Abstract
MicroRNAs (miRNAs) are short RNAs of about 22 nucleotides in length that post-transcriptionally regulate gene expression by binding to 3′ untranslated regions of mRNAs, thereby inducing translational silencing. Recently, more than 140 miRNAs have been identified in herpesviral genomes. Deciphering their role in viral biology requires the identification of target genes, a challenging task, since miRNAs require only limited complementarity. The subject of this review will be the herpesvirus miRNAs and their respective target genes that have been experimentally determined thus far. These miRNAs regulate fundamental cellular processes including immunity, angiogenesis, apoptosis, and key steps in the herpesvirus life cycle, latency and the switch from latent to lytic replication.
MicroRNAs regulate fundamental cellular processes in all metazoans
The first discovered microRNA (miRNA), lin-4 of Caenorhabditis elegans, was found because of its role in a developmental timing defect. Functional analysis of the lin-4 gene revealed that it does not encode a protein, but instead two short transcripts of 60 and 24 nucleotides in length. It was demonstrated that the lin-4 RNA was inducing post-translational silencing of lin-14, a developmental control gene whose protein product is involved in temporally regulating cell lineage patterning in C. elegans. Further work showed that the mechanism of this post-translational regulation was mediated through complementary binding of the lin-4 miRNA to sequences within the 3′UTR (untranslated region) of lin-14 [1-3]. This novel RNA-based inhibition was thought to be specific to C. elegans until the discovery of the let-7 miRNA, which was found to be conserved in many metazoans, including humans and flies [4-6]. To date, more than 900 human miRNAs have been identified (see http://microrna.sanger.ac.uk/sequences/) [7]. MicroRNAs have been isolated from every metazoan and plant species examined thus far, and around 30% of all metazoan miRNAs are conserved between species (for review see [8]).
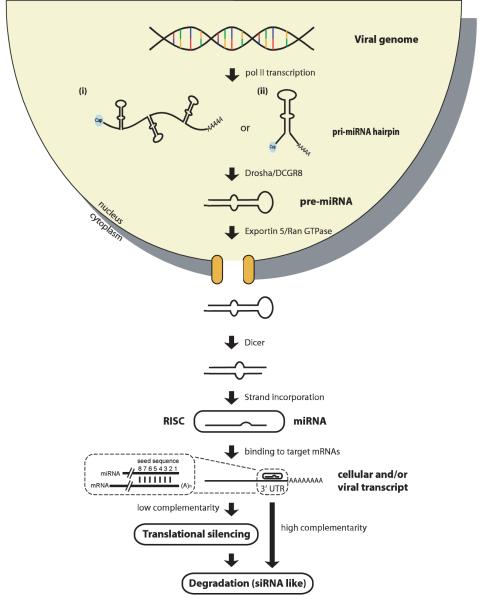
In general, miRNAs are derived from hairpins within transcripts (usually synthesized by RNA polymerase II, or polII) by two RNA endonuclease cleavage reactions before they are incorporated into the RNA-induced silencing complex (RISC; see Figure 1) [9]. The major function of miRNAs appears to be regulation of gene expression through translational inhibition and mRNA degradation (for review see [10]). In rare cases, miRNAs can also act as positive regulators when bound together with other 3′ UTR binding complexes [11].
Figure 1.
Biogenesis of miRNAs. Genes encoding miRNAs are generally transcribed from polII promoters. The majority of miRNAs are encoded in introns, but a small percentage are encoded in exons of protein coding genes. MicroRNA genes can occur either as (i) clusters of multiple hairpins or as (ii) a single hairpin structure. The hairpins in primary transcripts (pri-miRNAs) are recognized by Drosha/DGCR8, a RNase III type endonuclease that cleaves off the 5′ and 3′ ends, leaving a two-nucleotide 3′ overhang. The 60-80 nt hairpin, termed pre-miRNA, is rapidly exported from the nucleus to the cytoplasm via the Exportin5/RAN-GTPase pathway. The pre-miRNA is now recognized by a cytoplasmic RNase III type endonuclease, Dicer, which is also known to cleave dsRNA to create siRNA. Dicer cleaves off the bulged end of the hairpin now forming a short dsRNA with each end having a two nucleotide 3′ overhang. The final step in miRNA biogenesis is the incorporation of one strand of the short RNA duplex into the RNA Induced Silencing Complex (RISC) to form a mature miRNA. Both strands can be incorporated into RISC and as a consequence many miRNA genes encode two mature miRNAs. Once the mature miRNA is incorporated into RISC, it targets the 3′ UTR of mRNAs that contain complementary sequences. It has been observed that positions 2-8 of the miRNA are most important for targeting of mRNAs; this site is referred to as the miRNA seed sequence (for review see [8, 9]).
The herpesvirus family
The herpesvirus family (Herpesviridae) includes large enveloped viruses containing 120 to 240 kbp double stranded DNA. Based on their host and tissue specificities, together with their replication characteristics, the family is divided into three subfamilies: Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae. Evolutionarily, herpesviruses are old viruses with all three subfamilies present in mammals, birds, fish, and reptiles. To date, eight human herpesviruses are known: three alpha-, three beta-, and two gammaherpesviruses.
A hallmark of herpesvirus biology is their ability to establish and maintain latent infections in which the viral genome circularizes and persists as an episome with very limited viral gene expression occurring. Herpesviruses establish infections that persist for the life of the host; therefore, an intricate balance exists between host immune surveillance and viral immune evasion [12]. In general, latent infection has no immediate negative consequences for the infected cell. However, latency in cells of lymphoid, epithelial, and endothelial origin can be associated with tumorigenesis for two human gammaherpesviruses: Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV). EBV and KSHV are the causative agents of several malignancies including Burkitt’s lymphomas (BL), non-Hodgkin’s disease, and nasopharyngeal carcinoma (NPC) for EBV, and Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD) for KSHV. In contrast, human alpha- and betaherpesviruses are not associated with cancer but instead their pathogenesis is due to lytic replication (a viral lifecycle characterized by uncontrolled viral replication) and subsequent immune responses that result in tissue destruction. An example of this is the encephalitis caused by herpes simplex virus 1 (HSV-1).
A typical herpesvirus genome encodes between 85 and 100 open reading frames, which are classified as either latent genes (expressed during latency) or lytic genes (expressed during productive infection). Temporally, genes are further subdivided into three types: (i) immediate early (IE) genes that regulate viral reactivation from latency, (ii) early genes, most of which encode proteins required for viral DNA replication, and (iii) late genes that code for structural proteins required for viral morphogenesis [12]. To aid lifelong persistence, it has been estimated that 25% of all herpesvirus genes modulate and/or abrogate host cellular responses to viral infection (for review see [13]). However, the recent discovery of herpesvirus-encoded miRNAs suggests that this percentage might be significantly higher.
Herpesviruses express multiple miRNAs during latency and lytic replication
In 2004 Tuschl and colleagues reported the molecular cloning of five EBV miRNAs from Burkitt’s lymphoma (BL) cells, a finding that started a new field in virology [14]. Since then, more than 140 herpesvirus miRNAs have been identified (Table 1, Figure 2). Initially, three EBV miRNAs were found located within the BHRF gene, and two within the BART gene (BHRF and BART stand for ‘BamHI fragment H rightward open reading frame 1′ and ‘BamHI-A region rightward transcript’). Bioinformatic approaches, in combination with the use of tiled arrays and molecular cloning, revealed 17 additional miRNA genes in the BART region of EBV; these genes are located within a 12 kbp region that was absent in the EBV strain studied in the original report [15, 16]. Recently, two additional BART miRNA genes were identified in EBV-positive NPC tissue samples, bringing the total number of miRNA genes to 25 [17]. BART and BHRF miRNAs are differentially expressed in lymphoid and epithelial cells and furthermore under different programs of viral latency. BART miRNAs are expressed more abundantly in epithelial cells than in B cells, whereas BHRF miRNAs have only been detected during type III latency (when all known latency-associated genes are expressed). Induction of lytic replication in latently-infected BL cell lines leads to induction of a subset of EBV miRNAs not expressed during latency [15, 18-20]. EBV miRNAs are also expressed early after de novo infection of primary B cells, which might suggest roles in the establishment of latency [21]. In the EBV-related lymphocryptovirus (LCV) of the rhesus macaque, 16 miRNAs were identified, eight of which show sequence homology to EBV miRNAs, suggesting conservation of miRNAs in this subfamily [15].
Table 1.
Number of miRNA genes in different herpesviruses. Note that many miRNA genes can give rise to two miRNAs
| Virus | Number of miRNAs |
|---|---|
| HSV-1 | 7 |
| HSV-2 | 3 |
| MDV-1 | 14 |
| MDV-2 | 17 |
| HCMV | 11 |
| MCMV | 18 |
| EBV | 25 |
| LCV | 16 |
| RRV | 7 |
| KSHV | 12 |
| MHV-68 | 9 |
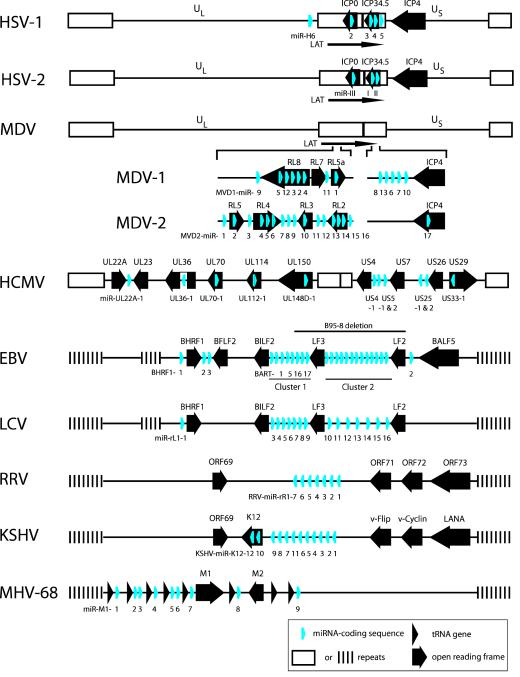
Figure 2.
Location of miRNA genes within herpesvirus genomes. Genomes are represented for alphaherpesviruses (HSV-1, HSV-2, MDV-1, MDV-2), a betaherpesvirus (HCMV), and gammaherpesviruses (EBV, LCV, RRV, KSHV, MHV-68). MDV-1 and MDV-2 were drawn as one complete genome with the respective miRNA encoding regions depicted in more detail. Genomes are not drawn to scale. Figure was compiled from data published in Refs. [14, 16, 22-24, 27, 32, 34, 35, 37, 38]. Abbreviations: US, unique short; UL, unique long; LAT, latency associated transcript.
After the identification of EBV miRNAs, four independent groups cloned KSHV miRNAs from PEL-derived cell lines. In total, 12 genes were identified, all of them within the major latency-associated region of the genome, giving rise to at least 17 mature miRNAs. Ten of the 12 genes formed a single cluster [16, 22-24], whereas the remaining two were located within the K12 open reading frame [16]. Expression of the primary transcripts (pri-miRNAs) is controlled by three promoters, one latent and two lytic. In PEL cells all miRNAs are highly expressed during latency, and (with the exception of miR-K10a and miR-K10b) induction of lytic replication has only moderate effects on viral miRNA expression [25, 26].
The genome of rhesus rhadinovirus (RRV), a gammaherpesvirus related to KSHV, encodes seven miRNAs [27]. Like in KSHV, these miRNAs are encoded within the latency-associated region of the genome; however, their sequences are not homologous to those of KSHV.
Murine gammaherpesvirus type 68 (MHV68) encodes nine miRNAs that are embedded within tRNA-like genes at the 5′ end of the genome; these genes have been suggested to be transcribed by RNA polymerase III (polIII) [28].
The presence of miRNA genes in betaherpesviruses is, so far, restricted to cytomegaloviruses (CMV). Nine miRNA genes, were initially found in human cytomegalovirus (HCMV) scattered throughout the viral genome and are expressed from multiple promoters [28]. This number was later expanded to 11, when two additional HCMV miRNAs were identified both by cloning and bioinformatic prediction of hairpins conserved between HCMV and chimpanzee CMV [29, 30]. HCMV miRNAs are readily detectable by Northern blot after de novo infection of epithelial, endothelial, and neuronal cells, even in the presence of cycloheximide, indicating that HCMV miRNAs are expressed as immediate-early gene transcripts [29]. The genome of murine cytomegalovirus (MCMV) encodes 18 miRNA genes. Quantitative analysis of viral miRNA expression after MCMV infection revealed that at early time points post infection the majority of expressed miRNAs were of viral origin [31]. CMV latency in vivo affects multiple tissues, including bone marrow; unfortunately, due to the lack of latent tissue culture models, miRNA expression during CMV latency has not been investigated.
Among alphaherpesviruses, miRNAs have been identified in herpes simplex viruses 1 and 2 (HSV-1 and -2), and Marek’s disease viruses 1 and 2 (MDV-1 and -2) [32-39]. Interestingly, like in KSHV, alphaherpesvirus miRNA genes are located within a region expressed during latency. HSV latency is characterized by the expression of the latency associated transcript (LAT), a non-coding transcript that is antisense to two lytic genes: ICP0, a transcriptional regulator, and ICP34.5, a neurovirulence factor (see [40] for a review on LAT). Recently, four miRNAs (miR-H2 to miR-H5) were cloned from a variety of sources including: (i) HEK 293 cells (a cell line derived from human embryonic kidney) ectopically expressing LAT, (ii) productively infected Vero cells (a cell line derived from African green monkey kidney), and (iii) latently infected trigeminal ganglia in mice [32]. One additional miRNA gene (miR-H6) was located upstream of LAT in HSV-1, and 11 miRNAs were predicted to be encoded elsewhere in the viral genome but to date have not been cloned [32, 33]. Most recently, Umbach and colleagues confirmed the expression of miR-H2 to miR-H6 in human trigeminal ganglia, and also identified two novel miRNAs (miR-H7 and miR-H8) also located within LAT [41]. The genome of HSV-2 encodes three miRNAs within LAT that are positionally conserved, as compared to its close relative, HSV-1 [34, 35].
Burnside and colleagues used 454 deep sequencing to identify 13 miRNAs expressed from the genome of MDV-1. These miRNAs were mapped to the inverted repeat short and long regions (IRs and IRL) of the MDV-1 genome. Eight of these miRNA genes are located within the meq oncogene region, whereas the others map to the LAT region [36, 37]. In the closely related MDV-2 virus, conventional cloning techniques identified 17 miRNAs which, like those of MDV-1, were mapped to the IRs and IRL genomic regions [38, 39].
To date, with the exception of varicella zoster virus (VZV) [41], all herpesviruses examined,express miRNAs (Table 1, Figure 2). However, the use of mass parallel sequencing to analyze small RNA libraries from virus-infected cells might uncover new, less abundantly expressed, miRNAs.
Herpesviral miRNAs closely resemble their host cellular counterparts
With respect to gene organization, viral miRNA genes recapitulate their cellular counterparts. They are organized either as single genes (e.g. in CMV) or in clusters (e.g. in alphaherpesviruses and gammaherpesviruses), the latter allowing for co-regulated expression (Figure 2). To date there is no evidence that herpesviral proteins are involved in viral miRNA maturation, which is strictly dependent on Drosha/DGCR8 and Dicer processing (Figure 1). Viral miRNA and host miRNA sequences can be located within introns or exons of protein encoding genes. The relative genomic location of the pre-miRNA and surrounding splice-donor/acceptor sites might lead to competition between miRNA maturation and mRNA splicing, like it occurs for cellular genes. For example, the EBV BART miRNAs, which are located within introns of a multiple spliced transcript, are processed prior to splicing, thereby suppressing the usage of surrounding exons. It is not clear whether a single BART transcript can gives rise to an intron-encoded miRNA and a fully processed mRNA [19].
One hallmark of cellular miRNAs is that ~30% are highly conserved across species. For instance, eight of 29 EBV miRNAs showed sequence similarity to those of its close relative LCV [15]. In contrast, no homology was noted between KSHV and RRV miRNAs [27]. In this case, one possible explanation is that, in rhesus macaques and chimpanzees two different rhadinoviruses exist (RRV1 and RRV2), while to date only one human rhadinovirus strain has been identified (KSHV).
If miRNA function is important for viral biology then the corresponding sequences will likely co-evolve with their respective host target sequences [42]. Hence, answering the question whether viral miRNAs are conserved will be greatly aided by understanding their targets and function. Thus far, sequence analysis of both EBV and KSHV miRNA gene loci from a large number of cell lines and primary isolates revealed very few polymorphisms, which suggest in vivo selection for intact miRNA genes; albeit indirect, this constitutes a genetic argument for biological function [43].
What genes are regulated by herpesvirus miRNAs?
Understanding the functions of viral miRNAs requires the determination of target genes, which can be viral and/or cellular. While the initial reports identifying viral miRNAs predicted many gene targets [14, 22, 23], the number of experimentally determined targets is still modest. The currently validated targets were identified by combining bioinformatics and experimental approaches (see Box 1 for a summary of the most commonly used techniques).
Box 1. Identifying miRNA targets by bioinformatic and experimental approaches.
Unlike siRNAs, which bind to their targets with 100% complementarity, a single miRNA can bind multiple targets with differing degrees of complementarity. The “seed-sequence”, nucleotides 2 to 8 of a miRNA, is a major predictor for targeting. Furthermore, additional base pairing, location within the 3′UTR and AU-richness surrounding the seed sequence contribute to targeting efficiency [63]. Based on these criteria, several open access software programs have been developed for prediction of potential miRNA targets: Pic-Tar (see http://pictar.mdc-berlin.de/), Target-Scan (http://www.targetscan.org/), miRanda (http://www.microrna.org/). While the latest revised algorithms are valuable tools, it is important to note that miRNA targeting is dependent on a number of factors that include the expression levels of miRNA and target genes. For example, a certain gene might have HSV miRNA binding sites but because it is only expressed in cells non-susceptible to virus infection it is not relevant as viral miRNA target. Also, a potential viral miRNA target might be highly expressed in lymphocytes but the corresponding viral miRNA is expressed at low levels. Hence, bioinformatic prediction does not replace the need to experimentally determine targets in the context of the appropriate biological systems.
All miRNA targets described in this review have been experimentally confirmed. Based on the realization that miRNA-dependent inhibition of translation often entails transcript degradation, many laboratories have successfully utilized genome-wide transcriptome analysis under conditions where miRNAs are either ectopically expressed or are inhibited by sequence-specific inhibitors called antagomirs. It is desirable to analyze miRNA targets under conditions where miRNA maturation takes place (i.e using polII expression vectors or retroviral vectors), since transfection of miRNA mimics might increase off-target effects. Genes showing changed expression levels are routinely confirmed for targeting by cloning their respective 3′UTRs down-stream of a reporter gene and subsequently by introducing seed-sequence mutations, which should abrogate miRNA-dependent inhibition. Finally, if antibodies specific for the target protein are available, the strongest experimental evidence for miRNA regulation is to demonstrate decreased target protein expression by Western blot analysis. Very recently, high throughput proteomics analysis has been used to examine the entire proteome in response to miRNA expression [74, 75]; however such approaches have not yet been reported for any viral miRNAs. To prove biological function, some virologists have created recombinant viruses with altered miRNA expression repertoires, as in the studies of HCMV miR-UL112-1 [46] as discussed in the main text of this article. The combination of recombinant viruses and appropriate animal models, where available, will likely produce further insights into the role of viral miRNAs in viral persistence and latency in vivo.
Regulation of alphaherpesvirus latency by LAT-encoded miRNAs
During latency, HSV-1 appears to expresss only non-coding RNA transcripts. The most abundant of these transcripts is the 8.3 kbp LAT, which after splicing gives rise to a 2 kbp stable intron. Many genetic studies have been aimed at determining LAT’s function and its role in latency. Mutations of different LAT regions within recombinant viruses affect the ability of the virus to establish latency and to reactivate, and in addition sensitizes latently infected cells to apoptosis inducers (recently reviewed in [40]). However, the mechanisms governing these LAT phenotypes are still not fully understood.
Recently, four miRNAs were identified in HSV-1 LAT and three in HSV-2 LAT [32, 34, 35]. Most of these miRNA sequences overlap with lytic genes in an antisense fashion. For instance, in HSV-1, miR-H2 is antisense relative to ICP0, whereas both miR-H3 and miR-H4 are antisense relative to ICP34.5. Based on this antisense positioning, it was predicted that LAT-encoded miRNAs might target these genes, but only miR-H2 has been shown experimentally to repress ICP0 protein production [32]. Targeting of ICP0 in HSV-2 was also experimentally confirmed for miR-III, which is antisense relative to ICP0 [35]. The genomes of both HSV-1 and HSV-2 also encode two miRNAs whose sequences are antisense relative to the lytic gene ICP34.5. The ability of these antisense miRNAs to target and repress ICP34.5 expression was experimentally validated for the HSV-2 miRNAs, miR-I and miR-II [34, 35]. These data suggest that miR-H2, miR-H3 and miR-H4 (of HSV-1), and miR-I, miR-II and miR-III (of HSV-2) contribute to the maintenance of latency by shutting off translation of ICP0 and ICP34.5. Whether these miRNAs also target cellular genes in a seed-sequence specific fashion is currently not known. The fact that the relative locations, but not the sequences, of LAT miRNAs are conserved between HSV-1 and HSV-2 suggests antisense positioning as the main mechanism for HSV miRNA targeting. However, it is not currently known if these miRNAs also target cellular genes in the same manner.
Umbach and collaborators also identified a fifth HSV-1 miRNA, miR-H6, expressed from a separate novel transcript upstream of LAT [32]. Bioinformatic analysis revealed a miR-H6 seed-sequence within the 3′UTR of ICP4 that was experimentally confirmed to mediate ICP4 protein downregulation. ICP4, located downstream of LAT, is the major viral transactivator required for productive infection and reactivation [32, 40]. There appears to be no miR-H6 counterpart in HSV-2, but MDV-2 encodes a ICP4 antisense miRNA [35]. Recently, it was shown that ICP4 negatively regulates LAT expression in HSV-2, leading to the suppression of LAT encoded miRNAs. This suggests that transcription factors and miRNAs encoded in the LAT region of alphaherpesviruses are inversely regulated and thereby function as a molecular switch [35]. Further indication for the importance of these miRNAs in latency control is the fact that LAT miRNAs are expressed at several hundreds of thousands copies per cell in latently infected trigeminal ganglia, whereas only hundreds of copies per cell are detected during productive infection of Vero cells [32].
In summary, the presence of similar miRNAs in the LAT regions of all alphaherpesviruses examined strongly suggests that a main function for the LAT region is to express miRNAs, which contribute to the establishment and maintenance of latency by targeting ICP0, ICP34.5, and ICP4 via siRNA- and miRNA-mediated inhibition. There are, however, still open questions. This model would postulate a mechanism for reactivation that relies on the inhibition of LAT-encoded miRNA expression and/or activity. Therefore, only careful genetic studies which analyze the contribution of each LAT miRNA in vivo will ultimately determine whether LAT harbors additional activities.
One HCMV miRNA regulates both cellular and viral target genes
A total of 11 miRNAs are encoded at different locations within the HCMV genome [28-30]. Bioinformatic analyses have predicted potential host and viral gene targets but, currently, experimental validation is limited to targets for only one miRNA, miR-UL112-1 [44-46].
Target prediction revealed a miR-UL112-1 seed match in the 3′UTR of a host gene coding for major histocompatibility complex class I-related chain B (MICB), a cellular ligand essential for NK cell killing of virally infected cells [46]. Luciferase reporters and Western blot analysis in transfected cells clearly showed that MICB protein levels are regulated by miR-UL112-1. Furthermore, Stern-Ginnossar and colleagues generated a miR-UL112-1 knock-out virus which resulted in increased expression of MICB in infected cells and lead to increased susceptibility to NK cell killing, providing direct evidence that miR-UL112-1 plays an important role in immune evasion. Interestingly, MICB surface expression is also targeted by the viral protein UL16 which induced endocytosis and subsequently degradation of MICB [47, 48]. This study is the first example in which a miRNA knock-out recombinant virus was used to study viral miRNA function during infection [46].
In addition to host gene targeting, miR-UL112-1 has also been reported to regulate viral genes encoded in the major immediate early (MIE) region of HCMV [44, 45]. Two of these potential targets, UL112/113 and IE72 (also termed IE1) encode proteins that promote viral DNA replication, while the third UL120/121 has a currently unknown function [49]. The ability of miR-UL112-1 to directly target this set of MIE genes was experimentally validated using luciferase constructs [44, 45]. To demonstrate that miR-UL112-1 could regulate IE72 expression during viral infection, a synthetically produced miR-UL112-1 mimic was transfected into cells prior to infection with HCMV. This resulted in a reduction in IE72 protein expression but also caused the reduction of additional MIE gene products, suggesting that miR-UL112-1 mediated regulation was not solely IE72 specific. Furthermore, DNA replication seemed to be inhibited in response to transfection of miR-UL112-1 mimics [44]. Murphy and colleagues used a novel algorithm and predicted that miRNAs of alpha-, beta-, and gammaherpesviruses would target their respective IE transactivators and moreover also predicted IE72 of HCMV to be targeted by miR-UL112-1. In addition to reporter assays, experiments using recombinant viruses lacking either miR-UL112-1 or the target site in IE72 demonstrated miR-UL112-1-dependent regulation of IE72 protein levels during viral infection [45]. However, in contrast to the previous study [44], mutant viruses showed no differences in replication compared to the wild type strain, suggesting that miRNA regulation of the IE region seems to play a role in viral latency and reactivation [45]; unfortunately, these processes cannot be easily studied for HCMV in tissue culture models. It is noticeable that miR-UL112-1 provides the first example in which a single miRNA targets both cellular and viral gene expression contributing to immune evasion and potentially reactivation from latency.
EBV miRNAs target cellular genes and appear to regulate viral genes during latent and lytic replication
The EBV genome includes 25 miRNA genes giving rise to a total of 39 mature miRNAs, all being expressed from two primary transcripts, BART and BHRF1. One of the miRNAs, miR-BART2, is located in an antisense position relative to the 3′UTR of the early lytic gene BALF5, which encodes the viral DNA polymerase [14]. This was an important finding because it represents the first example of a viral antisense miRNA. Because the sequence of miR-BART2 is completely complementary to the sequence of BALF5 mRNA, it was predicted that miR-BART might cleave BALF5 mRNA using a siRNA-like mechanism. Indeed, BALF5 cDNA clones containing unusual 3′ ends that matched the predicted miR-BART2 cleavage site had previously been described, but the mechanism governing this 3′ end processing was unknown [50]. Barth and colleagues confirmed that miR-BART2 is required for the cleavage of BALF5 in EBV infected cells, and this process is reduced after lytic induction [51]. Whether miR-BART2 regulation of BALF5 plays a role in maintaining latency or whether the viral polymerase is temporally regulated during lytic replication by this mechanism is currently unknown.
Bioinformatic prediction of potential targets for BART miRNAs revealed that the 3′UTR of the transcript encoding viral latent membrane protein 1 (LMP1) contains partial sequence similarity to a number of BART miRNAs [52]. LMP1 is a viral oncoprotein which is required for EBV-dependent immortalization of human primary B cells. LMP1 protein synthesis was experimentally shown to be modulated by three different BART cluster 1 miRNAs, (miR-BART16, miR-BART17-5p, and miR-BART1-5p). In part, LMP1-dependent transformation involves activation of the nuclear factor-kappa B (NF-κB) pathway, which promotes cellular proliferation and survival [53]. Using NF-κB reporters, it was demonstrated that BART mediated repression of LMP1 affects NF-κB activity. In addition, miR-BART regulation of LMP1 also significantly decreased cell sensitivity to apoptosis. Because over-expression of LMP1 leads to increased sensitivity to apoptosis and reduced NF-κB activity, it appears that BART miRNAs contribute to cellular survival during EBV latency by tightly regulating LMP1 expression levels [52].
To date, only two cellular genes have been found to be targeted by EBV BART miRNAs: those coding for p53 up-regulated modulator of apoptosis (PUMA) and CXCL-11, an IFN-inducible T-cell attracting chemokine. Bioinformatic analysis predicted the PUMA gene to be targeted by miR-BART5, and this was confirmed by 3′UTR reporter assays, and antagomir inhibition experiments [54]. PUMA is a pro-apoptotic protein whose expression is [54] transcriptionally upregulated by the tumor suppressor p53 [55, 56]. Choy and colleagues found that PUMA expression is low in about 60% of primary NPC tissue samples studied, in contrast to p53 which is upregulated in EBV-associated NPCs. Furthermore, inhibition of miR-BART5 with antagomirs in an EBV-positive NPC cell line (which expresses low amounts of PUMA) induced apoptosis, demonstrating for the first time inhibition of apoptosis through viral miRNA targeting of cellular pro-apoptotic proteins [54].
Analysis of several EBV-associated primary tumors revealed a high miR-BART expression, whereas BHRF miRNAs were only detected in AIDS diffuse large B cell lymphomas (AIDS-DLBCL) during type III latency, a gene expression program in which all latent genes are expressed [20, 57]. Based on work by Pfeffer and colleagues, who predicted a binding site for miR-BHRF1-3 within the 3′UTR of CXCL-11 [14], Xia and collaborators subsequently showed that miR-BHRF1-3 expression is inversely correlated with CXCL-11 expression during type III latency [57]. Targeting was experimentally confirmed via antagomir-based inhibition of miR-BHRF1-3. Inhibition of CXCL-11, a secreted T-cell attractant, could potentially protect EBV-infected tumor cells from immune surveillance. This study [57] and a recent miRNA profiling study on primary NPC biopsies [18] underlines the importance of analyzing miRNA expression in primary tissues rather than in established cell lines, since it appears that many EBV-infected cell lines express significantly reduced levels of BART miRNAs [21].
Identification of KSHV miRNA targets by gene expression profiling
The KSHV genome contains a total of 12 miRNA genes, all located in the KSHV latency associated region (KLAR). Targets for these miRNAs have largely been determined by unbiased gene expression profiling studies rather than bioinformatic prediction [58-61]. The first published cellular target of KSHV miRNAs was the host gene coding for thrombospondin 1 (THBS1), a tumor suppressor and anti-angiogenic factor, reported to be down-regulated in KS lesions [59]. Samols and colleagues generated HK 293 cells expressing 10 KSHV miRNAs and found 65 genes that showed decreased mRNA levels compared to vector controls. The 3′UTRs of altered genes had a higher frequency of seed-sequence matches, including that of THBS1, which contained 34 potential binding sites for multiple KSHV miRNAs. By using 3′UTR luciferase reporter assays and Western blot analysis, direct targeting and repression of THBS1 expression for several KSHV miRNAs (miR-K12-1, miR-K12-3-3p, miR-K12-6-3p and miR-K12-11) was demonstrated. THBS1 is the first cellular gene targeted by multiple viral miRNAs. Because THBS1 activates latent TGFβ, Samols and colleagues used TGFβ responsive reporter assays to demonstrate that KSHV miRNA repression of THBS1 translates into decreased TGFβ activity. Given that angiogenesis is a hallmark of KS, the finding that KSHV miRNAs target a strong inhibitor of angiogenesis, suggest that KSHV miRNAs contribute to pathogenesis [59].
Recently, the Ganem group reported a highly comprehensive tandem-array approach to identify miRNA targets, utilizing gene expression profiling in endothelial cells after de novo infection, and in B cells that ectopically expressed individual, or various sets of, miRNAs. For a gene to be recognized as a potential target, its expression had to be reduced in the ectopic miRNA expressing cells and increased in latently-infected PEL cells transfected with the corresponding antagomir [61]. For each KSHV miRNA, about 10 to 30 cellular genes passed these criteria. Analysis of miR-K5 revealed 11 gene targets, including bcl-2-associated factor (BCLAF1), which was studied in more detail. BCLAF1 functions as a transcriptional repressor and when over-expressed can mediate apoptosis [62]. In addition to miR-K5, both miR-K12-9 and miR-K12-10b were also found to target and regulate the expression of BCLAF1 [61]. Ziegelbauer and colleagues demonstrated that transfection of miR-K5, miR-K12-9 or miR-K12-10b into human umbilical vein endothelial cells (HUVEC) were able to inhibit etoposide-induced caspase activation, thereby suggesting that miRNA repression of BCLAF1 inhibits apoptosis [61]. Interestingly, increased etoposide-induced apoptosis was observed when HUVEC cells were plated at a lower density and then transfected with the same KSHV miRNAs, indicating that BCLAF1 has anti-apoptotic activity during certain growth conditions. Although suggesting an anti-apoptotic function for BCLAF1 was counterintuitive at first, the researchers went on to find that BCLAF1 expression in latently infected PEL cells inhibits lytic viral replication. In addition, inhibiting KSHV miRNA targeting of BCLAF1 with antagomirs resulted in decreased lytic reactivation in KSHV-infected endothelial cells (SLK). Together, these data suggest that targeting BCLAF1 sensitizes latently-infected cells to signals that induce reactivation from latency [61]. Hence, miR-K5 provides the first example by which targeting of a host cellular gene contributes to latency control (Figure 3).
Figure 3.
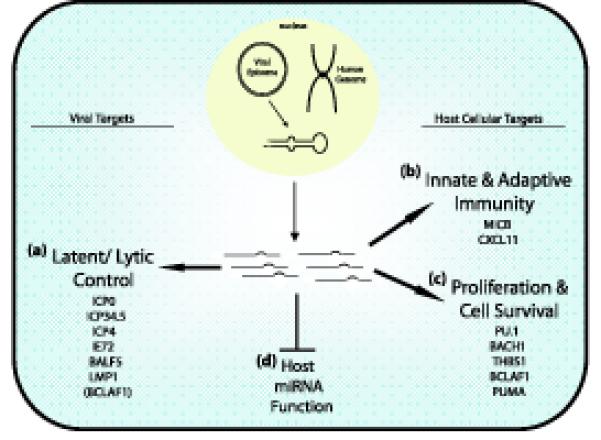
Themes of herpesvirus miRNA regulation. Both viral and host cellular genes are targeted. (a) One main theme is the tight control of latency by inhibition of immediate early or early genes as shown for ICP0, ICP4, ICP34.5, and IE72. Additionally, EBV LMP1 expression is fine tuned by viral miRNAs to enhance cell survival during latency. Targeting of BCLAF1, a cellular gene, contributes to latent/lytic replication by sensitizing cells to reactivation. (b) Targeting of antiviral responses (MICB and CXCL11). (c) Modulation of cellular proliferation and survival. Target genes have anti-angiogenic (THBS1), or pro-apoptotic activity (BCLAF-1, PUMA, and Bach-1). (d) Impairing host cellular miRNA function. Seed sequence-independent modes of viral miRNA function have also been suggested. One alternative model is based on de novo infection experiments with MCMV, which demonstrated that early after infection viral miRNA synthesis completely overtakes host cellular miRNA production [73]. As a consequence host miRNA function might be impaired leading to a global de-repression of the cell transcriptome. This could be due to hijacking of either Drosha/DGCR8 processing or RISC loading.
Data from our lab and the Cullen group revealed that one KSHV miRNA, miR-K12-11, shares 100% seed sequence homology with a human miRNA, miR-155 [58, 60]. Because the seed sequence is the most important parameter in mRNA target recognition (Box 1) [63], it was predicted that both miRNAs might target an overlapping set of host genes. Gene expression profiling in both HK 293 cells and BJAB cells (an EBV negative Burkitt’s lymphoma cell line) stably expressing either miR-155 or miR-K12-11 identified a common set of genes that were downregulated in the presence of either miRNA. Further computational analysis found that one gene, BACH1, contained four target sites within its 3′UTR [58, 60]. Targeting and inhibition of BACH1 by both miRNAs was validated by 3′ UTR reporter assays, mutagenesis and Western blot analysis. Importantly, PEL-derived cell lines that express high levels of miR-K12-11, but not miR-155, expressed very low BACH1 levels. BACH1 is a transcriptional repressor that has been shown to repress expression of heme-oxygenase 1 (HMOX1), a protein that enhances cell survival and proliferation [64]. Because KSHV has been reported to directly increase HMOX1 levels during endothelial cell infection, these studies suggest that viral miRNAs could contribute to HMOX1 upregulation by inhibiting expression of its transcriptional repressor BACH1 [60, 65]. Interestingly, based on its overexpression pattern in a number of B cell lymphomas, miR-155 has been characterized as an oncomir, i.e. a miRNA with tumorigenic activity [66]. Also, it has been shown that miR-155 mediated regulation is essential for B-cell function and differentiation into plasma cells [67]. Because KSHV infected PEL cells are tumorigenic B-cells that do not express miR-155 and appear to be blocked from fully differentiating into plasma cells, it has been proposed that the mimicking of miR-155 regulatory pathways by viral miR-K12-11 could be a contributing component to KSHV transformation in this lymphoma [58, 60].
Further experimentation is needed to identify the cellular miR-K12-11 targets whose dysregulation during B cell development might directly contribute to KSHV pathogenesis.
Concluding remarks and future directions
Thus far, it appears that there are several unifying themes of miRNA regulation utilized by herpesviruses including latent/lytic control, immune evasion, and cell survival and proliferation (Figure 3). One unique feature of viral miRNA regulation, to our best knowledge not described for metazoan cells, is the expression of antisense transcripts that serve as pre-miRNAs to block IE gene expression in alphaherpesviruses. Although it has yet to be demonstrated, a number of genes within HCMV (UL70, US29, and UL150), EBV and LCV (LF2 and LF3, both of which might function in reactivation) could potentially be regulated by this antisense mechanism. The importance of miRNAs targeting antiviral responses is further supported by the observation that not only CMV, but also KSHV and EBV miRNAs target the cellular ligand MICB [68]. Furthermore, MICB is targeted by the UL16 protein of HCMV [47, 48] and by the MIR1 and MIR2 proteins of KSHV [13] and it will be interesting to see whether a similar dichotomy of regulation exists in other herpesviruses. Gammaherpesvirus miRNA-dependent regulation of cell survival and proliferation certainly points to a role in de-regulating cell growth. A key question to be answered is whether viral miRNAs directly contribute to EBV and KSHV-associated malignancies. Thus far the strongest candidate for such a role in KSHV is the discovery of miR-K12-11, which mimics the oncomir miR-155. Interestingly, MDV-1, an avian tumorigenic virus, also encodes a miR-155 mimic [69, 70], and EBV strongly up-regulates miR-155 in BLs [71, 72]. As is the case with other viral latency-associated genes, future studies will depend on the availability of robust genetic systems and appropriate in vitro and in vivo models (Box 2).
Box 2. Future research topics and challenges.
Experimental identification and validation of targets for all herpesviral miRNAs
Mutation of miRNA genes within context of viral genome
Analysis of cell type-specific miRNA regulation (i.e. EBV lymphoid vs. epithelial)
Functional studies on gammaherpesvirus miRNAs in the context of transformation
In vivo studies using appropriate animal models, where available (i.e. RRV and MHV-68)
Table 2.
Viral and cellular targets of herpesviral miRNAs
| Herpesvirus | miRNA | Target | Function | Reference |
|---|---|---|---|---|
| Viral targets | ||||
| HSV-1 | miR-H2-3p | ICP0 | Immediate Early | [32] |
| transactivator | ||||
| miR-H3 | ICP34.5 | Neuro-virulence | [32] | |
| miR-H4 | factor | [32] | ||
| miR-H6 | ICP4 | [32] | ||
| Immediate Early | ||||
| transactivator | ||||
| HSV-2 | miR-I | ICP34.5 | Neuro-virulence | [34,35] |
| miR-II | ICP34.5 | factor | [34-35] | |
| miR-III | ICP0 | Neuro-virulence | [34,35] | |
| HCMV | miR-UL112-1 | IE72 | factor | [46] |
| Immediate Early | ||||
| transactivator | ||||
| Immediate Early | ||||
| transactivator | ||||
| EBV | miR-BART2 | BALF5 | DNA polymerase | [14] |
| miR-BART1-5p | LMP1 | Viral oncogene | [52] | |
| miR-BART16 | [52] | |||
| miR-BART17- 5p |
[52] | |||
| Cellular Targets | ||||
| MDV-1 | mdv1-miR- | PU.1 | Hematopoietic | [70] |
| HCMV | M4a | MICB | transcription | [46] |
| KSHV | miR-UL112-1 | BACH1 | factor | [58,60] |
| miR-K12-11a | THBS1 | NK cell ligand | [59] | |
| miR Cluster | BCLAF1 | Transcriptional | [61] | |
| miR-K12-5 | suppressor | |||
| Angiogenesis | ||||
| inhibitor | ||||
| Pro-apoptotic | ||||
| factor | ||||
| EBV | miR-BART5 | PUMA | Pro-apoptotic | [54] |
| factor | ||||
| miR-BHRF1-3 | CXCL11 | Chemokine, T-cell | [57] | |
| attractant | ||||
Shown to have seed sequence homology with human miR-155
Glossary
- Antagomir
a 2′O-methylated RNA oligonucleotide that is complementary to a specific miRNA. It is used to inhibit miRNA function.
- miRNA mimic
a synthetic miRNA, delivered by transfection.
- Oncomir
a miRNA with transforming activity and/or upregulated in human cancer (e.g. miR-155 or miR-16).
- siRNA
a small interfering RNA that binds with complete sequence complementarity to mRNAs and induce RISC-dependent degradation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Ruvkun G, et al. Dominant gain-of-function mutations that lead to misregulation of the C. elegans heterochronic gene lin-14, and the evolutionary implications of dominant mutations in pattern-formation genes. Dev Suppl. 1991;1:47–54. [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5(4):659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 12.Roizman B, Sears AE. Herpes Simplex viruses and their replication. In: Roizman B, Whitley RJ, Lopez C, editors. The Human Herpesviruses. Raven Press; New York, NY: 1993. pp. 11–68. [Google Scholar]
- 13.Areste C, Blackbourn DJ. Modulation of the immune system by Kaposi’s sarcoma-associated herpesvirus. Trends Microbiol. 2009;17(3):119–29. doi: 10.1016/j.tim.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 15.Cai X, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006;12(5):733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu JY, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83(7):3333–41. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosmopoulos K, et al. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol. 2009;83(5):2357–67. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. 2008;82(18):9094–106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81(18):9967–75. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratt ZL, et al. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386(2):387–97. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai X, et al. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102(15):5570–5. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2(4):269–76. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 24.Samols MA, et al. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79(14):9301–5. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai X, Cullen BR. Transcriptional origin of Kaposi’s sarcoma-associated herpesvirus microRNAs. J Virol. 2006;80(5):2234–42. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J Virol. 2005;79(22):14457–64. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer A, et al. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology. 2007;364(1):21–7. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 29.Dunn W, et al. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;7(11):1684–95. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 30.Grey F, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79(18):12095–9. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeffer S. Identification of virally encoded microRNAs. Methods Enzymol. 2007;427:51–63. doi: 10.1016/S0076-6879(07)27003-X. [DOI] [PubMed] [Google Scholar]
- 32.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–3. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui C, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80(11):5499–508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang S, et al. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105(31):10931–6. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83(3):1433–42. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burnside J, et al. Marek’s disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J Virol. 2006;80(17):8778–86. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burnside J, et al. Deep sequencing of chicken microRNAs. BMC Genomics. 2008;9:185. doi: 10.1186/1471-2164-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y, et al. Marek’s disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J Virol. 2007;81(13):7164–70. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao Y, et al. MicroRNA profile of Marek’s disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J Virol. 2008;82(8):4007–15. doi: 10.1128/JVI.02659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloom DC. HSV LAT and neuronal survival. Int Rev Immunol. 2004;23(12):187–98. doi: 10.1080/08830180490265592. [DOI] [PubMed] [Google Scholar]
- 41.Umbach JL, et al. Analysis of human {alpha}-herpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009 doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sood P, et al. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103(8):2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall V, et al. Conservation of virally encoded micrornas in Kaposi sarcoma--associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric castleman disease. J Infect Dis. 2007;195(5):645–59. doi: 10.1086/511434. [DOI] [PubMed] [Google Scholar]
- 44.Grey F, et al. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy E, et al. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105(14):5453–8. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cosman D, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14(2):123–33. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 48.Dunn C, et al. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197(11):1427–39. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87(Pt 7):1763–79. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 50.Furnari FB, Adams MD, Pagano JS. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc Natl Acad Sci U S A. 1993;90(2):378–82. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barth S, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lo AK, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104(41):16164–9. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izumi KM, Kieff ED. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci U S A. 1997;94(23):12592–7. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choy EY, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205(11):2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han J, et al. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci U S A. 2001;98(20):11318–23. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7(3):683–94. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 57.Xia T, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68(5):1436–42. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gottwein E, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450(7172):1096–9. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samols MA, Hu J, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007 doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skalsky RL, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81(23):12836–45. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41(1):130–4. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasof GM, Goyal L, White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol. 1999;19(6):4390–404. doi: 10.1128/mcb.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8(12):107–18. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 65.McAllister SC, et al. Kaposi sarcoma-associated herpesvirus (KSHV) induces heme oxygenase-1 expression and activity in KSHV-infected endothelial cells. Blood. 2004;103(9):3465–73. doi: 10.1182/blood-2003-08-2781. [DOI] [PubMed] [Google Scholar]
- 66.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15(4):352–8. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 67.Baltimore D, et al. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9(8):839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 68.Nachmani D, et al. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5(4):376–85. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 69.Morgan R, et al. Sequence conservation and differential expression of Marek’s disease virus microRNAs. J Virol. 2008;82(24):12213–20. doi: 10.1128/JVI.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Y, et al. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J Virol. 2009;83(1):489–92. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gatto G, et al. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36(20):6608–19. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin Q, et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82(11):5295–306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dolken L, et al. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81(24):13771–82. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]