Abstract
Antibiotic-resistant strains of pathogenic bacteria are increasingly prevalent in hospitals and the community. New antibiotics are needed to combat these bacterial pathogens, but progress in developing them has been slow. Historically, most antibiotics have come from a small set of molecular scaffolds whose functional lifetimes have been extended by generations of synthetic tailoring. The emergence of multidrug resistance among the latest generation of pathogens suggests that the discovery of new scaffolds should be a priority. Promising approaches to scaffold discovery are emerging; they include mining underexplored microbial niches for natural products, designing screens that avoid rediscovering old scaffolds, and repurposing libraries of synthetic molecules for use as antibiotics.
There is a perpetual need for new antibiotics: Whereas most drugs will be just as effective in the future as they are today, the inevitable rise of resistance will erode the utility of today’s antibiotics (1). Two factors exacerbate this supply problem by creating unique dis-incentives for antibiotic development (2). First, antibiotics are used in smaller quantities than other drugs. Prescriptions for chronic illnesses can last years or decades, whereas a standard course of antibiotics lasts only weeks; therefore, antibiotics yield lower revenues than most drugs. Second, whereas most newly approved drugs can be prescribed to all who would benefit, the use of a newly approved antibiotic may be restricted to the treatment of serious bacterial infections. The result is a quandary: Resistance is on the rise while antibiotic discovery and development are on the decline (3, 4).
The unfavorable economics of antibiotic development have had a chilling effect on industrial discovery programs, and policy-based efforts to reverse this decline deserve attention (3). This perspective focuses on a different, yet no less formidable, challenge: finding new classes of antibiotics.
On the face of it, antibiotic discovery would seem to be straightforward. The goal is to kill an organism that is only distantly related to humans; unique, essential targets should be abundant, and novel antibiotics with low toxicity should be easy to find. Yet, the history of antibiotic development suggests otherwise. Since the early 1960s, only four new classes of antibiotics have been introduced, and none of these have made a major impact yet; the ~$30 billion global antibiotics market is still dominated by antibiotic classes discovered half a century ago. Since then, most “new” antibiotics have been chemically tailored derivatives of these well-worn scaffolds. In this review, we argue that the rise of resistant pathogens should redouble our focus on discovering not just new antibiotics, but new classes of antibiotics. We then highlight some promising approaches to scaffold discovery: mining underexplored microbial niches for natural products, designing screens that avoid rediscovering old scaffolds, and repurposing libraries of synthetic molecules for use as antibiotics.
A new generation of resistant pathogens
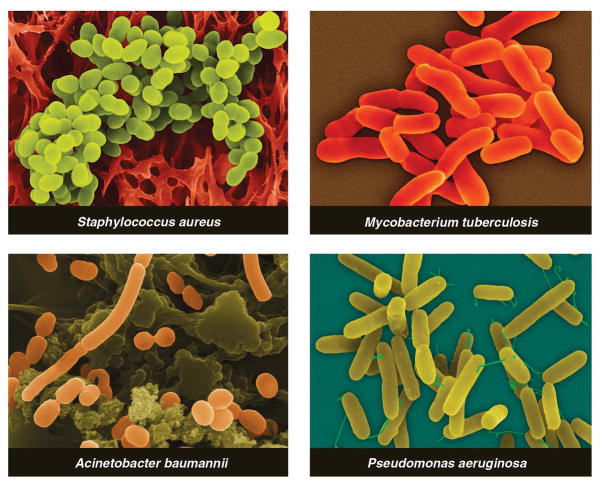
Three classes of antibiotic-resistant pathogens are emerging as major threats to public health (Fig. 1). First, methicillin-resistant Staphylococcus aureus (MRSA) is estimated to cause ~19,000 deaths per year in the U.S. (5). Apart from their high mortality rate, MRSA infections lead to an estimated $3 to 4 billion of additional health care costs per year. Furthermore, the rising prevalence of MRSA increases the likelihood that vancomycin-resistant S. aureus (VRSA) (6)—just as deadly as MRSA but more challenging to treat—will become a new scourge in hospitals.
Fig. 1.
Multidrug-resistant strains of these bacterial pathogens are on the rise.
Pathogens from the second class, multidrug-resistant (MDR) and pan-drug-resistant (PDR) Gram-negative bacteria, are less prevalent than MRSA, but they pose the grave threat of infections that are truly untreatable (7). These strains of Acine-tobacter baumannii, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa are resistant to some (MDR) or all (PDR) of the antibiotic classes commonly used to treat Gram-negative bacteria: penicillins, cephalosporins, carbapenems, monobactams, quinolones, aminoglycosides, tetracyclines, and polymyxins (7). Prospects for finding new antibiotics for Gram-negative pathogens are especially poor: Their outer membrane blocks the entry of some antibiotics, and efflux pumps expel many of the remainder.
The third class comprises MDR and extensively-drug-resistant (XDR) strains of Mycobacterium tuberculosis (MDR-TB and XDR-TB), which are a rising threat in the developing world (8). MDR-TB treatment requires a two-year course of antibiotics with serious side effects; XDR-TB is even more difficult to cure and often fatal (9). Cases of MDR-TB and XDR-TB have been reported in the U.S. and other developed countries.
In spite of the rise of resistant pathogens, the rate of new antibiotic approvals is dropping. Where will new antibiotics come from? In the past, this question has mostly been answered through synthetic tailoring of a small group of “scaffolds.”
Few scaffolds, many generations of tailoring
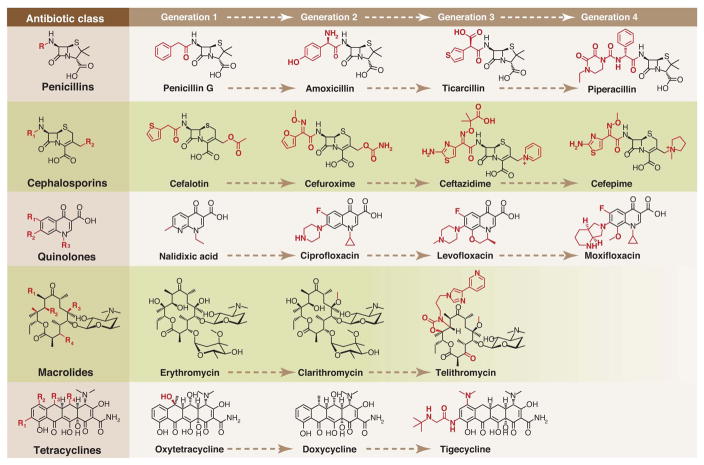
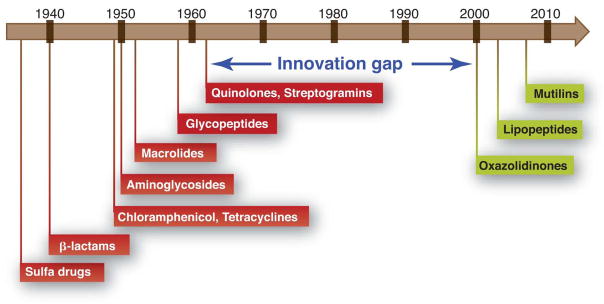
Members of each antibiotic class share a common core structure, or scaffold. For example, the cephalosporins share a β-lactam embedded in a fused 4,6-ring system (Fig. 2). Most chemical scaffolds from which today’s antibiotics are derived were introduced between the mid-1930s and the early 1960s (Fig. 3). Aside from the introduction of carbapenems in 1985, all antibiotics approved for clinical use between the early 1960s and 2000 were synthetic derivatives of existing scaffolds. Just four such scaffolds—cephalosporins, penicillins, quinolones, and macrolides—account for 73% of the antibacterial new chemical entities filed between 1981 and 2005 (10).
Fig. 2.
Synthetic tailoring is widely used to create successive generations of antibiotic classes. Scaffolds are colored black; peripheral chemical modifications are colored red. The quinolone scaffold is synthetic, while the other scaffolds are natural products.
Fig. 3.
Between 1962 and 2000, no major classes of antibiotics were introduced.
During synthetic tailoring (Fig. 2), the core of the antibiotic is left intact, preserving its activity, but the chemical groups at its periphery are modified to improve the drug’s properties. New generations are often designed to be active against pathogens that have become resistant to the previous generation. For example, second- (11) and third-generation (12) cephalosporins like cefaclor and ceftazidime are more resistant to destruction by the resistance enzyme beta-lactamase, and they can penetrate the Gram-negative outer membrane more effectively. When new beta-lactamases emerged that can cleave third-generation cephalosporins, pharmaceutical companies developed fourth-generation molecules like cefepime, which are less susceptible to cleavage by these enzymes (13). Cephalosporins and other semisynthetic antibiotics account for 64% of the new chemical entities filed between 1981 and 2005 (10), suggesting that incremental synthetic tailoring of natural scaffolds has become the predominant mode of antibiotic discovery. The most useful scaffolds have therefore been those that are easy for medicinal chemists to tailor; this allows many derivatives to be synthesized and tested for improved properties.
Organic synthesis plays two other key roles in antibiotic discovery. First, scaffolds like the quinolones and oxazolidinones are derived entirely from chemical synthesis; these fully synthetic scaffolds account for an additional 25% of the antibiotic NCEs. Second, some natural scaffolds like carbapenems can now be produced entirely by organic synthesis, expanding the scope of accessible scaffold modifications.
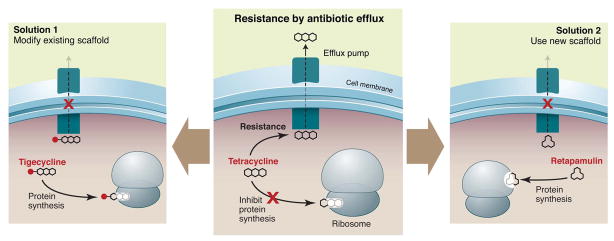
The interplay between semisynthesis and total synthesis—and the ability of synthetic modifications to unlock the therapeutic potential of a scaffold—are exemplified by the tetracyclines. Resistance to this class of 30S-targeting antibiotics is mediated in part by a widely distributed gene encoding an efflux pump. Semisynthetic modifications to the tetracycline scaffold yielded the glycylcycline tigecycline (14). This third-generation molecule (Fig. 2) is no longer a substrate for the efflux pump, restoring its activity against tetracycline-resistant pathogens. A fully synthetic route to the tetracyclines (15) makes it possible to modify scaffold positions that difficult to modify semisynthetically, further broadening the range of accessible derivatives.
Making incremental improvements to existing scaffolds is a good short-term strategy for refilling the antibiotic pipeline, but a presumably more sustainable way to combat resistance is to discover new scaffolds. Their utility will depend on three criteria: spectrum of activity against Gram-positive and Gram-negative pathogens, lack of cross-resistance to existing drugs, and amenability to generations of synthetic tailoring.
Next-generation scaffolds: Natural products
More than two-thirds of clinically-used antibiotics are natural products or their semisynthetic derivatives (10). It is therefore troubling that natural product discovery efforts have waned in recent years (16); this decline is due in part to a rising rate of scaffold rediscovery (17) and the accompanying difficulty in finding new antibiotics. Recent efforts to search new modalities—underexplored ecological niches, unmined bacterial taxa, and the genomes of even well-studied bacteria—have yielded novel molecules, while new screening strategies have begun to circumvent the time-consuming problem of rediscovery (18).
New places to look
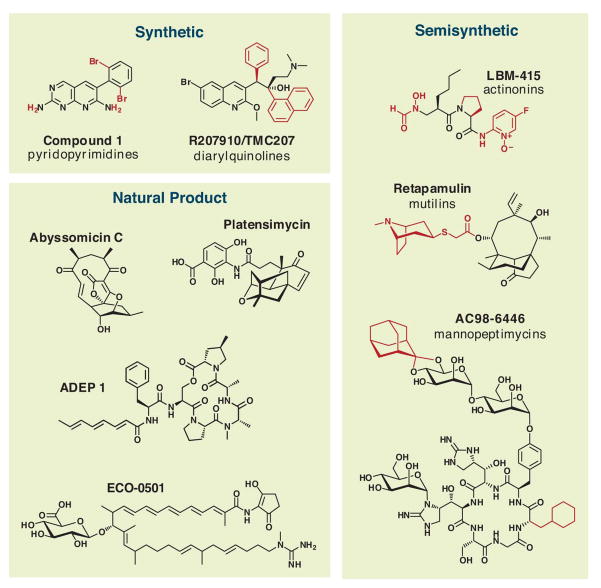
Most natural product antibiotics have come from soil actinomycetes, reflecting the historical bias of pharmaceutical screening programs toward these easily collected and cultured bacteria (1). Searches of underexplored ecological niches and bacterial taxa have revealed new molecules. Marine niches are particularly promising; for example, a deep-sea sediment sample yielded an actinomycete that produces the abyssomicins (19), a new antifolate scaffold (Fig. 4). Terrestrial and marine symbioses are also promising ecological niches; recent efforts to study bacterial symbionts of insects, ascidians, and fungi have yielded many new natural products (20–23). Among underexplored bacterial taxa, myxobacteria are particularly prolific natural product producers, and their continued mining holds much promise for the discovery of new antibiotic scaffolds (24).
Fig. 4.
The chemical structures of new and underexplored antibiotic scaffolds mentioned throughout the text are organized by type into three categories: synthetic, semisynthetic, and natural product. For synthetic and semisynthetic scaffolds, core scaffolds are shown in black and variable positions are shown in red.
The genome sequences of a handful of actinomycetes and myxobacteria have revealed that these bacteria generally harbor >25 gene clusters encoding secondary metabolites. Given that only one to four natural products are known from a typical bacterium under various culture conditions, researchers may as yet have discovered only 10% of natural products from screened strains and just 1% of molecules from the global consortium of microbial producers (25). Taking this lesson to heart, several industrial and academic groups have carried out bioinformatics-based efforts to mine bacterial genomes for new natural products (26, 27). Ecopia Biosciences (now Thallion Pharmaceuticals) has had particular success with their genome-scanning approach, including the discovery of ECO-0501, a new antibiotic scaffold (28) (Fig. 4). If the throughput of these genomics-based approaches to natural product discovery can be scaled up efficiently, their contribution to antibiotic discovery will be increasingly important.
Finally, some promising candidate scaffolds for development may already be known. The founding members of the three most recently introduced antibiotic classes—mutilins, lipopeptides, and oxazolidinones—were each discovered at least two decades before they were introduced. Old patent literature seems a good place to start; based on a 1985 patent from Eli Lilly, a group from Bayer recently isolated a series of peptide antibiotics that activate the bacterial chambered protease ClpP, leading to trolled proteolysis and cell death (29) (Fig. 4). Focusing development efforts on known but underexplored scaffolds can mitigate the risk of a costly and time-consuming de novo discovery program.
Combating rediscovery
Out of 1000 randomly selected actinomycetes, approximately ten will produce streptomycin and four will produce tetracycline (30). If extracts from these strains are screened against an indicator organism, most hits from the screen will be unhelpful rediscoveries. Two new screening strategies are beginning to circumvent the problem of rediscovery (16). First, researchers at Cubist have developed a strain of Escherichia coli that harbors resistance genes for the fifteen most commonly rediscovered antibiotics (31). Hits from their screening efforts are therefore pre-selected to be members of novel classes.
Second, a group at Merck has reported a bacterial antisense technology that allows them to knock down the expression level of a given S. aureus gene, decreasing the level of the encoded protein to the point that, in principle, it is present in growth-limiting quantities (32). Using this approach, they discovered platensimycin, the founding member of a new class of fatty acid biosynthesis inhibitors (33) (Fig. 4), as well as several new protein synthesis inhibitory scaffolds..
Next-generation scaffolds: Synthetic molecules
Fully synthetic molecules are a crucial component of the current antibiotic arsenal: The quinolones are highly effective broad-spectrum antibiotics, and the oxazolidinones are of increasing importance in the treatment of Gram-positive pathogens, including MRSA. However, recent efforts to discover and develop new synthetic scaffolds—based largely on high-throughput screens of novel targets identified by bacterial genomics—have not yet been successful (34).
Historically, synthetic scaffolds have originated outside of antibiotic discovery programs. The first drug in the sulfa class of antibiotics, Prontosil, was originally developed as a dye at Bayer, and the first quinolone was nalidixic acid, an intermediate in the synthesis of chloroquine. The oxazolidinones were discovered at DuPont as antibacterials, but were originally developed to treat foliage diseases of plants.
Since the late 1990s, the rise of bacterial genomics held the promise of rejuvenating the discovery of synthetic antibiotics (35). The genome sequences of pathogens like Haemophilus influenzae, Staphylococcus aureus, Streptococcus pneumoniae, and Escherichia coli made it possible to identify conserved enzymes that are essential for bacterial growth. These novel targets served as the basis for high-throughput screens of synthetic compound libraries, an approach that has been fruitful in other therapeutic areas. However, while genomics-based technologies have accelerated the process of identifying targets of existing drugs(36), they have not yet yielded new antibiotics (34, 37).
Use external libraries and a whole-cell screen
The success of repurposing synthetic molecules from other development programs (38), and the failure of other approaches, holds two important lessons for developing new synthetic antibiotics. First, look outside antibacterial development programs for synthetic libraries to screen. Most pharmaceutical companies have invested considerable resources in synthesizing small molecule libraries for other therapeutic areas. Given the current level of uncertainty about which targets are relevant in an infected host (39) and how antibiotics get into bacterial cells (40), libraries developed for other therapeutic areas maybe just as likely to harbor hits as compound libraries developed for antibacterial screening.
Second, unbiased whole-cell screens have fewer pitfalls than other assays. The advantages of target-based screening—knowledge of the target and ease of optimization using a biochemical screen—are outweighed by the disadvantage of having to engineer cell permeability into a scaffold at a subsequent stage of the development process. Technologies like genome-wide expression profiling (36) and whole-genome resequencing of resistant mutants (41) have accelerated the target identification process; the latter was used to identify ATP synthase as the target of the new antimycobacterial agent R207910, the product of a whole cell screen (41).
Even with a strong hit in a whole-cell antibacterial assay, testing candidates in the right animal model early in development is crucial. In vitro kill assays fail to recapitulate key elements of a bacterial infection, such as the hypoxia and oxidative stress that Mycobacterium tuberculosis experiences in a host (42). A recent report has cast doubt on whether lipid synthesis is a viable target for Gram positive pathogens; its authors argue that most models of infection fail to account for the fact that lipids in human serum can circumvent the inhibition of fatty acids synthesis (39). While future experiments will help resolve whether lipid synthesis inhibitors will be useful as drugs for Staphylococcus and Streptococcus, the mycolic acid pathway is already a well-validated target for Mycobacterium tuberculosis. Any identified fatty acid synthesis inhibitors should therefore be tested against TB, rather than being shelved for lack of efficacy against other Gram-positive pathogens.
A recent example of success
A recent report from Pfizer demonstrates the utility of repurposing external compound libraries by screening them in a whole-cell antibacterial assay (38). Miller and coworkers screened a one-million-compound library developed for eukaryotic protein kinase inhibition in an assay of E. coli killing, predicting that the low molecular weight ATP-mimetic molecules in the library might inhibit an essential bacterial enzyme and therefore exhibit antibacterial activity. They identified a set of pyridopyrimidines (Fig. 4) that are subnanomolar inhibitors of the biotin carboxylase subunit of acetyl-CoA carboxylase (ACC), acting as competitive inhibitors of ATP binding. These molecules are selective for bacterial ACC over eukaryotic protein kinases and have potent activity against Gram-negative bacteria in vitro and in vivo. Similar efforts using other existing libraries could uncover new targets and scaffolds.
Is there still a role for target-based antibiotic discovery?
The failure of bacterial genomics to validate novel targets or yield new antibiotics has cast doubt on the utility of target-based discovery programs (34, 37). Nevertheless, retooled target-based strategies can play an important role in discovery. Examples include developing novel scaffolds for old targets and grouping new targets by inhibitor class.
A new look at old targets
Most clinically-used antibiotics inhibit enzymes from pathways that have been known for decades: peptidoglycan synthesis, ribosomal protein synthesis, folate synthesis, and nucleic acid synthesis and topoisomerization. Future generations of existing scaffolds should continue to have success in the clinic, and these classical targets will thus remain useful. However, a complementary and perhaps more promising strategy is to develop new scaffolds for these targets, thereby avoiding cross-resistance with existing drugs.
For example, the recently introduced mutilin retapamulin (Fig. 4) targets the 50S subunit of the bacterial ribosome, but is unaffected by resistance to other 50S-targeting classes like macrolides (43). Another target that deserves renewed focus is Lipid II; the success of glycopeptide antibiotics like vancomycin bodes well for other Lipid II-binding molecules like the mannopeptimycins (Fig. 4) and lantibiotics (44).
Grouping targets by inhibitor scaffold
To identify new targets, candidates are often grouped by a functional criterion, such as membership in a validated pathway or essentiality for growth in the laboratory. The attendant dangers of single-target bias (34) argue in favor of a strategy that begins with a wider funnel at its early stages.
A different way of grouping targets—by a common inhibitor scaffold rather than by pathway—may not only reveal new targets, but also clues about how to inhibit them. For example, ATP-binding enzymes are a group of targets that can be inhibited by ATP-mimetic scaffolds, and they deserve particular attention for two reasons.
First, bacterial genomes encode hundreds of ATP-binding proteins. They include well-validated targets like DNA gyrase, the target of the quinolones, and a host of new or underexplored targets: the chambered protease ClpP (29), ATP synthase (41), aminoacyl-tRNA synthetases, and acyl-CoA carboxylase. The sensor kinase PhoQ is essential for the virulence of Salmonella (45), and several widely-conserved essential genes encode proteins of unknown function that are predicted to bind ATP (46), suggesting that this class might include a particularly broad range of relevant targets. Insights from outside the antibiotic arena are also important for antibiotics; the observation that Zn-dependent hydrolases are efficiently inhibited by small molecules with Zn-chelating groups has led to the development of inhibitors for a broad range of enzymes, including angiotensin-converting enzyme, histone deacetylases, and matrix metalloproteases. Indeed, semisynthetic derivatives of actinonin—a Zn-chelating natural product that inhibits the Zn-dependent bacterial enzyme peptide deformylase—have been considered as antibiotic candidates (47) (Fig. 4).
Second, Miller and coworkers have demonstrated the feasibility of finding molecules from libraries of ATP-mimetic molecules that are selective for bacterial targets over human targets (48). Screening these libraries in whole-cell assays could simultaneously identify new targets and new lead compounds with scaffolds that can be optimized synthetically.
A more inclusionary approach?
In the heyday of antibiotic discovery, the pool of lead compounds was large enough for pharmaceutical companies to focus on broad-spectrum antibiotics for use as single-agent therapies and shelve compounds that failed these high therapeutic barriers. Today’s greater need for new antibiotics may encourage the development of lead molecules with characteristics that, until recently, have been seen as liabilities: narrow activity spectra and high intrinsic resistance rates.
The rule for antibacterial activity spectrum has been ‘broader is better’. However, the challenge of finding new broad-spectrum antibiotics and the rising threat from specific pathogens like MRSA have led to the development and approval of more agents with a narrower spectrum of activity, particularly those that kill Gram-positive but not Gram-negative bacteria. Extending this trend to near its logical limit, two groups recently reported Staphylococcus-selective antibiotics: One group used a repurposed series of eukaryotic cholesterol synthesis inhibitors to block the production of the gold pigment staphyloxanthin (49), from which the species name aureus is derived; the other group identified inhibitors of the tubulin-like protein FtsZ to block cell division (50). It remains to be seen whether compounds with a spectrum this narrow find a therapeutic niche; one prerequisite for their use would be the availability of rapid diagnostics to identify the etiological agent of infection (51). Such genus-selective agents may have the benefit of sparing more of the endogenous microflora than conventional antibiotics, thereby avoiding complications like secondary Clostridium difficile infections.
Most bacterial infections are treated with a single antibiotic, ruling out the use of molecules with high intrinsic resistance rates. However, pairing these compounds into additive or synergistic combinations could rescue candidates formerly thought to be untenable for development. While development of combination therapies carries the risk of unforeseen toxicity, precedents like amoxicillin-clavulanate and isoniazid-rifampicin-pyrazinamide-ethambutol all argue that antibacterial combination therapies can be quite successful, especially in suppressing the development of resistance. Whether natural or synthetic, broad-spectrum or narrow, single agents or combinations, new scaffolds will be an essential component of a sustainable plan for combating resistance.
Fig. 5.
Contributor Information
Michael A. Fischbach, Email: fischbach@fischbachgroup.org.
Christopher T. Walsh, Email: christopher_walsh@hms.harvard.edu.
References
- 1.Walsh C. Antibiotics: Actions, Origins, Resistance. ASM Press; Washington, DC: 2003. [Google Scholar]
- 2.Nathan C, Goldberg FM. Nature reviews. 2005 Nov;4:887. doi: 10.1038/nrd1878. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C. Nature. 2004 Oct 21;431:899. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 4.von Nussbaum F, Brands M, Hinzen B, Weigand S, Habich D. Angewandte Chemie (International ed) 2006 Aug 4;45:5072. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]
- 5.Klevens RM, et al. Jama. 2007 Oct 17;298:1763. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 6.Weigel LM, et al. Science (New York, NY) 2003 Nov 28;302:1569. [Google Scholar]
- 7.Falagas ME, et al. BMC infectious diseases. 2005;5:24. doi: 10.1186/1471-2334-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman SE, Chaisson RE. Nature medicine. 2007 Mar;13:295. doi: 10.1038/nm0307-295. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, et al. American journal of respiratory and critical care medicine. 2008 Nov 15;178:1075. doi: 10.1164/rccm.200801-132OC. [DOI] [PubMed] [Google Scholar]
- 10.Newman DJ, Cragg GM. Journal of natural products. Mar 2007;70:461. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 11.Neu HC, Fu KP. Antimicrobial agents and chemotherapy. 1978 Apr;13:584. doi: 10.1128/aac.13.4.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn GL. J Antimicrob Chemother. 1982 Nov;10(Suppl C):1. doi: 10.1093/jac/10.suppl_c.1. [DOI] [PubMed] [Google Scholar]
- 13.Garau J, Wilson W, Wood M, Carlet J. Clin Microbiol Infect. 1997;3:S87. [Google Scholar]
- 14.Noskin GA. Clin Infect Dis. 2005 Sep 1;41(Suppl 5):S303. doi: 10.1086/431672. [DOI] [PubMed] [Google Scholar]
- 15.Charest MG, Lerner CD, Brubaker JD, Siegel DR, Myers AG. Science (New York, NY) 2005 Apr 15;308:395. doi: 10.1126/science.1109755. [DOI] [PubMed] [Google Scholar]
- 16.Vederas Li. Science (New York, NY) 2009 [Google Scholar]
- 17.Baltz RH. J Ind Microbiol Biotechnol. 2006;33:507. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 18.Clardy J, Fischbach MA, Walsh CT. Nature biotechnology. 2006 Dec;24:1541. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 19.Bister B, et al. Angewandte Chemie (International ed) 2004 May 3;43:2574. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 20.Donia MS, Ravel J, Schmidt EW. Nature chemical biology. 2008 Jun;4:341. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Partida-Martinez LP, Hertweck C. Nature. 2005 Oct 6;437:884. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 22.Piel J. Natural product reports. 2009 Mar;26:338. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 23.Scott JJ, et al. Science (New York, NY) 2008 Oct 3;322:63. [Google Scholar]
- 24.Wenzel SC, Muller R. Current opinion in drug discovery & development. 2009 Mar;12:220. [PubMed] [Google Scholar]
- 25.Watve MG, Tickoo R, Jog MM, Bhole BD. Archives of microbiology. 2001 Nov;176:386. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 26.Challis GL. Journal of medicinal chemistry. 2008 May 8;51:2618. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 27.McAlpine JB. Journal of natural products. 2009 Feb 6; doi: 10.1021/np800742z. [DOI] [PubMed] [Google Scholar]
- 28.Banskota AH, et al. The Journal of antibiotics. 2006 Sep;59:533. doi: 10.1038/ja.2006.74. [DOI] [PubMed] [Google Scholar]
- 29.Brotz-Oesterhelt H, et al. Nature medicine. 2005 Oct;11:1082. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 30.Baltz RH. SIM News. 2005 Sep/Oct;55:186. [Google Scholar]
- 31.Gullo VP, McAlpine J, Lam KS, Baker D, Petersen F. J Ind Microbiol Biotechnol. 2006 Jul;33:523. doi: 10.1007/s10295-006-0107-2. [DOI] [PubMed] [Google Scholar]
- 32.Singh SB, Phillips JW, Wang J. Current opinion in drug discovery & development. 2007 Mar;10:160. [PubMed] [Google Scholar]
- 33.Wang J, et al. Nature. 2006 May 18;441:358. [Google Scholar]
- 34.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nature reviews. 2007 Jan;6:29. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 35.Rosamond J, Allsop A. Science (New York, NY) 2000 Mar 17;287:1973. doi: 10.1126/science.287.5460.1973. [DOI] [PubMed] [Google Scholar]
- 36.Freiberg C, Fischer HP, Brunner NA. Antimicrobial agents and chemotherapy. 2005 Feb;49:749. doi: 10.1128/AAC.49.2.749-759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mills SD. Biochem Pharmacol. 2006 Mar 30;71:1096. doi: 10.1016/j.bcp.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Boguski MS, Mandl KD, Sukhatme VP. Science (New York, NY) 2009 Jun 12;324:1394. doi: 10.1126/science.1169920. [DOI] [PubMed] [Google Scholar]
- 39.Brinster S, et al. Nature. 2009 Mar 5;458:83. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 40.Nikaido H. Microbiol Mol Biol Rev. 2003 Dec;67:593. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andries K, et al. Science (New York, NY. 2005 Jan 14;307:223. [Google Scholar]
- 42.Cho SH, et al. Antimicrobial agents and chemotherapy. 2007 Apr;51:1380. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidovich C, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007 Mar 13;104:4291. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breukink E, de Kruijff B. Nature reviews. 2006 Apr;5:321. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 45.Bader MW, et al. Cell. 2005 Aug 12;122:461. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 46.Gerdes SY, et al. Journal of bacteriology. 2003 Oct;185:5673. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen DZ, et al. Biochemistry. 2000 Feb 15;39:1256. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 48.Miller JR, et al. Proceedings of the National Academy of Sciences of the United States of America. 2009 Feb 10;106:1737. [Google Scholar]
- 49.Liu CI, et al. Science (New York, NY) 2008 Mar 7;319:1391. [Google Scholar]
- 50.Haydon DJ, et al. Science (New York, NY) 2008 Sep 19;321:1673. [Google Scholar]
- 51.Bootsma MC, Diekmann O, Bonten MJ. Proceedings of the National Academy of Sciences of the United States of America. 2006 Apr 4;103:5620. doi: 10.1073/pnas.0510077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Research in the authors’ laboratories is supported by the Department of Molecular Biology and the Center for Computational and Integrative Biology at Massachusetts General Hospital (M.A.F.) and NIH grants GM20011 and GM49338 (C.T.W.). C.T.W. is a member of the board of directors of Achaogen (South San Francisco, CA).