Abstract
Autophagy is an active homeostatic degradation process for the removal or turnover of cytoplasmic components wherein the LC3 ubiquitin-like protein undergoes an Atg7 E1-like enzyme/Atg3 E2-like enzyme-mediated conjugation process to induce autophagosome biogenesis1–4. Besides its cytoprotecive role, autophagy acts on cell death when it is abnormally upregulated. Thus, the autophagy pathway requires tight regulation to ensure that this degradative process is well balanced. Two death effector domains (DED1/2) containing cellular FLICE-like inhibitor protein (cFLIP) and viral FLIP (vFLIP) of Kaposi’s sarcoma-associated herpesvirus (KSHV), Herpesvirus saimiri (HVS), and Molluscum contagiosum virus (MCV) protect cells from apoptosis mediated by death receptors5,6. Here, we report that cellular and viral FLIPs suppress autophagy by preventing Atg3 from binding and processing LC3. Consequently, FLIP expression effectively represses cell death with autophagy, as induced by rapamycin, an mTor inhibitor and an effective anti-tumour drug against KSHV-induced Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL)7,8. Remarkably, either a DED1 α2-helix ten amino-acid (α2) peptide or a DED2 α4-helix twelve amino-acid (α4) peptide of FLIP is individually sufficient for binding FLIP itself and Atg3, with the peptide interactions effectively suppressing Atg3–FLIP interaction without affecting Atg3-LC3 interaction, resulting in robust cell death with autophagy. Our study thus identifies a checkpoint of the autophagy pathway where cellular and viral FLIPs limit the Atg3-mediated step of LC3 conjugation to regulate autophagosome biogenesis. Furthermore, the FLIP-derived short peptides induce growth suppression and cell death with autophagy, representing biologically active molecules for potential anti-cancer therapies.
During the process of autophagosome elongation, the LC3 ubiquitin-like protein is cleaved by the Atg4 cysteine protease to expose its car-boxy-terminal Gly, which is then activated by the Atg7 E1-like enzyme and transferred to the Atg3 E2-like enzyme. LC3 is then covalently conjugated to phosphatidylethanolamine and embedded into the lipid membranes for autophagic vesicle expansion9,10. The specific role of the LC3–Atg4–Atg3–Atg7 conjugation reaction is not yet known, but it is essential for autophagosome formation and may be the driving force for the deformation or curvature of the autophagic vesicle membrane.
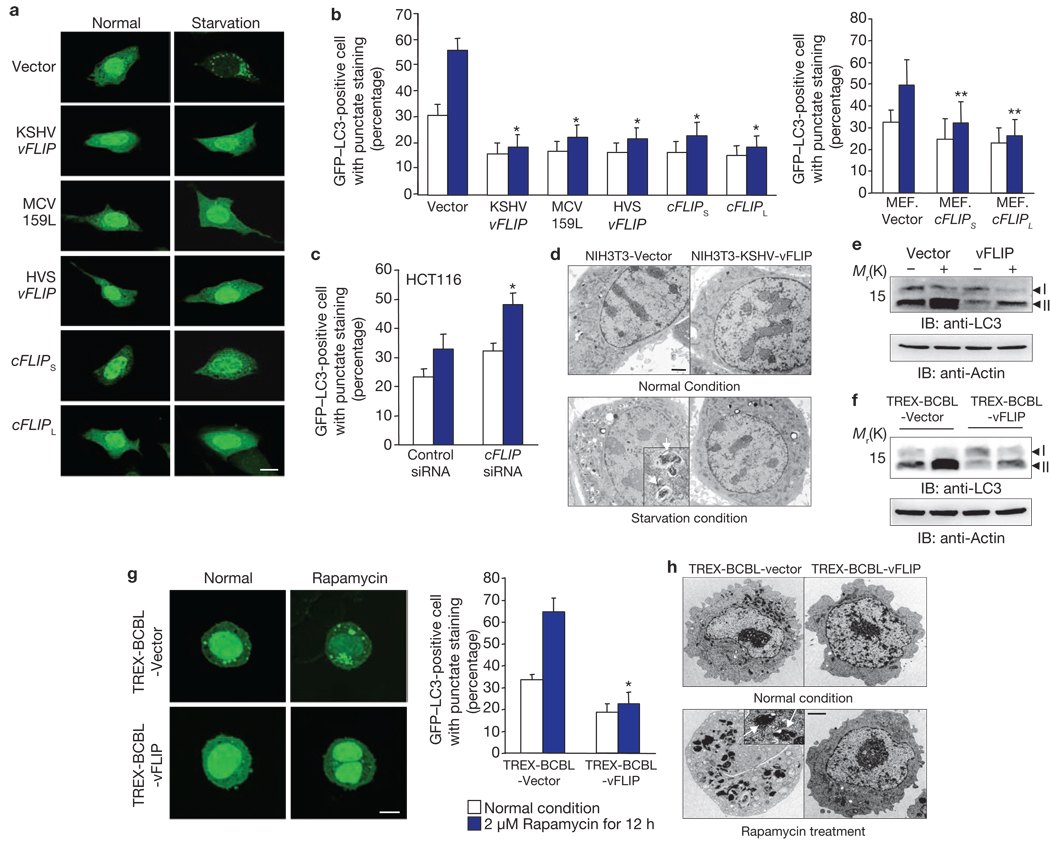
We and others have demonstrated that cellular and viral Bcl-2 inhibits autophagy through its direct interaction with the Beclin1 autophagy protein11–13. To identify additional anti-autophagic viral proteins, we screened a KSHV expression library using a GFP–LC3 staining pattern: on autophagic stimulation, GFP–LC3 redistributes from a diffused staining pattern throughout the cytoplasm and nucleus to a cytoplasmic punctate structure, specifically labelling preautophagosomal and autophagosomal membranes14. This study identified that vFLIP (also called K13) effectively suppressed autophagy induced by starvation or rapamycin, as shown by the reduction of GFP–LC3 puncta (Fig. 1a, b). As seen with KSHV vFLIP, the long and short forms of cellular FLIP (cFLIPL and cFLIPS), HVS vFLIP, and MCV 159L also suppressed autophagy induced by starvation or rapamycin, at levels comparable to KSHV vFLIP (Fig. 1a, b; Supplementary Information, Figs S1a, b, S2a). Conversely, FLIP siRNA significantly reduced endogenous cFLIP expression, subsequently increasing rapamycin-induced autophagy, whereas a control, scrambled siRNA or caspase 8-specific siRNA showed little to no effect on rapamycin-induced autophagy under identical conditions (Fig. 1c; Supplementary Information, Figs S1c, S2b, c). Electron microscopy showed that although small differences in the basal number of autophagic vacuoles per cell were observed under normal conditions, a number of autophagic vacuoles were detected in NIH3T3 vector cells but not in NIH3T3-KSHV-vFLIP cells under conditions of starvation (Fig. 1d; Supplementary Information, Fig. S3). Furthermore, a large portion of LC3-I was converted to LC3-II in NIH3T3 vector cells under conditions of starvation, whereas little processed LC3-II was detected in NIH3T3-KSHV-vFLIP, NIH3T3-MCV-159L, NIH3T3-HVS-vFLIP, and NIH3T3-cFLIPS cells (Fig. 1e; Supplementary Information, Fig. S4a). KSHV vFLIP effectively blocked autophagy in a number of cell types (Supplementary Information, Fig. S5).
Figure 1. FLIP suppresses autophagy.
(a–c) At 12–16 h post-transfection with GFP–LC3, NIH3T3 (a, b), MEF (b) or HCT116 (c) cells containing vector or the FLIP gene were treated with Hank’s solution for 2–4 h (a) or with 2 µM rapamycin for 1–3 h (b, c). Subsequently, GFP–LC3 was detected using an inverted fluorescence microscope. Scale bar, 10 µm (a). The number of GFP–LC3-positive dots per cell was counted using a fluorescence microscope. Data are mean ± s.e.m.; n = 200–300 cells from three independent experiments; *P < 0.01; **P < 0.05 (b, c). At 60 h post-transfection with control siRNA or cFLIP siRNA, MEF cells were transfected with GFP–LC3 (c). (d) Autophagosomes were visualized through scanning electron microscopy. Scale bar, 50 nm. The arrows indicate autophagosomes. Immunoblots (IB) of cFLIP and actin expression are shown in Supplementary Information, Fig. S1c. (e, f) NIH3T3 vector and NIH3T3-KSHV-vFLIP cells (e) or TREX-BCBL-Vector and TREX-BCBL-vFLIP cells (f) were treated with Hank’s solution or 2 µM rapamycin for 2–4 h, respectively, followed by IB with anti-LC3 and anti-actin antibody. (g) At 12–16 h post-transfection with GFP–LC3, TREX-BCBL-vector and TREX-BCBL-vFLIP cells were treated with doxycycline for 24 h, followed by incubation with 2 µM rapamycin for an additional 12 h. GFP–LC3 was detected using an inverted fluorescence microscope. Scale bar, 5µm. The number of GFP–LC3-positive dots per cell was counted using a fluorescence microscope. Data are mean ± s.e.m.; n = 200–300 cells from three independent experiments; *P < 0.01. (h) Autophagosomes were visualized by scanning electron microscopy. Scale bar, 50 nm.
To investigate whether vFLIP expression suppresses autophagy in virus-infected cells, we constructed a KSHV-infected primary infusion lymphoma (PEL) line, BCBL1 (TREX-BCBL), in which a Flag-tagged KSHV vFLIP gene was integrated into the chromosomal DNA under the control of a tetracycline-inducible promoter15 (Supplementary Information, Fig. S4b). At 12 h post-transfection with GFP–LC3, doxycycline-treated TREX-BCBL-vector and TREX-BCBL-vFLIP cells were incubated with rapamycin (2 µM) for an additional 12 h. GFP–LC3 puncta were then counted and LC3 processed, which showed that ectopic expression of vFLIP in TREX-BCBL cells extensively suppressed rapamycin-induced autophagy (Fig. 1f, g). Electron microscopy studies also showed that rapamycin treatment resulted in a significant increase in the number of autophagic vacuoles in TREX-BCBL vector cells but not in TREX-BCBL-vFLIP cells (Fig. 1h; Supplementary Information, Fig. S3). These results collectively indicate that cFLIP and vFLIP expression effectively blocks autophagy.
FLIPs interacts with the Fas-associated death-domain-containing protein (FADD), IKKαβγ complex, TRAFs and 14-3-3σ5,16–18 (J.S.L. and J.U.J., unpublished results). To determine the effects of the interactions between vFLIP, FADD, IKKαβγ and 14-3-3σ on anti-autophagic activity, we constructed NIH3T3 cells stably expressing various vFLIP mutants: m14-3-3σ lacking 14-3-3σ-binding; mTRAF2 lacking TRAF2-binding16; 67AAA and 58AAA with reduced NF-κB activation17; and mFADD lacking FADD-binding19. As found previously17, NF-κB activation was markedly reduced in the 67AAA and 58AAA mutants, whereas in the 67AAA, 58AAA and mFADD mutants, TNFα-induced apoptosis was poorly blocked or not blocked at all (Supplementary Information, Fig. S6a, b). However, all of the vFLIP mutants were still capable of blocking rapamycin-induced autophagy as efficiently as vFLIP wild-type cells (Supplementary Information, Fig. S6c). This result indicates that the anti-autophagic activity of KSHV vFLIP is genetically separable from its anti-apoptotic and NF-κB activation activities. Both KSHV vFLIP and cFLIPS could activate NF-κB in addition to their anti-apoptotic and anti-autophagy activity, whereas MCV 159L could not activate NF-κB but had anti-apoptotic and anti-autophagic activity; HVS vFLIP did not have detectable NF-κB activation and anti-apoptosis activity but had anti-autophagic activity (Supplementary Information, Fig. S6d, e). This indicates that anti-autophagic activity is primarily shared among cellular and viral FLIPs.
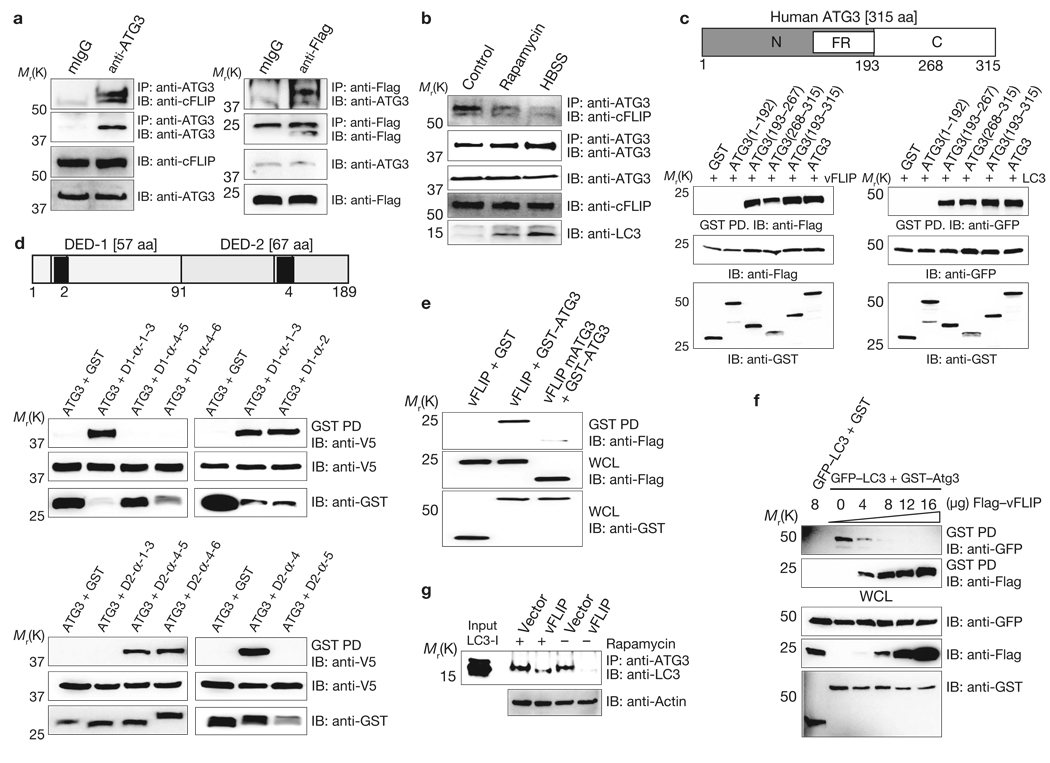
A yeast two-hybrid screen revealed the Atg3 E2-like enzyme of the LC3 conjugation system as a binding partner of vFLIP. Co-immunoprecipitation showed that cFLIPL and KSHV vFLIP interacted strongly with endogenous Atg3 (Fig. 2a). Additionally, cFLIPL, cFLIPS, KSHV vFLIP, MCV159L and HVS vFLIP efficiently interacted with GST–Atg3 or GFP–Atg3 mammalian fusions as well (Supplementary Information, Fig. S7a). Furthermore, the interaction between endogenous Atg3 and cFLIP seemed to be regulated by environmental conditions: high levels of cFLIP interacted with Atg3 under normal conditions, but this interaction seemed to decline under rapamycin-induced or starvation-induced autophagic conditions (Fig. 2b), indicating the differential binding of FLIP and Atg3. We constructed human Atg3 deletion mutants fused with a GST mammalian expression vector based on the yeast Atg3 structure (ref. 20): GST–Atg31–192, GST–Atg3193–267, GST–Atg3268–315, and GST–Atg3193–315. GST–PD showed that vFLIP independently bound either the 193–267 amino acid or 268–315 amino acid regions of the Atg3 C-terminal in a manner similar to the binding of LC3 and Atg3 (Fig. 2c). Furthermore, MCV 159L and HVS vFLIP showed Atg3 binding patterns similar to KSHV vFLIP, whereas cFLIPS bound only full-length Atg3 (Supplementary Information, Fig. S7b).
Figure 2. FLIP interacts with Atg3.
(a) BCBL cells were used for immunoprecipitation (IP) with mouse IgG or anti-Atg3, followed by immunoblotting (IB) with anti-cFLIP antibody (left panel). NIH3T3-KSHV-vFLIP cells were used for IP with mouse IgG or anti-Flag antibody, followed by IB with anti-Atg3 antibody (right panel). (b) FLIP and Atg3 interaction after autophagy induction. HCT116 cells were treated with or without 2 µM rapamycin or Hank’s solution (HBSS) for 4 h before IP with anti-Atg3 antibody, followed by IB with anti-cFLIP antibody. (c) Schematic diagram of human Atg3 (top). N, amino terminus; FR, flexible region; C, carboxy terminus. At 48 h post-transfection with GST or GST–Atg3 along with Flag–vFLIP (left) or GST or GST–Atg3 along with GFP–LC3 (right), HEK293T cells were used for GST pulldown, followed by IB with the indicated antibodies. (d) Schematic diagram of KSHV vFLIP (top). The black boxes indicate the K-α2 and K-α4 peptides. At 48 h post-transfection with V5-Atg3 and either GST–vFLIP DED1 mutants (middle) or GST–vFLIP DED2 mutants (bottom), HEK293T cells were used for GST pulldown, followed by IB with anti-V5 antibody. (e) At 48 h post-transfection with GST, GST–Atg3, and either Flag–vFLIP or Flag–vFLIP mAtg3, HEK293T cells were used for GST pulldown, followed by IB with anti-Flag antibody. (f) At 48 h post-transfection with GST–Atg3 and GFP–LC3 along with increasing amount of Flag–vFLIP, HEK293T cells were used for GST pulldown, followed by IB with anti-GFP or anti-Flag antibody. (g) NIH3T3 vector and NIH3T3 vFLIP cells were treated with 2 µM rapamycin for 4 h and used for IP with anti-Atg3 antibody, followed by IB with anti-LC3 antibody. Finally, whole-cell lysates (WCLs) were used for IB with the indicated antibodies to show expression (a–g).
Given the substantial similarities between DED1 and DED2 (refs 19, 21), Atg3 bound each DED of KSHV vFLIP with similar affinities (Supplementary Information, Fig. S7c). Remarkably, the DED1 α2 helix (10 amino acids) and the DED2 α4 helix (12 amino acids) of vFLIP were individually sufficient for Atg3 binding, whereas deletion of both helical sequences (vFLIP mAtg3) considerably abrogated Atg3 binding (Fig. 2d, e). Accordingly, vFLIP mutants that no longer interacted with 14-3-3σ, IKKγ or FADD but still bound Atg3 were as efficient as the wild-type in blocking rapamycin-induced autophagy, but the vFLIP mAtg3 mutant that no longer bound Atg3 did not block rapamycin-induced autophagy (Fig. 2e; Supplementary Information, Figs S6c, S7d). Although the vFLIP mAtg3 mutant lost its anti-apoptotic and NF-κB activation activities because of the removal of significant portions of its coding sequences, it still interacted with 14-3-3σ as efficiently as wild-type vFLIP (Supplementary Information, Figs S6a, b, S7e). These results demonstrate that vFLIP and Atg3 interact efficiently with each other using two independent binding regions: Atg3 uses the 193–267 and 268–315 amino acid regions, whereas vFLIP uses the DED1 α2 helix and the DED2 α4 helix. As the Atg3 binding patterns of FLIP and LC3 are similar, increasing vFLIP, cFLIPS, or cFLIPL showed increased FLIP–Atg3 interaction, leading to a marked decrease in LC3-Atg3 interaction (Fig. 2f ; Supplementary Information, Fig. S8). Furthermore, vFLIP expression resulted in a marked reduction of endogenous Atg3–LC3 interaction under normal conditions, although this interaction was weakly recovered after rapamycin treatment (Fig. 2g).
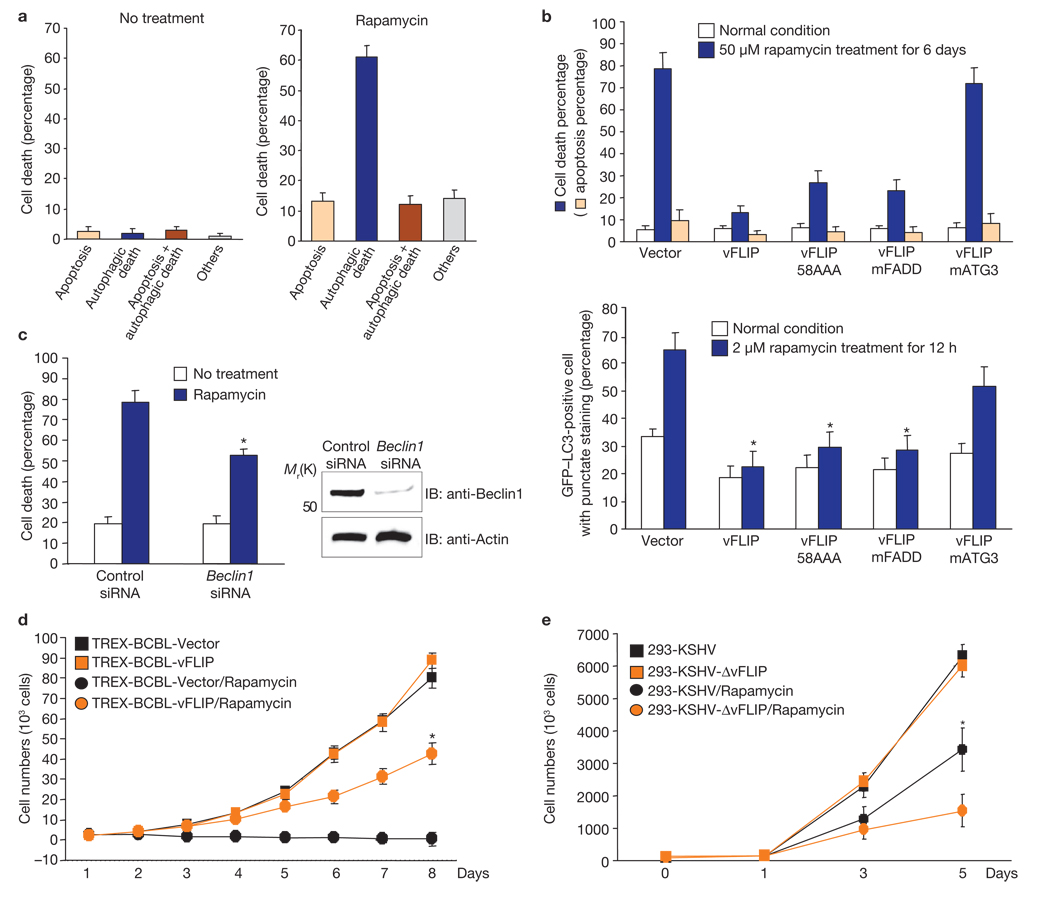
The anti-tumour and immunosuppressive potency of rapamycin in KSHV-related diseases has been well demonstrated7,8: it reduces KSHV-induced PEL cell growth in culture and inhibits the progression of dermal KS in kidney transplant recipients while providing effective immunosuppression. Rapamycin effectively induced growth suppression and death in KSHV-infected BCBL1 cells where its primary action seemed to be geared towards cell death with autophagy (Fig. 3a, b; Supplementary Information, Fig. S9a). Electron microscopy also indicated that approximately 60% of cells that were dead through rapamycin treatment showed characteristics of cell death with autophagy (Fig. 3a). Furthermore, Beclin1 siRNA significantly reduced endogenous Beclin1 expression and subsequently attenuated rapamycin-induced cell death, whereas a scrambled siRNA did not (Fig. 3c). On the other hand, ectopic expression of vFLIP wild-type, 58AAA or mFADD mutants markedly blocked rapamycin-induced growth suppression and death with autophagy, whereas the vFLIP mAtg3 mutant provided no protection for the BCBL1 cells from rapamycin-induced autophagic death (Fig. 3b, d; Supplementary Information, Fig. S9b). Furthermore, after rapamycin treatment, HEK293 cells carrying the KSHVΔvFLIP22 mutant showed detectably reduced growth rates, compared with HEK293 cells carrying wild-type KSHV (Fig. 3e). Ectopic expression of wild-type vFLIP readily suppressed rapamycin-induced growth inhibition of HEK293 cells carrying KSHVΔvFLIP, which was also seen with ectopic expression of the vFLIP 58AAA and mFADD mutants, albeit to a lesser extent. Finally, the vFLIP mAtg3 mutant expression had little or no effect on rapamycin-induced growth inhibition (Supplementary Information, Fig. S9c, d). These results further indicate a physiological role for vFLIP–Atg3 interaction in the inhibition of rapamycin-induced autophagy and growth inhibition.
Figure 3. vFLIP blocks growth suppression and cell death with autophagy induced by rapamycin.
(a) TREX-BCBL vector cells were treated or untreated with 50 nM rapamycin for 6 days in the presence of doxycycline and subjected to scanning electron microscopy. The morphologies of more than 100 dead cells were examined to quantify the levels of apoptosis, autophagic death, apoptosis/autophagic death, and others forms of cell death. (b) At 12–16 h post-transfection with GFP–LC3, doxycycline-treated TREX-BCBL vector, TREX-BCBL-vFLIP and TREX-BCBL-vFLIP mutant cells were incubated with or without 50 nM rapamycin for 6 days (top) or 2 µM rapamycin for 12 h (bottom). The percentage of cell death (trypan blue staining) and apoptosis level (annexin V staining) of rapamycin-treated cells was quantified as mean ± s.d. of three independent experiments. The number of GFP–LC3-positive dots per cell was counted using a fluorescence microscope. Data are mean ± s.e.m.; n = 200–300 cells from three independent experiments; *P < 0.01. (c) BCBL1 cells were transfected with control siRNA or Beclin1 siRNA and treated with or without rapamycin for 5 days. The percentage of cell death was determined by trypan blue staining. Data are mean ± s.e.m.; n = 200–300 cells from three independent experiments; *P < 0.05. (d) TREX-BCBL vector and TREX-BCBL-vFLIP cells were mock-treated or treated with 50 nM rapamycin for the indicated periods of time. The results were quantified as mean ± s.d. of the combined results from three independent experiments; *P < 0.001. (e) HEK293 cells carrying KSHV or KSHVΔvFLIP22 were treated with 500 nM rapamycin for 6 days. The results were quantified as mean ± s.d. of the combined results from three independent experiments; *P < 0.01. Trypan blue staining and a Beckman Coulter Z2 Particle Count and size analyzer (BC Z2 CS analyser) were used to determine cell death and cell growth analysis (b–e).
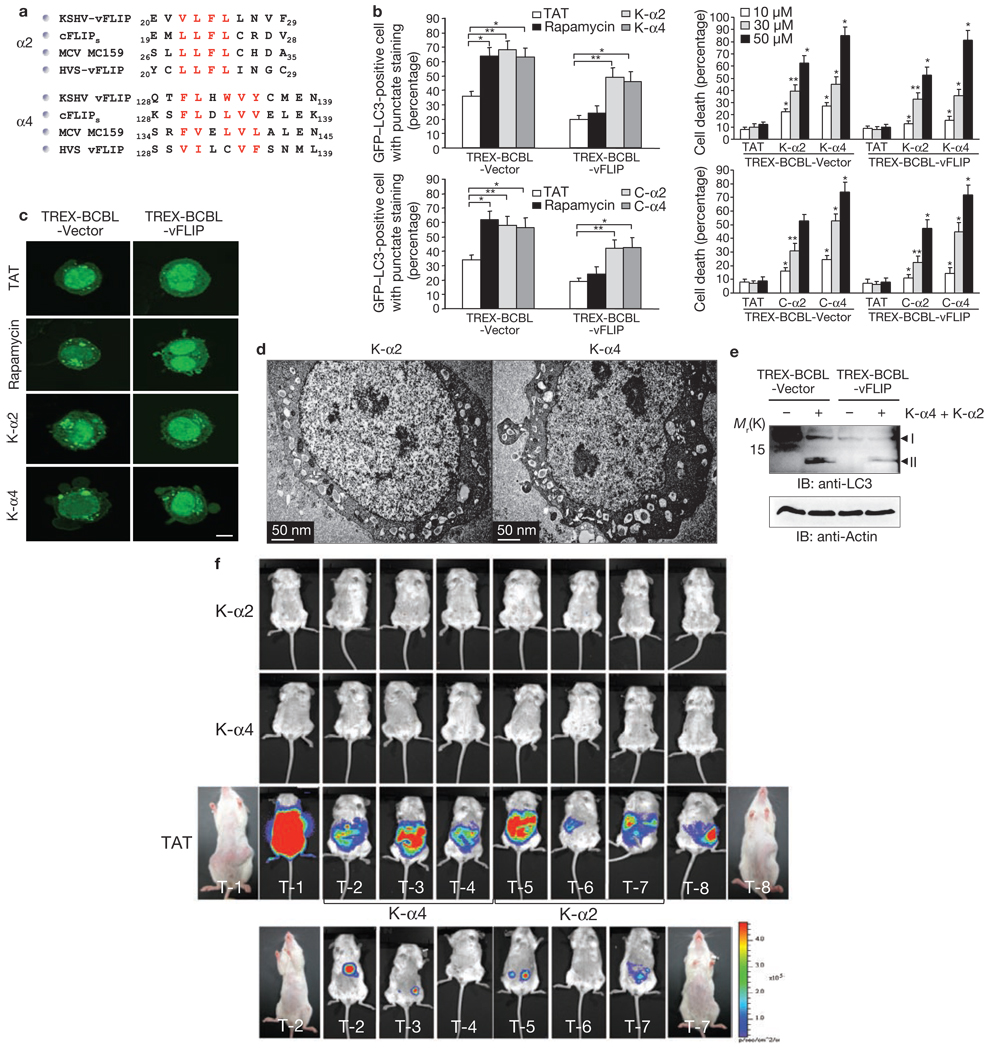
The KSHV vFLIP vFLIP α2 (amino acids 20–29) and α4 (amino acids 128–139) peptides that independently bound Atg3 at high levels were fused with the HIV-1 TAT protein transduction domain for intracellular delivery as retro-inverso versions to circumvent proteolytic degradation23–26 and tested for their potential effects on autophagy induction (Fig. 4a). We also included the mutant peptides K-α2m and K-α4m, where the hydrophobic core residues of the K-α2 (L21F22L23) and K-α4 (F130L-WVY139) peptides were replaced with alanines, as well as the C-α2 and C-α4 peptides containing the cFLIPs amino acid regions 19–28 and 128–139, respectively (Fig. 4a; Supplementary Information, Fig. S10a). This showed that either 30 or 50 µM of the K-α2, K-α4 C-α2, or C-α4 peptides were able to induce autophagy in TREX-BCBL-vector cells as effectively as rapamycin (2 µM; Fig. 4b). Furthermore, overnight incubation of BCBL1 cells with K-α2 and C-α2 peptides (30 or 50 µM) induced 50–60% cell death, and incubation with K-α4 and C-α4 peptides (30 or 50 µM) induced 75–90% cell death (Fig. 4b). In striking contrast, treatment with HIV-1 TAT only, the K-α2m, or the K-α4m mutant peptides showed no increase in autophagy or cell death (Fig. 4bc; Supplementary Information, Fig. S10bc). Electron microscopy studies showed severe autophagy in most (~80%) of the dead cells, and both autophagy and apoptosis in a small population (~10%) of dead cells or others, such as necrosis (~10%; Fig. 4d; Supplementary Information, Fig. S11). Moreover, the accelerated levels of LC3 processing and long-lived protein proteolysis were detected in K-α2- and K-α4-treated BCBL1 cells (Fig. 4e; Supplementary Information, Fig. S12).
Figure 4. FLIP α2 and α4 peptides induce cell death with autophagy.
(a) Sequence alignment of the FLIP α2 and α4 peptide sequences. The red letters correspond to hydrophobic core residues. (b) At 12–16 h post-transfection with GFP–LC3, TREX-BCBL vector and TREX-BCBL-vFLIP cells were treated with doxycycline for 24 h, followed by incubation with 30 µM of the TAT, K-α2 or K-α4 peptide (top left) or the TAT, C-α2, or C-α4 peptide (bottom left) for an additional 12 h. Subsequently, autophagy was quantified as means ± s.d. of the combined results from three independent experiments (2 µM rapamycin treatment was included as a control). TREX-BCBL vector and TREX-BCBL-vFLIP cells were treated with doxycycline for 24 h, followed by incubation with 0, 30, or 50 µM of the TAT, K-α2, or K-α4 peptide (top right) or the TAT, C-α2, or C-α4 peptide (bottom right) for an additional 12 h. Trypan blue staining was then used to determine cell death (as a percentage). Data are mean ± s.e.m.; n = 200–300; three independent experiments; *P < 0.01; **P < 0.05. (c) The details are identical to those described in b and GFP–LC3 puncta were subsequently detected using an inverted fluorescence microscope. Scale bar, 5 µm. (d) KSHV-infected BCBL1 cells were treated with 30 µM of the K-α2 or the K-α4 peptide for 12 h and subjected to scanning electron microscopy. Scale bar, 50 nm. (e) TREX-BCBL-Vector and TREX-BCBL-vFLIP cells were treated with the K-α and the K-α4 (30 µM each) peptide for 12 h, followed by immunoblotting (IB) with anti-LC3 and anti-actin antibody. (f) NOD/SCID mice received an injection of 5 × 106 BCBL1–Luc cells, followed by intraperitoneal injections with 300 µg the TAT, K-α2 or Kα-4 peptide for three weeks (top three panels). After three weeks, a group of three mice treated with the TAT peptide was challenged with 300 µg of the K-α2 or Kα-4 peptide for an additional three weeks (bottom panel, see Supplementary Information, Fig. S15). Tumours were measured by in vivo bioluminescence imaging.
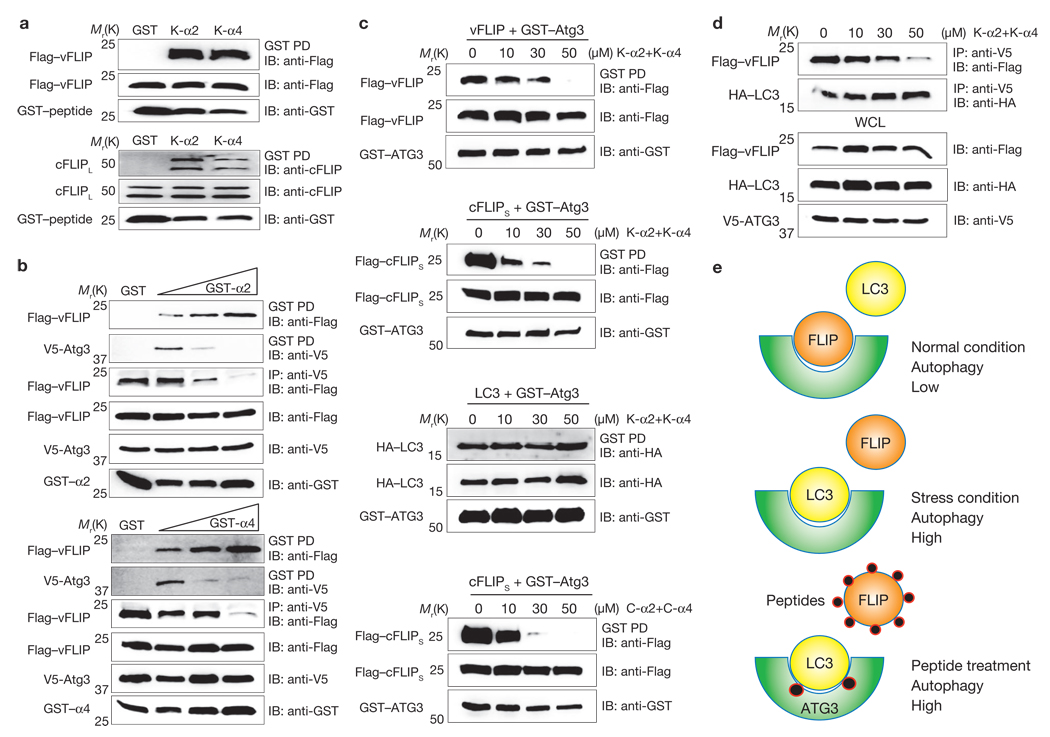
Binding studies showed that, in addition to being able to bind Atg3, the vFLIP α2 and α4 peptides efficiently interacted with the cFLIPL and vFLIP proteins (Fig. 5a). Furthermore, with co-expression of vFLIP and Atg3 alongside increasing amounts of the vFLIP peptides, the interaction between vFLIP and the K-α2/K-α4 peptides increased, whereas the interaction between Atg3 and the K-α2/K-α4 peptides seemed to decrease, indicating that the vFLIP peptides have higher binding affinities to vFLIP itself than to Atg3 (Fig. 5b). Under these conditions, Atg3–FLIP interaction, but not Atg3–LC3 interaction, was markedly reduced (Supplementary Information, Figs S13, S14). Thus, because of their high FLIP-binding affinities, the FLIP peptides may sequester FLIP to prevent Atg3–FLIP interaction, but have little or no effect on Atg3–LC3 interaction. Indeed, incubation with the α2 and/or α4 peptides markedly blocked the interactions of vFLIP, cFLIPS, or cFLIPL with Atg3 in a concentration-dependent manner without affecting Atg3–LC3 interaction (Fig. 5c, d; Supplementary Information, Figs S13, S14). On the basis of these results, we propose that the FLIP protein efficiently competes with LC3 for Atg3 binding under normal conditions, resulting in low levels of autophagy (Fig. 5e). However, under conditions of stress, FLIP dissociates from the Atg3 complex, allowing Atg3–LC3 interaction and resulting in high levels of autophagy. Finally, peptide treatment efficiently sequesters FLIP from Atg3 to facilitate Atg3–LC3 interaction, resulting in unbalanced, high levels of autophagy (Fig. 5e).
Figure 5. The vFLIP peptide binds FLIP at higher affinity and peptide binding blocks Atg3–FLIP interaction without affecting Atg3–LC3 interaction.
(a) At 16 h post-transfection with GST, GST-K-α2 or GST-K-α4 along with Flag–vFLIP (top) or Flag–cFLIPL (bottom), HEK293T cells were used for GST pulldown, followed by immunoblotting (IB) with anti-Flag or anti-FLIP antibody. Whole-cell lysates (WCLs) were used for IB with anti-GST, anti-Flag or anti-FLIP antibody. (b) At 16 h post-transfection with V5-Atg3 and Flag–vFLIP with increasing amounts of GST–K-α2 or GST–K-α4, HEK293T cells were used for GST pulldown, followed by IB with anti-Flag or anti-V5 antibody or for immunoprecipitation (IP) with anti-V5 antibody, followed by IB with anti-Flag antibody. WCLs were used for IB with anti-GST, anti-Flag or anti-V5 antibody. (c) At 12–16 h post-transfection with GST–Atg3 along with Flag–vFLIP (first), Flag–cFLIPS (second), or HA–LC3 (third) with increasing amounts of the K-α2+K-α4 peptides (top three panels) or C-α2+C-α4 peptides (bottom panel), HEK293T cells were used for GST pulldown, followed by IB with anti-Flag or anti-HA antibodies. WCLs were used for IB with anti-GST and anti-Flag antibody. (d) At 12–16 h post-transfection with Flag–vFLIP, HA–LC3 and V5-Atg3, HEK293T cells were treated with increasing amounts of the K-α2+K-α4 peptides and used for IP anti-V5 antibody, followed by IB with anti-Flag or anti-HA antibody. WCLs were used for IB with anti-V5, anti-HA and anti-Flag antibody. (e) Model of FLIP- and its peptide-mediated deregulation of autophagy.
We then used NOD/SCID xenografted mice that received an intrapenitoreal injection of BCBL1–Luc cells expressing the luciferase gene as a traceable bioluminescence reporter and evaluated these mice for the development of PELs, as previously shown27,28. After being injected with the tumour cells, the mice received intraperitoneal injections of TAT (300 µg), the K-α2 or the Kα-4 peptide on days 2, 4 and 6, and twice weekly for an additional two weeks. All of the mice that were injected with the TAT peptide developed PEL, with evident distention and ascites in the peritoneal cavity, as well as markedly increased luminescence (Fig. 4f). In contrast, the mice that received injections of the K-α2 or Kα-4 peptide never acquired lymphomas, with little or no traceable luminescence (Fig. 4f). A group of three mice that developed PEL after TAT peptide treatment was then challenged with 300 µg K-α2 or Kα-4 peptide for an additional three weeks. Treatment with the K-α2 or Kα-4 peptide led to a marked regression of tumours (Fig. 4f; Supplementary Information, Fig. S15) and subsequent autopsies showed no detectable damage in the tissues and organs of the mice injected with the K-α2 or Kα-4 peptide (data not shown). These results indicate that the FLIP-derived short peptides carry potent in vitro and in vivo activity for growth suppression and the autophagy-mediated death of tumour cells.
As seen with apoptosis, autophagy provides an important innate safeguard mechanism that protects the organism against invading pathogens and unwanted cancerous cells, and also keeps the organism healthy29,30. Although autophagy and apoptosis use fundamentally different molecular mechanisms to execute protective functions, they also seem to be highly interconnected31,32. Our study shows that, as seen with Bcl-2 (refs 11,13), cellular and viral FLIPs not only possess anti-apoptotic activity but additionally serve as anti-autophagy molecules through their inhibitory interaction with the E2-like enzyme, Atg3. This seems to suggest that, because of its importance in various biological processes, each step of the autophagy signal transduction pathway is under tight surveillance, for example, by the mTor kinase in autophagy induction33, by Bcl-2 in autophagosome generation, and by FLIP in autophagosome elongation. On the other hand, as permanent parasites, viruses have to constantly modify themselves to efficiently and comprehensively escape host immune responses. Indeed, with a large genetic capacity, γ-herpesviruses have evolved to acquire viral versions of Bcl2 and FLIP to avoid elimination by the host’s apoptotic and autophagy-mediated immune responses. Finally, the biological activity of FLIP-derived short peptides may have possible uses in autophagy-mediated clinical applications, potentially leading to a paradigm shift in targeted cancer therapy.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturecellbiology/
METHODS
Cell culture
HCT116, NIH3T3, MEF, HaCat, and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 1% penicillin-streptomycin (Gibco-BRL). Transient transfections were performed with FuGENE 6 (Roche), Lipofectamine 2000 (Invitrogen), or calcium phosphate (Clontech), according to the manufacturer’s instructions. HCT116, MEF, HaCat, and NIH3T3 stable cell lines were generated using a standard selection protocol with 2 µg ml−1 of puromycin. BCBL-1 was grown in RPMI 1640 supplemented with 10% fetal calf serum.
Plasmid construction
All constructs for transient gene expression in mammalian cells were derived from pcDNA5/FRT/TO (Invitrogen) or pEF-IRES-puro. DNA fragments corresponding to the coding sequences of the KSHV–vFLIP, human cFLIPs, human cFLIPL, MCV–MC159, and HVS–vFLIP genes were amplified by polymerase chain reaction (PCR) and subcloned into pcDNA5/FRT/TO between the AflII and BamHI restriction sites or pEF-IRES-puro between the AflII and XbaI sites and selected for stable transfectants. Flag-tagged KSHV-vFLIP 58AAA and 67AAA mutants were generated by PCR using oligonucleotide-directed mutagenesis, with the resultant PCR products then subcloned into pcDNA5/FRT/TO and pEF-IRES-puro. GST-tagged vFLIP and vFLIP mutant genes were cloned into a pcDNA5/FRT/TO derivative encoding an N-terminal GST epitope tag between the BamHI and NotI sites. GST-tagged hATG3 and hATG3 mutant genes were cloned into a pcDNA5/FRT/TO derivative encoding an N-terminal GST epitope tag between the BamHI and NotI sites. Truncated mutant constructs of KSHV–vFLIP and hATG3 were created by subcloning the PCR products of cDNA fragments containing each domain of the associated genes into pcNDA5/FRT/TO. All constructs were sequenced using an ABI PRISM 377 automatic DNA sequencer to verify 100% correspondence with the original sequence.
Yeast two-hybrid screen
Yeast transformations with a cDNA library were performed using a method described previously9. Library screening and recovery of plasmids were performed according to the manufacturer’s instructions (Clontech).
Immunoblot analysis and immunoprecipitation assay
For immunoblotting, polypeptides were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred onto a PVDF membrane (Bio-Rad). Immunodetection was achieved with anti-V5 (1:5000) (Invitrogen), anti-Flag (1:5000) (Sigma), anti-HA (1:5000), anti-GST (1:2000) (Santa Cruz Biotech), anti-tubulin (1:1000), anti-actin (1:1000) (Santa Cruz Biotech), anti-Beclin1 (1:500) (BD Bioscience), anti-cFLIP (Dave-2, NF6) (1:1000) (Alexis Biochemicals), anti-LC3 (1:1000) (Novus, Cosmo Bio) and anti-Atg3 antibody (1:500) (MBL). The proteins were visualized by a chemiluminescence reagent (Pierce) and detected by a Fuji Phosphor Imager.
For GST pulldowns, cells were collected and lysed in an NP40 buffer supplemented with complete protease inhibitor cocktail (Roche). Post-centrifugation supernatants were pre-cleared with protein A/G beads for 2 h at 4°C. Pre-cleared lysates were mixed with 50% slurry of glutathione-conjugated Sepharose beads (Amersham Biosciences) and the binding reaction incubated for 4 h at 4°C. Precipitates were then washed extensively with a lysis buffer. Proteins bound to glutathione beads were eluted by boiling them with an SDS loading buffer for 5 min.
For immunoprecipitation, cells were collected and lysed in an NP40 buffer supplemented with complete protease inhibitor cocktail (Roche). After pre-clearing with protein A/G agarose beads for 1 h at 4°C, whole-cell lysates were used for immunoprecipitation with the indicated antibodies. Generally, 1–4 µg of the commercial antibody was added to 1 ml of the cell lysate and incubated at 4°C for 8–12 h. After adding protein A/G agarose beads, incubation was continued for an additional 1 h. The immunoprecipitates were then washed extensively with a lysis buffer and eluted by boiling them with an SDS loading buffer for 5 min.
Autophagy analyses
For starvation studies, cells were washed three times with PBS and incubated in Hank’s solution (Invitrogen) for 2–4 h at 37°C. Cells were treated with Hank’s solution or with complete medium or serum-reduced medium containing 2 µM rapamycin (Sigma). Autophagy was assessed by GFP–LC3 redistribution, LC3 mobility shifts, and electron microcopy. For GFP–LC3 redistribution, HCT116, HaCat, MEF, NIH3T3, MCF-7 and BCBL1 cells were transfected with a GFP–LC3 expression plasmid. At 12–16 h post-transfection, GFP–LC3 was detected under normal, starvation or rapamycin-treatment conditions using an inverted fluorescence microscope. The percentage of GFP–LC3-positive cells with punctate staining was determined in three independent experiments. To quantify GFP–LC3-positive autophagosomes per transfected cell, six random fields representing 200–300 cells were counted. For the LC3 mobility shift assay, the cells were treated on ice for 30 min, lysed with 1% Triton X-100, and subjected to immunoblot analysis with an anti-LC3 antibody.
Small interfering RNA (siRNA) suppression of gene expression
All siRNAs were produced from Dharmacon Research. siRNAs specific to Beclin1 (5´AAGAUCCUGGACCGUGUCACC3´), cFLIP (5´GGAGCAGGGACAAGUUACA3´), caspase-8 (5´GAUCGAGGAUUAUGAAAGA3´) and a nonspecific, scrambled control siRNA were transfected using DharmaFect reagent (Dharmacon), according to the manufacturer’s instructions. At 48–72 h post-transfection, cells were analysed for autophagy.
Electron microscopy imaging
Cell pellets were fixed using a 0.1 M phosphate buffer (pH 7.4) containing 2% paraformaldehyde and 0.1% gluteraldehyde at room temperature for 1 h. The cell pellets were then post-fixed on ice for 2 h with 1% osmium tetroxide and rinsed three times with distilled water. The fixed cell pellets were dehydrated by an ethanol (EtOH) dilution series of up to 100% EtOH and then immersed in propylene oxide (PO) for 2 min, with the cycle performed three times. The pellets were then infiltrated in a 3:1 PO/eponate resin mixture overnight, embedded in 100% eponate resin (Ted Pell) in beam capsules, and allowed to harden overnight in a 65°C oven. After hardening, tissue blocks were sectioned to a thickness of 70 nm and placed on 300 mesh copper grids. The grids were then counter-stained with saturated uranyl acetate and lead citrate, and then viewed through a Zeiss EM 10 electron microscope (Zeiss). In order to assess different cell death levels, we counted the cells showing the features of autophagic death (the ultrastructures of cytoplasmic autophagy as double-membraned vacuoles with the intact nucleus), apoptotic death (the chromatin condensation and membrane blebbing with the intact cytoplasm), apoptosis and autophagic death (both features of apoptosis and autophagy), and others (necrosis and death with no specific features).
Cell cycle and apoptosis assays
5 × 106 cells were treated with the peptides, starvation conditions, rapamycin, cycloheximide (CHX), or CHX + TNFα for the indicated periods of time. After treatment, the cells were collected and fixed with 70% ethanol overnight. After staining the cells with propidium iodide for 30 min at room temperature, DNA content was measured by flow cytometry and analysed using Flow Jo. The cells were also stained with a FITC-conjugated annexin V antibody, followed by flow cytometry analysis with Flow Jo.
Cell proliferation and viability assays
BCBL1 cells were treated with the peptides, rapamycin for 12 h and their viability measured by trypan blue exclusion, followed by analyses using the Beckman Coulter Z2 particle count and size analyzer (BC Z2 CS Analyzer). A minimum of 100 cells per sample using triplicate samples was counted per condition per experiment.
Luciferase reporter assays
HEK 293T cells grown in six-well plates were transfected with 0.5 µg of the FLIP expression plasmids along with a NF-κB luciferase reporter construct (0.5 µg) and a constitutive β-gal expression construct (pGK-β-gal, 0.1 µg). At 1 day post-transfection, luciferase assays were performed in accordance with the manufacturer’s (Promega) protocols.
In vivo bioimaging
5 × 106 BCBL1-luciferase cells were injected intraperitoneally on day 0 of the experiment. The mice (n = 8) then received intraperitoneal injections on days 2, 4, 6, and twice weekly for an additional two weeks with the TAT, K-α2, or K-α4 peptide. A group of three mice that developed PEL after TAT peptide treatment was also challenged with 300 µg K-α2 or Kα-4 peptide for an additional three weeks. Mice were examined via in vivo imaging once a week thereafter. d-luciferin firefly potassium salt was purchased from Xenogen, a 30 mg ml−1 stock in PBS being filtered through 0.22-µm filters before use. d-luciferin (50 µl; 75 mg kg−1 body weight) was injected retro-orbitally and the mice imaged 1 min later using an IVIS imaging system (Xenogen). Data were acquired for 1 min. Gray-scale photographic images and bioluminescence colour images were superimposed using The Living Image, version 2.20 software overlay (Xenogen). Data were analysed with Igor Pro image analysis software (WaveMetrics). A region of interest (ROI) was manually selected over signal intensity and the area of the ROI was kept constant. Data are presented as average radiance (photons/s−1·cm−1sr−1 [steradian]) within the ROI. Finally, the mice were humanely killed by CO2 inhalation immediately after the development of PEL, as determined by a weight gain of greater than 10% total body mass in a 1-week period.
Supplementary Material
ACKNOWLEDGEMENTS
This work was partly supported by US. Public Health Service grants CA82057, CA91819, CA31363, CA115284, AI073099, RR00168, Hastings Foundation, Korean GRL Program K20815000001 (J.J.), AI083841, CA140964, the Lymphoma and Leukemia Society of USA, the Wright Foundation, and the Baxter Foundation (C.L.). We thank Don Ganem, Noboru Mizushima, Preet Chaudhary and Jeff Cohen for reagents, Ernesto Barron for EM analysis, and Steven Lee for manuscript preparation. Finally, we thank all of J.J.’s lab members for their discussions.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
J.S.L. performed all aspects of this study; Q.L. performed the initial study; J.Y.L, S.H.L, H.J, and H.R.L assisted with the experimental design and in collecting the data; F.C.Z and S.J.G. provided KSHV ΔvFLIP; C.L. assisted with the experimental design and interpretation; J.S.L. and J.J. organized this study and wrote the paper. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Sem. Cell Dev. Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Thome M, Tschopp J. Regulation of lymphocyte proliferation and death by FLIP. Nature Rev. Immunol. 2001;1:50–58. doi: 10.1038/35095508. [DOI] [PubMed] [Google Scholar]
- 6.Li FY, Jeffrey PD, Yu JW, Shi Y. Crystal structure of a viral FLIP. J. Biol. Chem. 2006;281:2960–2968. doi: 10.1074/jbc.M511074200. [DOI] [PubMed] [Google Scholar]
- 7.Izzedine H, Brocheriou I, Frances C. Post-transplantation proteinuria and sirolimus. N. Engl. J. Med. 2005;353:2088–2089. doi: 10.1056/NEJM200511103531922. [DOI] [PubMed] [Google Scholar]
- 8.Sin SH, et al. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines in vivo by inhibiting autocrine signaling. Blood. 2007;109:2165–2173. doi: 10.1182/blood-2006-06-028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 10.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;13:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Ku B, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell. Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 13.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura H, et al. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 2003;77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guasparri I, Wu H, Cesarman E. The KSHV oncoprotein vFLIP contains a TRAF-interacting motif and requires TRAF2 and TRAF3 for signalling. EMBO Rep. 2006;7:114–119. doi: 10.1038/sj.embor.7400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matta H, Chaudhary PM. Activation of alternative NF-κB pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein (vFLIP) Proc. Natl Acad. Sci. USA. 2004;101:9399–9404. doi: 10.1073/pnas.0308016101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matta H, Mazzacurati L, Schamus S, Yang T, Sun Q, Chaudhary PM. Kaposi’s sarcoma-associated herpesvirus (KSHV) oncoprotein K13 bypasses TRAFs and directly interacts with the IκB kinase complex to selectively activate NF-κB without JNK activation. J. Biol. Chem. 2007;24:24858–24865. doi: 10.1074/jbc.M700118200. [DOI] [PubMed] [Google Scholar]
- 19.Yang JK, et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol. Cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada Y, et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- 21.Bagneris C, et al. Crystal structure of a vFlip-IKKγ complex: insights into viral activation of the IKK signalosome. Mol. Cell. 2008;30:620–631. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Ye FC, et al. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-κB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 2008;82:4235–4249. doi: 10.1128/JVI.02370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gump JM, Dowdy SF. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol. Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Snyder EL, Meade BR, Saenz CC, Dowdy SF. Treatment of terminal peritoneal carcinomatosis by a transducible p53-activating peptide. PLos Biol. 2004;2:186–193. doi: 10.1371/journal.pbio.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 26.Hotchkiss RS, et al. TAT-BH4 and TAT-Bcl-xL peptides protect against sepsis-induced lymphocyte apoptosis in vivo. J. Immunol. 2006;176:5471–5477. doi: 10.4049/jimmunol.176.9.5471. [DOI] [PubMed] [Google Scholar]
- 27.Wu W, Rochford R, Toomey L, Harrington W, Jr, Feuer G. Inhibition of HHV-8/KSHV infected primary effusion lymphomas in NOD/SCID mice by azidothymidine and interferon-alpha. Leukemia Res. 2005;29:545–555. doi: 10.1016/j.leukres.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Keller SA, et al. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood. 2006;107:3295–3302. doi: 10.1182/blood-2005-07-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microb. 2009;18:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 32.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubinztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nature Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.