telomeres and the amount of telomeric 3′ overhangs continued to be reduced beyond those in the senescent vector control cells (Fig. 4g; compare Δ133p53 at day 96 and vector at day 35), indicating that the Δ133p53-induced extension of the replicative lifespan was not due to telomere stabilization.
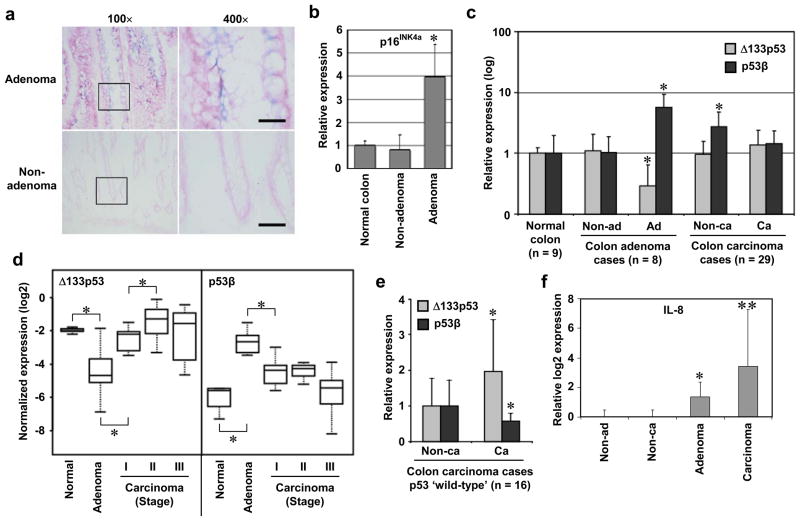
Colon adenomas are premalignant tumours associated with telomere shortening-induced replicative senescence30, 31 and oncogene-induced, interleukin-regulated premature senescence10, 11, 32. Consistently, we observed positive SA-β-gal staining in adenoma tissues (Fig. 5a). The expression of p16INK4A, an in vivo senescence marker33, was significantly more abundant in colon adenomas than in non-adenomas or normal colon tissues (Fig. 5b; Supplementary Information, Fig. S7a, c), as reported previously10, 32. Colon adenoma tissues expressed elevated levels of p53β and reduced levels of Δ133p53 compared with non-adenoma and normal colon tissues (Fig. 5c, d; Supplementary Information, Fig. S7). These results show that the senescence-associated p53 isoform expression signature (i.e., elevated p53β and reduced Δ133p53) occurs not only in cultured cells in vitro but also in humans in vivo, and in cells of different origins (i.e., both mesenchymal and epithelial origins).
Figure 5.
p53 isoform expression profiles in colon carcinogenesis in vivo. Elevated p53β and reduced Δ133p53 in colon adenomas with senescent phenotypes, but not in colon carcinomas. (a) SA-β-gal staining of non-adenoma and adenoma tissues. Case 7 is shown. The rectangular areas are enlarged in the right panels. Bars, 500 μm. (b) Nine normal colon tissues (Supplementary Table S1) and 8 matched pairs of non-adenoma and adenoma tissues (Supplementary Table S2) were examined for p16INK4a expression in immunoblot and quantitatively analyzed. The data (mean ± s.d.) are shown as the relative values to normal colon samples. *, p < 0.0001. (c) Expression levels of p53β and Δ133p53 were quantitatively examined in 9 normal colon tissues (Supplementary Table S1), 8 matched pairs of non-adenoma and adenoma tissues (Supplementary Table S2) and 29 matched pairs of non-carcinoma and carcinoma tissues (Supplementary Table S3). The data (mean and s.d.) are shown in a logarithmic scale as the relative values to normal colon samples. *, p < 0.05. (d) Expression levels of Δ133p53 and p53β in colon carcinomas were analyzed according to tumour stage: stage I (n = 8), stage II (n = 11) and stage III (n = 10). The data of normal colon and adenoma samples are same as those in c. The data normalized to β-actin levels were converted to log2 values and shown as box-and-whisker plots. *, p < 0.05. (e) Upregulation of Δ133p53 in colon carcinomas assumed to have ‘wild-type’ p53 (n = 16; Supplementary Table S3). The expression levels of Δ133p53 and p53β (mean and s.d.) in carcinoma tissues were expressed relative to those in non-carcinoma tissues. *, p < 0.05. The same data were analyzed by paired t-test in Supplementary Fig. S8. p53β was significantly less abundant in carcinoma tissues because of the marked increase in non-carcinoma tissues as shown in c. (f) IL-8 expression was examined by qRT-PCR in the same non-adenoma, adenoma, non-carcinoma and carcinoma tissues as above. The expression levels (mean and s.d.) are shown as relative log2 values to non-adenoma (defined as 0). *, p < 0.05. **, p < 0.001.
In contrast to colon adenomas, colon carcinoma tissues (Fig. 5c, bars “Ca”) did not show the senescence-associated p53 isoform expression signature, with Δ133p53 increased and p53β decreased back to similar levels to those in normal colons and non-adenoma tissues. Although the adjacent non-carcinoma tissues (Fig. 5c, bars “Non-ca”) expressed significantly elevated levels of p53β, its biological importance is currently unknown. The stage I carcinomas already failed to maintain the characteristics of adenomas, showing significantly increased Δ133p53 and decreased p53β compared with adenomas (Fig. 5d). These results suggest that the loss of the senescence-associated p53 isoform expression signature may signal an escape from the senescence barrier observed in premalignant tumours1, 3, 11, 32, 34. A further significant increase in Δ133p53 from stage I to II and a further decrease in p53β from stage II to III (Fig. 5d) might also play a role during stage progression. In the subgroup analysis based on p53 status, the expression levels of Δ133p53 were significantly higher in carcinoma tissues than in non-carcinoma tissues in p53 ‘wild-type’ cases (Fig. 5e; Supplementary Information, Fig. S8c), but not in p53 ‘mutant’ cases (Supplementary Information, Fig. S8c), in agreement with the ability of Δ133p53 to inhibit the full-length p53 function. Our overall in vivo data suggest that, although mutated p53 may have a predominant role over the p53 isoforms, altered expression of these isoforms contributes to the senescence phenotype in premalignant lesions, as well as the escape from the senescence barrier and the malignant progression especially in the cases without p53 gene mutations or at early carcinoma stages before the mutations occur.
Interleukin-8 (IL-8) was upregulated in colon adenoma tissues compared with adjacent non-adenoma tissues (Fig. 5f). The IL-8 signalling pathway is involved in both replicative senescence and oncogene-induced senescence in a p53-dependent manner34, which are observed in colon adenomas11, 30–32. However, it is unlikely that this cytokine-mediated mechanism for senescence primarily regulates, or is regulated by, the senescence-associated expression signature of the p53 isoforms, because colon carcinoma tissues without such signature (Fig. 5c) still expressed remarkably increased levels of IL-8 (Fig. 5f) and adjacent non-carcinoma tissues with elevated p53β (Fig. 5c) showed no increase in IL-8 expression (Fig. 5f). Considering our in vitro data that the senescence-associated p53 isoform expression signature is observed in replicative senescence but not in oncogenic Ras-induced senescence (Fig. 1b, c), a full malignant conversion from adenoma to carcinoma may require overcoming the senescence barriers by both p53 isoform-dependent (i.e., replicative senescence) and-independent (e.g., oncogene-induced, interleukin-regulated senescence) mechanisms. No significant difference in p53β or Δ133p53 expression between K-Ras wild-type (n = 19) and mutant (n = 4) colon carcinoma cases (data not shown) further suggested that the p53 isoforms were not primarily regulated by K-Ras.
p21WAF1 was upregulated commonly in replicative senescence (Fig. 1b), Δ133p53 knockdown-induced senescence (Fig. 3a; Supplementary Information, Fig. S3a) and p53β overexpression-induced senescence (Fig. 4a; Supplementary Information, Fig S5b). While MMP3 was similarly upregulated at both Δ133p53 knockdown- and p53β overexpression-induced senescence, the expression profiles of the other four senescence-associated p53 target genes in these two senescence states were not generally consistent with each other (Fig. 3e; Supplementary Information, Fig. S4). These findings on p53 target gene selectivity suggest that the molecular basis of Δ133p53 knockdown-induced senescence and p53β overexpression-induced senescence are overlapping but distinct from each other. Our data suggest that the repression of miR-34a, leading to continued expression of cell cycle progression genes5–7, is causally involved in a p53 inhibition-induced extension of cellular replicative lifespan (Fig. 4f). However, the induction of senescence by either Δ133p53 knockdown or p53β overexpression occurred without upregulating miR-34a (Supplementary Information, Fig. S2c). We speculate that human fibroblasts may become committed for miR-34a expression only at a later stage of their replicative lifespan, where the Δ133p53 overexpression experiment was carried out. Differential levels of p53 post-translational modifications14, such as pS15-p53 (Fig. 1b), and/or the concomitant occurrence of decreased Δ133p53 and increased p53β (Fig. 1b) may be involved in this commitment.
The diminished Δ133p53 protein or the induced p53β protein at replicative senescence was not due to a regulation at the transcriptional or mRNA splicing level (Supplementary Information, Fig. S9a). Treatment with a proteasome inhibitor MG132 showed that full-length p53 and p53β, but not Δ133p53, are subject to proteasomal degradation (Supplementary Information, Fig. S9b). Overexpressed full-length p53 led to upregulated p53β and downregulated Δ133p53 (Supplementary Information, Fig. S9c), both of which could in turn activate the function of full-length p53. The positive feedback regulation between the elevated activity of full-length p53 and the upregulated p53β or downregulated Δ133p53 may be essential for induction, maintenance and irreversibility of the senescence state of human cells. The molecular details of the protein expression and degradation of the p53 isoforms await further investigation.
In summary, based on the in vitro expression and functional analyses of endogenous proteins, which are supported by the overexpression experiments, this study provides the first evidence for the physiological regulation of replicative cellular senescence by natural p53 isoforms. With in vivo evidence that the establishment and loss of the specific expression profile of the p53 isoforms constitute the induction of and escape from the senescence state, respectively, this study may open up a new p53-based, senescence-mediated strategy to manipulate carcinogenesis and ageing.
METHODS
Cells
CC1, a human choriocarcinoma cell line expressing Δ133p53 due to the genomic rearrangement deleting the exons 2, 3 and 436, was a gift from Dr. Mitsuo Oshimura (Tottori University, Japan). Normal human fibroblast strains (MRC-5 and WI-38), H1299, RKO and 293T were obtained from American Type Culture Collection (Manassas, VA). hTERT/NHF, an hTERT (human telomerase reverse transcriptase)-immortalized human fibroblast cell line, was previously described37. MDAH041 was kindly provided by Dr. Michael Tainsky (Case Western Reserve University, Cleveland, OH). The treatment with Nutlin-3a was as described21.
Plasmid constructs
To generate the retroviral expression vectors of human p53 isoforms, FLAG-tagged p53β and FLAG-tagged Δ133p53 were PCR-amplified using pSVp53β and pSVDNp534, respectively, as the templates, and then inserted into Not I and Eco RI sites of pQCXIN vector (BD Biosciences, San Jose, CA). The full-length p53 cDNA was similarly cloned into the same vector. These constructs were verified by DNA sequencing. The retroviral construct pLPC-Myc-TRF2ΔBΔM was a gift from Dr. Titia de Lange (Rockefeller University, NY). The retroviral expression vector for H-RasV12 was a gift from Dr. Manuel Serrano (Spanish National Cancer Research Center). The shRNA knockdown vectors targeting p53 and POT1 were previously described15. The pCMVβ plasmid vector (Clontech, Mountain View, CA) and its derivatives containing the full-length p53 cDNA (wild-type and 273H mutant) were previously described38. The luciferase reporter plasmids containing the miR-34a promoter fragments (wild-type and mutated at the p53 binding site)7 were kindly provided by Dr. Greg Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). The pMIR-REPORT Luciferase vector (Ambion, Austin, TX) with the 3′-UTR (3′-untranslated region) of the MYCN gene containing two miR-34a binding sites (pMIR-MYCN-WT) and its counterpart with the miR-34a binding sites mutated (pMIR-MYCN-MT1&2)20 were kindly provided by Dr. Javed Khan (National Cancer Institute, Gaithersburg, MD).
Retroviral vector transduction
The retroviral constructs were transfected into Phoenix packaging cells (Orbigen, Inc., San Diego, CA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Vector supernatants were collected 48 h after transfection and used to infect cells in the presence of 8 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO). Two days after infection, the infected cells were selected with 600 μg/ml of G418 (Sigma-Aldrich), 2 μg/ml of puromycin (Sigma-Aldrich) or 1 mg/ml of zeocin (Invitrogen).
siRNA and antisense oligonucleotides
A stealth siRNA duplex oligoribonucleotide targeting Δ133p53 mRNA (Δ133si-1, 5′-UGU UCA CUU GUG CCC UGA CUU UCA A-3′), its scrambled control, and a standard siRNA duplex oligoribonucleotide targeting Δ133p53 mRNA (Δ133si-2, 5′-CUU GUG CCC UGA CUU UCA A[dT][dT]-3′) were synthesized at Invitrogen. Both Δ133si-1 and Δ133si-2 were designed to target the sequences that are present in Δ133p53 mRNA as 5′ untranslated region but spliced out of full-length p53 mRNA as intron 4. The following antisense 2′-O-methyl oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA): 5′-AAC AAC CAG CUA AGA CAC UGC CA-3′ for inhibiting miR-34a; and 5′-AAG GCA AGC UGA CCC UGA AGU-3′ as a control, which is complementary to the enhanced green fluorescence protein (EGFP). The LNA (locked nucleic acid) antisense oligonucleotide for miR-34a knockdown and the control oligonucleotide were purchased from Exiqon (Woburn, MA). siRNA, LNA antisense and 2′-O-methyl antisense oligonucleotides were transfected at the final concentration of 12 nM, 15 nM and 40 nM, respectively, into MRC-5 and WI-38 fibroblasts by using the Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the supplier’s protocol. In the experiments where cellular replicative lifespan was examined, the transfection was repeated every 4 days.
Cell proliferation assay, senescence-associated-β-galactosidase (SA-β-gal) staining, examination of cellular replicative lifespan, and bromo-deoxyuridine (BrdU) incorporation assay
For cell proliferation assay, 2.4 × 105 cells per well were plated into 12-well plates. These cells were collected and counted daily for a week using a hematocytometer. The experiments were performed at least twice and data at each time point were in triplicate. For examining cellular replicative lifespan, the number of cells was counted at each passage, and the number of population doublings (PDL) achieved between passages was determined by log2 (number of cells obtained/number of cells inoculated). SA-β-gal staining was performed as previously described15, 21. For BrdU incorporation assay, cells were incubated with 10 μM of BrdU for 24 h. The incorporated BrdU was detected using an anti-BrdU monoclonal antibody (Amersham Biosciences) and observed with a fluorescent microscope. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Immunoblot analysis
Cells or tissues were lysed in RIPA buffer [10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA, 1% NP-40, complete protease inhibitors (Roche, Indianapolis, IN), phosphatase inhibitor cocktail 1 and 2 (Sigma-Aldrich)]. SDS-PAGE, transfer to nitrocellulose or PVDF membranes (Bio-Rad, Hercules, CA), incubation with antibodies, and signal detection followed the standard procedures using ECL detection (Amersham Biosciences, Piscataway, NJ) or SuperSignal West Dura Extended Duration system (Pierce Biotechnology, Rockford, IL). The quantitative analysis of the immunoblot data was performed using the ImageJ 1.40g software (http://rsb.info.nih.gov/ij/).
Antibodies
A polyclonal antibody specifically recognizing Δ133p53 (MAP4) was raised at Moravian Biotechnology (Brno, Czech Republic) in rabbits injected with a mixture of peptides, MFCQLAKTC and FCQLAKTCP, which were synthesized as Multiple Antigenic Peptide by Dr. G. Bloomberg (University of Bristol, Bristol, UK). The other primary antibodies used were: TLQ404 for p53β; CM14, DO-12 (Millipore, Billerica, MA) and DO-1 (Santa Cruz Biotechnology, Santa Cruz, CA) for p53; anti-pS15-p53 (Cell Signaling Technology, Danvers, MA); EA10 (Carbiochem, San Diego, CA) for p21WAF1; SMP14 (Santa Cruz Biotechnology) for MDM2; 8G10 (Cell Signaling Technology) for caspase-3; M2 monoclonal antibody (Sigma-Aldrich) for FLAG tag; AC-15 (Sigma-Aldrich) for β-actin; G175-1239 (BD Pharmingen, San Diego, CA) for p16INK4a; anti-PARP (Cell Signaling Technology); anti-SIRT1 (Santa Cruz Biotechnology); and anti-E2F1 (Santa Cruz Biotechnology). Horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies (Santa Cruz Biotechnology) were used as secondary antibodies in immunoblots.
Real-time qRT-PCR for quantification of microRNA expression
RNA samples were prepared by using Trizol (Invitrogen). Reverse transcriptase reactions were performed using TaqMan microRNA reverse transcription kit (Applied Biosystems, cat. no. 4366596) and a miR-34a- or miR-34b-specific primer. The TaqMan microRNA assay kit for miR-34a (Applied Biosystems, cat. no. 4373278) or miR-34b (Applied Biosystems, cat. no. 4373037) was used according to the supplier’s protocol. Real-time PCR reactions were performed in triplicate. RNU66 (Applied Biosystems, cat. no. 4373382) was used as a control for quantification. Normalized expression of miR-34a or miR-34b was calculated by using the ΔΔCt method according to the supplier’s protocol (protocol no. 4310255B and User Bulletin no. 4303859B at http://www.appliedbiosystems.com/index.cfm).
Real-time qRT-PCR for quantification of mRNA expression
For quantitative measurement of PAI-1 mRNA, the SYBR Green PCR Master Mix (Applied Biosystems) was used with the following primers: 5′-CTC CTG GTT CTG CCC AAG T-3′ and 5′-CAG GTT CTC TAG GGG CTT CC-3′ for PAI-1; and 5′-TTC TGG CCT GGA GGC TAT C-3′ and 5′-TCA GGA AAT TTG ACT TTC CAT TC-3′ for β-2-microglobulin as an internal control. For IGFBP7, MMP3, BUB1, CDC20, IL-8 and IL-8R, the Taqman Universal PCR Master Mix (Applied Biosystems) was used with the following sets of probe and primers purchased from Applied Biosystems: IGFBP7 (Hs00944483_m1); MMP3 (Hs00968308_m1); BUB1 (Hs01557701_m1); CDC20 (Hs00426680_mH); IL-8 (Hs00174103_m1); and IL-8R (Hs001174304_m1). The endogenous control was β-2-microglobulin (cat. no. 4333766). Quantitative data analysis followed the ΔΔCt method, as above.
RT-PCR for p53β and Δ133p53
The primers specifically detecting the alternative splicing that generates β isoforms were: 5′-CTT TGA GGT GCG TGT TTG TGC-3′ and 5′-TTG AAA GCT GGT CTG GTC CTG A-3′. The primers specifically amplifying Δ133p53 mRNA transcribed from the promoter in intron 4 were: 5′-TGG GTT GCA GGA GGT GCT TAC-3′ and 5′-CCA CTC GGA TAA GAT GCT GAG G-3′. The general amplification of p53 mRNA (not in an isoform-specific manner) was performed with 5′-CTC ACC ATC ATC ACA CTG GAA-3′ and 5′-TCA TTC AGC TCT CGG AAC ATC-3′. GAPDH was amplified as a control by using 5′-CCA TCT TCC AGG AGC GAG A-3′ and 5′-TGT CAT ACC AGG AAA TGA GC-3′.
Luciferase reporter assay of miR-34a gene promoter
p53-null MDAH041 fibroblasts were retrovirally transduced with p53β, Δ133p53 or vector control, selected for G418 resistance, plated into 24-well plates, and then transfected with 0.2 μg/well of the full-length p53 expression plasmid (wild-type or 273H mutant) or the empty vector, together with 0.3 μg/well of the wild-type or mutant miR-34a promoter-luciferase construct and 3 ng/well of pRL-SV40 control plasmid (Promega, Madison, WI). Firefly luciferase activity (from miR-34a promoter) and Renilla luciferase activity (from pRL-SV40) were measured using the Dual-luciferase Reporter system (Promega).
Luciferase reporter assay of antisense knockdown of miR-34a
MRC-5 fibroblasts on 24-well plates were transfected with 0.3 μg/well of pMIR-MYCN-WT or pMIR-MYCN-MT1&2, 15 nM of LNA oligonucleotides, and 5 ng/well of pRL-SV40 (Promega) using the Lipofectamine 2000 transfection reagent (Invitrogen). Measurement of activities was performed as above.
Measurement of telomeric 3′ overhang and telomere length
In-gel Southern hybridization, under native and denatured conditions, was performed as previously described15. Genomic DNA samples were digested with Hinf I and electrophoresed through 0.7% agarose gel. After drying at 25°C for 30 min in a Bio-Rad model 583 gel dryer, the gel was hybridized with 32P-labeled [CCCTAA]4 oligonucleotide, followed by washing and signal detection using the Typhoon 8600 system (Molecular Dynamics, Sunnyvale, CA). The amounts of telomeric 3′ overhangs, normalized with loaded DNA amounts detected with ethidium bromide (EtBr) staining of the gel, were quantitated by using the ImageQuant version 5.2 software (Molecular Dynamics). After alkali denaturation (0.5M NaOH/1.5M NaCl) and neutralization (2.5M NaCl/0.5M Tris-HCl, pH 7.5) of the dried gel, the same procedures were repeated to examine telomere length, which was indicated as a peak TRF (terminal restriction fragment) length.
Human colon tissues
Normal colon tissues were obtained from immediate autopsy at Baltimore area hospitals in Maryland39. Eight pairs of colon adenoma and adjacent non-adenoma tissues were from the University of Maryland Medical Center and the Cooperative Human Tissue Network. Twenty-nine pairs of colon carcinoma and adjacent non-carcinoma tissues were from the University of Maryland Medical Center and Baltimore Veterans Affairs Medical Center40. All tissues were flash frozen immediately after resected. Tumour histopathology was classified according to the World Health Organization Classification of Tumour system. This study was approved by the Institutional Review Board of the National Institutes of Health. Supplementary Tables S1, S2 and S3 summarize information on tissue samples used in this study.
Statistical analyses
Statistical analysis was carried out with the paired or unpaired Student’s t test as appropriate.
Supplementary Material
Acknowledgments
We thank M. Serrano, M. Tainsky, M. Oshimura, G. Hannon, T. de Lange and J. Khan for cells and reagents, X. Wang for helpful discussion, E. Spillare for continuous support, and E. Michalova for technical assistance. This research was supported in part by the Intramural Research Program of the NIH, NCI. B.V. was supported by the grants from GACR (number 301/08/1468) and IGA MZ CR (number NS/9812-4). J.C.B. was supported by Breast Cancer Campaign, Cancer-Research UK and the “Institut National de la Sante et de la Recherche Medicale” (Inserm). D.L. is a Gibb fellow of the CRUK. H. J. participated in the NIH Summer Internship Program.
Footnotes
Author contributions
K.F., A.M.M., I.H., G.H.N., K.K., J.J.S., E.D.B., A.J.S., S.R.P. and H.J. performed experiments. E.A.M. provided expertise on statistical data analysis. B.V., J.-C.B. and D.P.L. provided essential reagents and suggestions. K.F., I.H. and C.C.H. coordinated the study and wrote the manuscript. C.C.H. was responsible for the overall project. All authors discussed the results and commented on the manuscript.
References
- 1.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 2.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 4.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bommer GT, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 6.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai CY, et al. p16INK4a expression begins early in human colon neoplasia and correlates inversely with markers of cell proliferation. Gastroenterology. 2000;119:929–942. doi: 10.1053/gast.2000.17952. [DOI] [PubMed] [Google Scholar]
- 11.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 12.Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, et al. p53 isoform Δ113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 2009;23:278–290. doi: 10.1101/gad.1761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webley K, et al. Posttranslational modifications of p53 in replicative senescence overlapping but distinct from those induced by DNA damage. Mol Cell Biol. 2000;20:2803–2808. doi: 10.1128/mcb.20.8.2803-2808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q, et al. Functional diversity of human protection of telomeres 1 isoforms in telomere protection and cellular senescence. Cancer Res. 2007;67:11677–11686. doi: 10.1158/0008-5472.CAN-07-1390. [DOI] [PubMed] [Google Scholar]
- 16.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 17.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 18.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21CIP1, but not p16INK4a. Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 19.Yamakuchi M, Ferlito M, Lowenstein C. J miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei JS, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumamoto K, et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gjoerup OV, et al. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc Natl Acad Sci USA. 2007;104:8334–8339. doi: 10.1073/pnas.0703164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kidokoro T, et al. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene. 2008;27:1562–1571. doi: 10.1038/sj.onc.1210799. [DOI] [PubMed] [Google Scholar]
- 27.Tang X, Milyavsky M, Goldfinger N, Rotter V. Amyloid-beta precursor-like protein APLP1 is a novel p53 transcriptional target gene that augments neuroblastoma cell death upon genotoxic stress. Oncogene. 2007;26:7302–7312. doi: 10.1038/sj.onc.1210542. [DOI] [PubMed] [Google Scholar]
- 28.Rozan LM, El-Deiry W. S p53 downstream target genes and tumor suppression: a classical view in evolution. Cell Death Differ. 2007;14:3–9. doi: 10.1038/sj.cdd.4402058. [DOI] [PubMed] [Google Scholar]
- 29.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 30.Hastie ND, et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 31.O’Sullivan J, et al. Telomere length in the colon declines with age: a relation to colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2006;15:573–577. doi: 10.1158/1055-9965.EPI-05-0542. [DOI] [PubMed] [Google Scholar]
- 32.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 33.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acosta JC, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 35.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 36.Horikawa I, Suzuki M, Oshimura M. An amino-terminally truncated p53 protein expressed in a human choriocarcinoma cell line, CC1. Hum Mol Genet. 1995;4:313–314. doi: 10.1093/hmg/4.2.313. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta S, et al. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blagosklonny MV, et al. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995–11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- 39.Autrup H, Harris CC, Schwartz RD, Trump BF, Smith L. Metabolism of 1,2-dimethylhydrazine by cultured human colon. Carcinogenesis. 1980;1:375–380. doi: 10.1093/carcin/1.5.375. [DOI] [PubMed] [Google Scholar]
- 40.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.