The crystal structure of human glycolate oxidase in complex with an inhibitor is described. A comparison with complexes with other inhibitors and with structures of homologous enzymes is given.
Keywords: glycolate oxidase; 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole; peroxisomal flavoenzymes; hyperoxaluria
Abstract
Glycolate oxidase, a peroxisomal flavoenzyme, generates glyoxylate at the expense of oxygen. When the normal metabolism of glyoxylate is impaired by the mutations that are responsible for the genetic diseases hyperoxaluria types 1 and 2, glyoxylate yields oxalate, which forms insoluble calcium deposits, particularly in the kidneys. Glycolate oxidase could thus be an interesting therapeutic target. The crystal structure of human glycolate oxidase (hGOX) in complex with 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole (CCPST) has been determined at 2.8 Å resolution. The inhibitor heteroatoms interact with five active-site residues that have been implicated in catalysis in homologous flavodehydrogenases of l-2-hydroxy acids. In addition, the chlorophenyl substituent is surrounded by nonconserved hydrophobic residues. The present study highlights the role of mobility in ligand binding by glycolate oxidase. In addition, it pinpoints several structural differences between members of the highly conserved family of flavodehydrogenases of l-2-hydroxy acids.
1. Introduction
Glycolate oxidase (EC 1.1.3.15; GOX), an FMN-dependent enzyme in peroxisomes, oxidizes glycolate to glyoxylate at the expense of oxygen, with the production of hydrogen peroxide. It exists in both plants and higher organisms. In plants, it is involved in photorespiration by oxidizing glycolate derived from the phosphoglycolate generated by the oxygenase reaction of Rubisco. Net photosynthesis is reduced owing to this pathway of carbon metabolism (Miziorko & Lorimer, 1983 ▶). In the peroxisomes of higher organisms the glycolate oxidase product glyoxylate is also converted to glycine by alanine-glyoxylate aminotransferase (AGT). Part of it can be reduced back to glycolate by glyoxylate/hydroxypyruvate reductase (GR/HPR) or oxidized to oxalate by NAD+-dependent lactate dehydrogenase in the cytosol. Human genetic deficiencies of AGT or GR/HPR lead to hyperoxaluria types 1 and 2, respectively. These severe diseases are particularly characterized by the pathological formation of kidney stones owing to the deposition of insoluble calcium oxalate (Danpure, 2001 ▶). Thus, glycolate oxidase may be an interesting target for inhibitors in both plants and animals.
GOX belongs to the evolutionary family of l-2-hydroxy acid dehydrogenases, which includes oxidases (Ghisla & Massey, 1991 ▶; Lindqvist, 1992 ▶; Maeda-Yorita et al., 1995 ▶) and electron transferases (Illias et al., 1998 ▶; Lederer, 1991 ▶; Lehoux & Mitra, 1999 ▶). In the latter enzymes the flavin is reoxidized by monoelectronic acceptors instead of oxygen. All family members adopt the β8α8-barrel fold with a highly conserved active site (Cunane et al., 2005 ▶; Leiros et al., 2006 ▶; Lindqvist, 1989 ▶; Lindqvist et al., 1991 ▶; Sukumar et al., 2004 ▶; Umena et al., 2006 ▶; Xia & Mathews, 1990 ▶). The common mechanism for substrate oxidation is generally believed to proceed by way of a carbanion intermediate generated by proton abstraction from the carboxylate α-carbon by an invariant histidine (Dewanti & Mitra, 2003 ▶; Gaume et al., 1995 ▶; Ghisla & Massey, 1980 ▶; Gondry et al., 2001 ▶; Lederer, 1991 ▶; Lederer et al., 2005 ▶; Rao & Lederer, 1998 ▶; Sobrado et al., 2001 ▶). The best known plant enzyme is the spinach enzyme (sGOX; Lindqvist, 1992 ▶). Wild-type and mutant forms have been characterized both at the enzymatic and structural levels (Lindqvist, 1989 ▶; Macheroux et al., 1991 ▶, 1992 ▶, 1993 ▶; Stenberg et al., 1995 ▶; Stenberg & Lindqvist, 1996 ▶, 1997 ▶). The enzymatic properties of the pig liver enzyme have also been studied in detail (Schuman & Massey, 1971a ▶,b ▶). The human enzyme (hGOX) was first purified in 1979 (Fry & Richardson, 1979 ▶; Schwam et al., 1979 ▶). Its gene was independently cloned by two groups (Jones et al., 2000 ▶; Williams et al., 2000 ▶) and an initial characterization of the recombinant enzyme has recently been described (Vignaud et al., 2007 ▶).
Several series of compounds designed as inhibitors of glycolate oxidase have been synthesized (Randall et al., 1979 ▶; Rooney et al., 1983 ▶; Williams et al., 1983 ▶). Stenberg & Lindqvist (1997 ▶) determined the crystal structure of sGOX in complex with two inhibitors, one of which was obtained from one of these series. In the present work, we describe the crystal structure of hGOX in complex with another inhibitor, which was identified by screening a chemical library. While this work was in progress, an independent structure of hGOX in complex with glyoxylate was deposited in the PDB (PDB code 2nzl; E. Ugochukwu, K. Kavanagh, E. Pilka, G. Berridge, J. É. Debreczeni, E. Papagrigoriou, A. Turnbull, F. Niesen, O. Gileadi, F. Von Delft, M. Sundstrom, C. H. Arrowsmith, J. Weigelt, A. Edwards & U. Oppermann, unpublished work) and the structure of hGOX in complex with glyoxylate and two other ligands was independently published (Murray et al., 2008 ▶).
2. Materials and methods
2.1. Protein preparation
hGOX, which was expressed in Escherichia coli using the plasmid described in Williams et al. (2000 ▶), was purified and the tag was cleaved following the procedures described in Vignaud et al. (2007 ▶). In the resulting protein, the N-terminal methionine is preceded by the four-residue sequence SAVK–. The discovery of the inhibitor 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole (CCPST) by screening a chemical library will be described elsewhere. Its IC50 was 1 µM.
2.2. Crystallization and data collection
Prior to crystallization, the enzyme was filtered through a PD10 26/10 desalting column in order to replace the phosphate buffer from the last purification step with 100 mM HEPES buffer pH 7.5. The protein was then concentrated to 10 mg ml−1. Its final concentration was adjusted to 7 mg ml−1 after adding FMN and the inhibitor in molar excesses of 1 and 10, respectively.
Crystallization was performed by the hanging-drop vapour-diffusion method using a precipitant solution containing 200 mM NaCl, 100 mM MMT (malic acid, MES and Tris) buffer pH 7.5 and 15%(w/v) polyethylene glycol 1000. Drops containing 2 µl protein mixture were mixed with 1 µl precipitant solution.
All data were collected to a resolution of 2.8 Å in a nitrogen stream at 110 K on beamline ID14-3 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. The crystal was cryoprotected in 25% glycerol and flash-frozen directly in the nitrogen stream. Data were processed and scaled using XDS (Kabsch, 1993 ▶).
2.3. Structure solution by molecular replacement
The structure of hGOX in complex with a glyoxylic acid ligand has recently been deposited in the PDB (PDB code 2nzl). This structure was used as a search model. Nevertheless, in order not to bias the result of molecular replacement, the ligand and FMN were removed from the search model. Molecular replacement was used as implemented in the program MOLREP (Murshudov et al., 1997 ▶), which gave a clear solution with four subunits in the asymmetric unit in space group P4212.
The model was improved by manual inspection and rebuilding using Coot (Emsley & Cowtan, 2004 ▶; Lohkamp & Emsley, 2005 ▶) and was refined using restrained refinement in REFMAC5 (Collaborative Computational Project, Number 4, 1994 ▶; Murshudov et al., 1997 ▶). NCS restraints were applied, except in the final round of refinement. FMN and inhibitor molecules were added manually using both 2F o − F c and F o − F c maps. All refinement excluded 5% of the reflections, which were set aside for R free calculations. The inhibitor library for REFMAC5 was created using the SKETCHER program from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶).
2.4. Structure analysis
The quality of the refined structure was checked with Coot (Emsley & Cowtan, 2004 ▶) and PROCHECK (Laskowski et al., 1993 ▶). The refined parameters are summarized in Table 1 ▶. Superposition of the Cα traces and calculation of the root-mean-square deviations were performed with the MSDFOLD server (Krissinel & Henrick, 2004 ▶).
Table 1. Data-collection and refinement summary.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P4212 |
| Unit-cell parameters () | a = b = 138.7, c = 186.8 |
| No. of molecules in the ASU | 4 |
| Resolution () | 2.8 |
| R merge | 9.3 (36) |
| I/(I) | 17.32 (3.56) |
| Completeness | 99.6 (98.2) |
| Refinement | |
| Refinement program | REFMAC5 |
| Reflections in the working set | 41393 |
| Reflections in the test set | 2179 |
| R factor (%) | 20.8 |
| R free (%) | 25.6 |
| No. of atoms modelled | 10565 |
| No. of waters | 33 |
| Average B factor (2) | 38.1 |
| Average B factor, solvent (2) | 25 |
| R.m.s.d. from ideal | |
| Bond lengths () | 0.009 |
| Angles () | 1.18 |
| Ramachandran plot | |
| Most favoured (%) | 91.6 |
| Additionally allowed (%) | 8.1 |
| Generously allowed (%) | 0.3 |
| Disallowed (%) | 0 |
All the protein representations in the figures were drawn using PyMOL (DeLano, 2002 ▶). The structure was deposited in the Protein Data Bank with code 2w0u.
3. Results
3.1. Quality of the structure
The final hGOX model at 2.8 Å resolution consists of four peptide chains refined to an R factor of 21% and an R free of 26% (Table 1 ▶). The electron density corresponding to residues 173–206 is particularly weak or absent in all monomers. In this region, the average temperature factor for built residues in all subunits is 66 Å2, which can be compared with average B factors of 38 Å2 for the rest of the four peptide chains. Chain A contains residues 5–178 and 204–362, chain B contains residues 5–172 and 204–363, chain C contains residues 5–172 and 207–363 and chain D contains residues 4–175 and 205–363. In all chains, interpretable electron densities were present for one FMN molecule as well as one CCPST molecule. The structural comparison between each pair of hGOX monomers shows an r.m.s. deviation of about 0.1 Å for all Cα atoms.
3.2. Quaternary structure
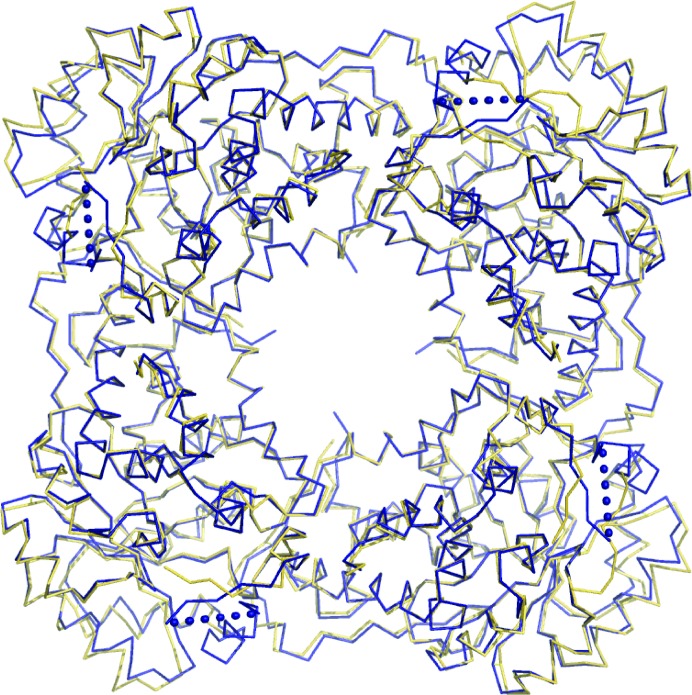
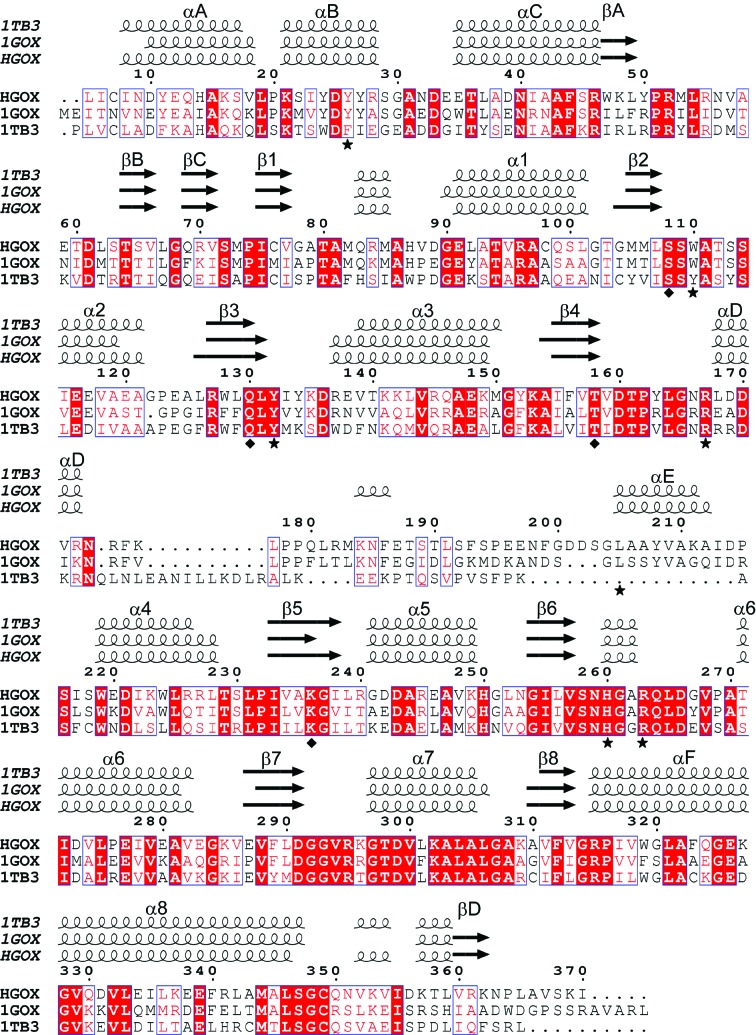
The asymmetric unit contains four monomers, with FMN and the inhibitor bound at the C-terminal end of a β8α8-barrel (Fig. 1 ▶). Superposition of the Cα trace of an individual hGOX subunit with one subunit of the model used for structure solution matches 335 positions with an r.m.s. deviation of less than 0.5 Å. Superposition with an sGOX subunit, with a sequence identity of 57%, matches 325 residues with an r.m.s. deviation of less than 1 Å (PDB code 1gox; Lindqvist, 1989 ▶). A similar superposition with the structure of a rat kidney LCHAO subunit (PDB code 1tb3; Cunane et al., 2005 ▶) also displays high structural similarity, with an r.m.s. deviation of less than 1 Å and a sequence identity of 44% [LCHAO is classified as an isozyme (isozyme B) of glycolate oxidase; Fig. 2 ▶].
Figure 1.
Cartoon representation of the hGOX asymmetric unit showing, from left to right, chains A, B, C and D. The helices are in blue and the β-strands are in orange. FMN and inhibitor are in ball-and-stick representation with atom-type colouring.
Figure 2.
Superposition of the Cα trace of the deduced oligomer structure of hGOX (blue ribbon) with the asymmetric unit of rat kidney LCHAO (yellow ribbon; PDB code 1tb3; Cunane et al., 2005 ▶). Dotted lines schematically represent the disordered loops in hGOX.
In the present crystal form the four subunits in the asymmetric unit are arranged as pearls on a string (Fig. 1 ▶). This arrangement clearly does not correspond to the functional molecule since hGOX is active as a tetramer in solution (Vignaud et al., 2007 ▶). Indeed, each subunit in the asymmetric unit and its respective crystallographic symmetry mates form a tetramer with C4 symmetry, which is the minimum biological entity expected from the structures of other family members. Superposition of the Cα traces of these C4 tetramers with those from sGOX gives an r.m.s. deviation of 1.20 Å over 1291 residues. Similarly, in the comparison with rat kidney LCHAO the r.m.s. deviation is lower than 1 Å over 1256 residues (Fig. 2 ▶). Each subunit in the tetramer buries a surface of about 1460 Å2, which can be compared with the 1375 Å2 of buried surface determined for the LCHAO tetramer.
3.3. Flavin interactions with the protein
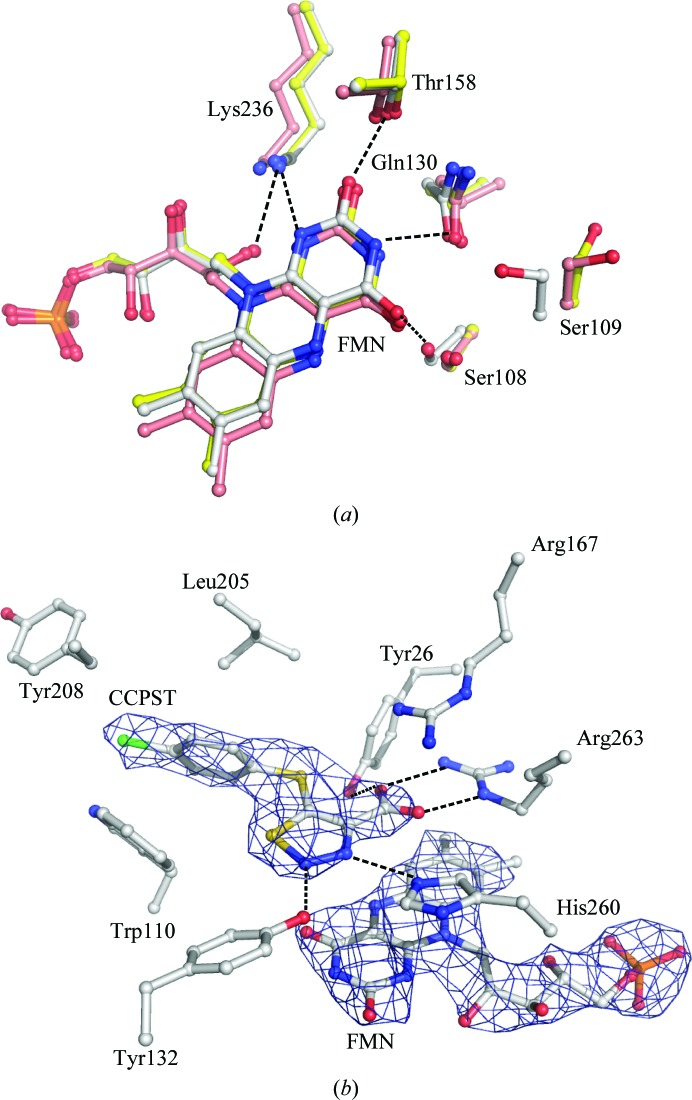
In the present structure, the flavin heteroatoms are involved in hydrogen bonds to loop residues at the C-terminal end of the barrel (Fig. 3 ▶ a). The invariant Lys236 lies within hydrogen-bonding distance of FMN N1, O2 and the ribityl 2′-OH. Hydrogen bonds are also formed between the side chain of Thr158 and FMN O2, the side chain of Gln130 and FMN N3, and the side chain of Ser108 and FMN O4. These residues are invariant in all the family members characterized to date. In addition, an extended piece of peptide chain (residues 79–81, not shown) lies behind the central isoalloxazine ring; the peptide carbonyl of Ala79 and the peptide amide of Ala81 form hydrogen bonds to the ribityl 2′-OH and FMN N5, respectively, thus blocking access to the flavin re side. These interactions are conserved in all the structures of family members, with a few exceptions that will be examined in §4.
Figure 3.
(a) Superposition of the flavins and flavin-binding residues of hGOX in complex with CCPST (white) and with glyoxylate (PDB code 2nzl; yellow) and of LCHAO (PDB code 1tb3; pink). The figure results from the superposition of the Cα traces of the three proteins. The numbering is based on the hGOX sequence. Dashed lines indicate hydrogen bonds. (b) hGOX active site showing the electron density (a 2F o − F c composite OMIT map at the 2σ level) around a stick representation of FMN and CCPST. Amino acids are also represented as sticks. The atoms are coloured white for carbon, red for oxygen, blue for nitrogen, yellow for sulfur and green for chloride.
Finally, Fig. 3 ▶(a) shows that in the CCPST complex the Ser109 hydroxyl, which does not interact with the ligand, points toward the flavin, while it is oriented at nearly 180° in the complex with glyoxylate. To validate this observation, the conformation of Ser109 was checked in the OMIT map (data not shown). This residue, which is adjacent to the invariant Ser108, can also be a threonine or an alanine in homologous enzymes. In all structures examined its side chain points away from the flavin, except in the present hGOX–CCPST structure and two sGOX structures, one without any ligand and one in complex with TACA. In these three structures, the Ser109 (Ser107 in sGOX) hydroxyl forms a hydrogen bond to the side-chain amide carbonyl of Gln130 (Gln127 in sGOX). In the other GOX structures the serine hydroxyl interacts with the peptide carbonyl of this same glutamine, which has not changed position. The reason for this orientation change between the complexes is unclear.
3.4. Inhibitor-binding site
In all monomers, an interpretable electron density was obtained that fits the 4-carboxy-5-(4-chlorophenyl)sulfanyl-1,2,3-thiadiazole (CCPST) structure (Fig. 3 ▶ b). The inhibitor is stabilized in the catalytic site by a network of hydrogen bonds, as illustrated in Fig. 3 ▶(b). Thus, in addition to an electrostatic interaction with the guanidinium group of the invariant Arg167, its carboxylate is engaged in hydrogen bonds to Arg263. The N atom at position 2 of the 1,2,3-thiadiazole ring forms a hydrogen bond to the hydroxyl group of Tyr132. The other ring N atom, at position 3, lies 3.0 Å away from N∊ of the catalytic base His260, in a correct orientation for a hydrogen bond (see below). The chlorophenyl ring forms a stacking interaction with Trp110 and is protected on the other side by Leu205. Access to the binding cavity is restricted but not completely prevented by Tyr208.
4. Discussion
4.1. Comparison of the overall protein structure with those of other family members: structure of loop 4
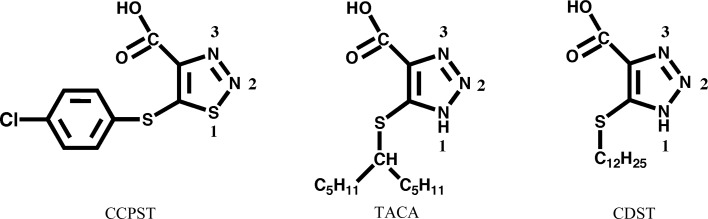
Not unexpectedly, superposition of the peptide chain of the CCPST–hGOX complex gives an excellent match with other hGOX structures: the structure of hGOX in complex with glyoxylate which was used for molecular replacement (PDB code 2nzl) and three other recent hGOX structures in complex with sulfate, glyoxylate and CDST, an inhibitor with general similarity to CCPST and TACA (Fig. 4 ▶; PDB codes 2rdw, 2rdu and 2rdt, respectively; Murray et al., 2008 ▶). The match is somewhat lower for other homologues such as Fcb2 (PDB code 1fcb; Xia & Mathews, 1990 ▶), mandelate dehydrogenase from Pseudomonas putida (MDH; PDB code 1p4c; Sukumar et al., 2004 ▶) and lactate oxidase from Aerococcus viridans (LOX; PDB codes 2j6x, 2du2 and 2e77; Leiros et al., 2006 ▶; Umena et al., 2006 ▶; Li et al., 2007 ▶), which present lower sequence identity to hGOX than sGOX and LCHAO.
Figure 4.
The chemical structures of CCPST, CDST and TACA.
The main difference between family members in the peptide-chain course lies in the region between barrel strand β4 and helix α4, commonly called loop 4, which is poorly conserved in terms of length and sequence, as exemplified by the sequence alignment in Fig. 5 ▶. In most structures of the homologous FMN-dependent l-2-hydroxy acid oxidases, segments of length between a few and two dozen residues within loop 4 are completely disordered. The few exceptions are the LOX structures (Leiros et al., 2006 ▶; Li et al., 2007 ▶; Umena et al., 2006 ▶) and the more recent structure of hGOX in complex with sulfate and with glyoxylate (Murray et al., 2008 ▶). In the high-resolution structure of MDH loop 4 is also entirely visible but is a part of sGOX loop 4 that was grafted onto the protein instead of the original loop (Sukumar et al., 2004 ▶). In these cases, loop 4 is seen to occlude access to the active site. However, in contrast to the hGOX–glyoxylate complexes, in the structure of the complex with CDST the density for segment 175–204 is missing. The authors ascribed the mobility of the loop to the necessity of making room for the hydrophobic substituent of the inhibitor (Murray et al., 2008 ▶). In the present crystal structure the minimal missing fragment encompasses residues 179–203 in chain A. The amino-terminal end at Pro179 in this chain is approximately 16 Å away from the carboxy-terminus of Ser203, which would allow the missing part to coil over the nearby protein surface. The role of loop 4 in catalysis is unclear. Replacing the original MDH loop residues 177–215, which anchor the protein to the membrane, by sGOX residues 176–195 rendered the enzyme soluble but hardly altered its catalytic properties (Xu & Mitra, 1999 ▶). In contrast, a single proteolytic cleavage in the corresponding loop in Fcb2 decreased the specific activity by two thirds and modified the affinity of a number of ligands (Ghrir & Lederer, 1981 ▶). With LCHAO, a three-residue insertion at the edge of the invisible part altered the pK a of the flavin N3—H position without seriously affecting the catalytic parameters (Belmouden & Lederer, 1996 ▶).
Figure 5.
Structure-based sequence alignment performed by the MSDFOLD server including hGOX, sGOX (PDB code 1gox; Lindqvist, 1989 ▶) and rat kidney LCHAO (PDB code 1tb3). Invariant residues are coloured white on a red background and conservative substitutions are coloured red on a white background. The numbering is that of hGOX. The secondary-structure elements derived from each crystal structure are drawn above the alignment. The helices are represented as spirals and β-strands are represented as arrows. The hGOX residues involved in the interaction with the inhibitor are marked by stars. The residues involved in the interaction with the flavin ring are marked by diamonds. This figure was generated using ESPript (Gouet et al., 2003 ▶).
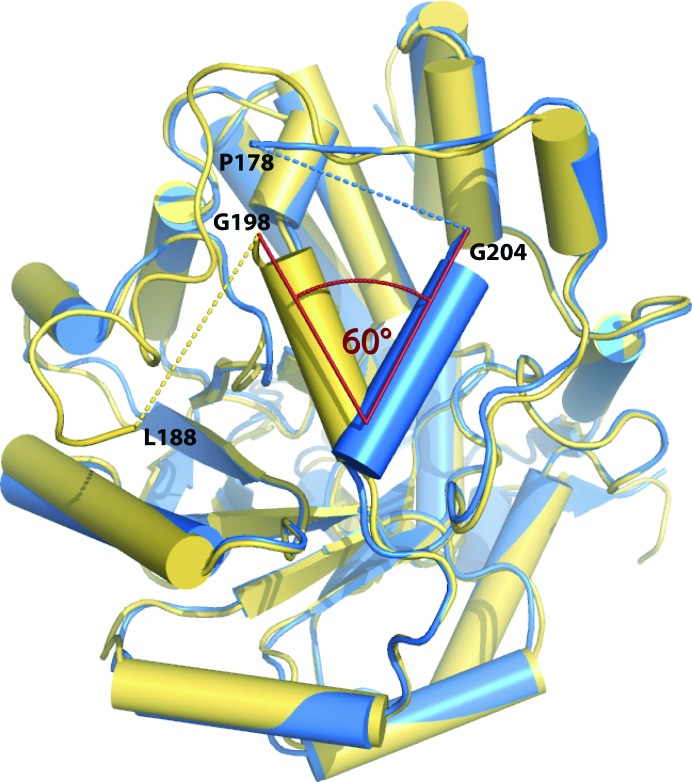
As previously remarked by several authors, the visible parts of loop 4 differ in conformation among the various structures (Cunane et al., 2002 ▶, 2005 ▶; Murray et al., 2008 ▶; Sukumar et al., 2004 ▶). Interestingly, the segment 198–206 is an α-helix in sGOX (αE, according to the nomenclature of Lindqvist, 1989 ▶), as is the corresponding segment in hGOX (203–211; Fig. 5 ▶). However, these helices do not structurally superimpose but point in two different directions and form an angle of about 60° (Fig. 6 ▶), as already noted for other hGOX complexes (Murray et al., 2008 ▶). With the helix orientation in the spinach enzyme, the cavity of the empty active site is open and accessible to solvent. However, in the present structure of hGOX in complex with CCPST, the α-helix 203–211 limits entry to the active site. This helix is perfectly superimposable onto the corresponding helix in the other recently published hGOX structures, whatever the ligand and whether disorder is present or absent in loop 4 (Murray et al., 2008 ▶; PDB entry 2nzl). An equivalent of helix αE with a slightly variable length exists in the crystal structures of Fcb2 and LOX. Whatever the active-site occupancies in several structures, the Fcb2 helix αE is in the same orientation as in sGOX, while that of LOX has the same orientation as in hGOX. In LCHAO the corresponding segment is disordered. The grafted region in MDH has yet another orientation. Overall, it would thus appear that the orientation of the αE helix may depend more on interactions with structural elements that remain to be defined rather than on the presence or absence of ligands in the active sites.
Figure 6.
Superposition of the Cα trace of an hGOX subunit (blue) with the structure of sGOX (yellow; PDB code 1gox). The disordered parts are indicated with dashed lines. The angle formed between the α-helices αE (198–206 in sGOX and 203–211 in hGOX) is indicated.
4.2. Flavin binding
The hydrogen-bonding pattern of the flavin to the protein described above is conserved in most homologous enzymes, with a few exceptions. In all the hGOX structures determined to date (Murray et al., 2008 ▶, PDB entry 2nzl and the present work), the distance between N5 and the Ala91 peptide amide lies between 2.9 and 3.2 Å, suggesting the existence of a hydrogen bond as mentioned above. In these cases FMN O4 is at the correct distance to form a hydrogen bond to the Ser108 side chain. However, in the crystal structure of the spinach enzyme in the absence of ligand (PDB code 1gox; Lindqvist, 1989 ▶), a different orientation of the isoalloxazine ring plane is observed. In this case, while the interactions of the ribityl 2′-OH, of N1 and of O2 are identical to those in all other structures (Fig. 3 ▶ a), FMN N5 lies 5.2 Å away from the backbone N atom of Ala79 (Ala81 in hGOX), which rules out a hydrogen bond. In addition, FMN O4 interacts with Tyr129 (Tyr132 in hGOX) instead of Ser106 (Ser108 in hGOX) and the latter interacts with a water molecule on the re side of the flavin. It was proposed that this water molecule mimics the oxygen molecule which reoxidizes the reduced flavin during the catalytic cycle (Lindqvist et al., 1991 ▶). Subsequent structures have suggested that binding of the substrate, or a ligand, would suppress the pocket owing to a movement of the flavin towards the backbone (Stenberg & Lindqvist, 1997 ▶). This hypothesis has not yet been verified in so far as no structure yet exists for comparison with another unliganded oxidase of the same family. Interestingly, these changes in the FMN hydrogen-bonding network do not seem to be induced or accompanied by a major structural reorganization of the protein backbone.
4.3. Comparison of glycolate oxidase–inhibitor complexes
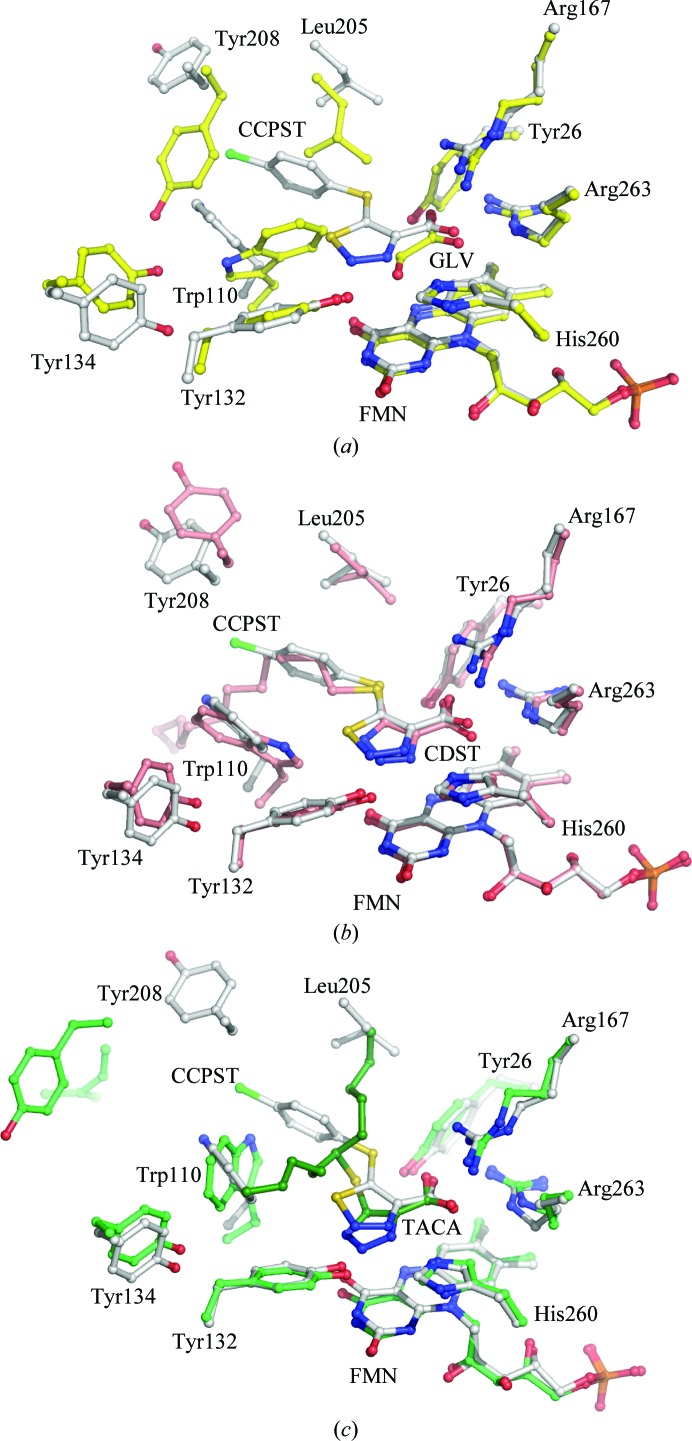
The carboxylate groups of glyoxylate, CCPST and CDST in hGOX and of TACA in sGOX superpose very well, as illustrated in Fig. 7 ▶(a) for CCPST and glyoxylate, Fig. 7 ▶(b) for CCPST and CDST and Fig. 7 ▶(c) for CCPST and TACA. They all interact with the same invariant active-site residues Arg167, Arg263 and Tyr26 (hGOX numbering) as highlighted in Fig. 3 ▶(b).
Figure 7.
(a) Superposition of the Cα trace of the hGOX active site in complex with CCPST (white) and with glyoxylate (GLV; yellow; PDB code 2nzl; Murray et al., 2008 ▶). (b) Superposition of the Cα trace of the hGOX active site in complex with CCPST (white) and with CDST (pink; PDB code 2rdt; Murray et al., 2008 ▶). (c) Superposition of the Cα trace of the hGOX active site in complex with CCPST (white) and of sGOX in complex with TACA (green) (PDB code 1al7; Stenberg & Lindqvist, 1997 ▶).
The five-membered rings of CCPST and CDST superimpose reasonably well and the chlorophenyl group occupies the exact same location as the first C atoms of the CDST aliphatic chain. In contrast, the orientations of the CCPST and TACA rings differ significantly, possibly owing to the necessity of accommodating the bifurcated TACA substituent. The three inhibitors are hydrogen bonded to Tyr132 (Tyr129 in sGOX) via the ring N2. Despite the somewhat different orientations of the planes of the three five-membered rings, the N atoms at position 3 of the rings superimpose well and are correctly oriented and at a suitable distance to form a hydrogen bond to N∊ of the catalytic histidine. The latter must be neutral in the oxidized enzyme, owing to its function as an active-site base as in other family members (Ghisla & Massey, 1991 ▶; Lederer, 1991 ▶; Lindqvist, 1992 ▶), with its Nδ proton hydrogen bonded to the invariant Asp170. It is believed to be protonated in the reduced enzyme (Rao & Lederer, 1998 ▶). For the triazoles TACA and CDST, the hydrogen-bond donor could be a tautomer of the structures shown in Fig. 4 ▶. However, the thiadiazole ring of CCPST does not carry a proton; neither does the glyoxylate keto group. This configuration thus suggests that binding of these two compounds elicits protonation of His260. A precedent for this situation exists with an enzyme of the same evolutionary family. Indeed, with lactate monooxygenase it was shown that binding of oxalate to the active site elicited the uptake of a proton from the solution (Ghisla & Massey, 1977 ▶). In view of the sequence and structure similarities between family members, this proton must be taken up by the homologue of His260 (hGOX).
The largest differences between the various complexes arise from the necessity of accommodating the five-membered ring substituents. In addition to a displacement of Leu205, Murray et al. (2008 ▶) underlined a concerted movement of Trp110, Tyr208 and Tyr134 between the complex with glyoxylate and that with CDST, leading to mobility of loop 4. Fig. 7 ▶(a) shows that CCPST binding similarly induces movements of large amplitude relative to the glyoxylate complex. Compared with CDST binding, CCPST binding (Fig. 7 ▶ b) induces only a slight shift in the orientation of the Trp110 indole-ring plane and of Tyr132 but a noticeable displacement of Tyr134 and Tyr208. For example, while the Tyr208 Cα atoms lie within 0.9 Å of one another in the superposition, the phenol groups are separated by 2.7 Å. Finally, in the TACA complex the tryptophan side chain has yet another orientation (Fig. 7 ▶ c). In order to accommodate the five-carbon branch of the substituent, the indole ring has moved by 3.5 Å and is rotated by about 115°. The mobility of the tryptophan side chain in sGOX, depending on the active-site occupancy and the nature of the ligand, has been underlined previously (Stenberg & Lindqvist, 1996 ▶). There is no real structural equivalent of Leu205 and Tyr208 in the TACA–sGOX complex owing to the different orientation of helix αE.
In conclusion, the structure of the CCPST–hGOX complex compared with those of other complexes of hGOX and sGOX suggests a two-tier binding site. One tier is composed of a number of invariant catalytic residues that do not appreciably change their orientation and provide electrostatic interactions as well as hydrogen bonds to the carboxylate and five-membered ring and the other tier is composed of a highly flexible series of residues that provide an essentially hydrophobic environment to the ring substituent without necessarily preventing contact with the solvent.
Supplementary Material
PDB reference: glycolate oxidase–4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole complex, 2w0u, r2w0usf
Enhanced figure: interactive version of Fig. 1
Acknowledgments
The authors acknowledge access to beam time at the European Synchrotron Research Facility, Grenoble, France and at MAX-lab, Lund, Sweden. They are indebted to Dr F. S. Mathews for critical reading of the manuscript. This study was supported by grants from the Swedish Research Council (YL) and by fellowships from the CNRS Institut de Chimie des Substances Naturelles to CV and NP. The authors are grateful to Professor J.-Y. Lallemand for his interest in and support of the project.
References
- Belmouden, A. & Lederer, F. (1996). Eur. J. Biochem. 238, 790–798. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cunane, L. M., Barton, J. D., Chen, Z.-W., Lê, K. H. D., Amar, D., Lederer, F. & Mathews, F. S. (2005). Biochemistry, 44, 1521–1531. [DOI] [PubMed]
- Cunane, L. M., Barton, J. D., Chen, Z. W., Welsh, F. E., Chapman, S. K., Reid, G. A. & Mathews, F. S. (2002). Biochemistry, 41, 4264–4272. [PubMed]
- Danpure, C. J. (2001). The Metabolic and Molecular Bases of Inherited Disease, edited by C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, K. W. Kinzler & B. Vogelstein, pp. 3323–3367. New York: McGraw–Hill.
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, California, USA.
- Dewanti, A. R. & Mitra, B. (2003). Biochemistry, 42, 12893–12901. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fry, D. W. & Richardson, K. E. (1979). Biochim. Biophys. Acta, 568, 135–144. [DOI] [PubMed]
- Gaume, B., Sharp, R. E., Manson, F. D. C., Chapman, S. K., Reid, G. A. & Lederer, F. (1995). Biochimie, 77, 621–630. [DOI] [PubMed]
- Ghisla, S. & Massey, V. (1977). J. Biol. Chem. 252, 6729–6735. [PubMed]
- Ghisla, S. & Massey, V. (1980). J. Biol. Chem. 255, 5688–5696. [PubMed]
- Ghisla, S. & Massey, V. (1991). Chemistry and Biochemistry of Flavoenzymes, edited by F. Müller, Vol. 2, pp. 243–249. Boca Raton: CRC Press.
- Ghrir, R. & Lederer, F. (1981). Eur. J. Biochem. 120, 279–287. [DOI] [PubMed]
- Gondry, M., Dubois, J., Terrier, M. & Lederer, F. (2001). Eur. J. Biochem. 268, 4918–4927. [DOI] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Illias, R. M., Sinclair, R., Robertson, D., Neu, A., Chapman, S. K. & Reid, G. A. (1998). Biochem. J. 333, 107–115. [DOI] [PMC free article] [PubMed]
- Jones, J. M., Morrell, J. C. & Gould, S. J. (2000). J. Biol. Chem. 275, 12590–12597. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst. 26, 795–800.
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993). J. Mol. Biol. 231, 1049–1067. [DOI] [PubMed]
- Lederer, F. (1991). Chemistry and Biochemistry of Flavoenzymes, edited by F. Müller, Vol. 2, pp. 153–242. Boca Raton: CRC Press.
- Lederer, F., Amar, D., Ould Boubacar, A. K. & Vignaud, C. (2005). Flavins and Flavoproteins 2005, edited by T. Nishino, R. Miura, M. Tanokura & K. Fukui, pp. 193–204. Tokyo: ArchiTect Inc.
- Lehoux, I. E. & Mitra, B. (1999). Biochemistry, 38, 5836–5848. [DOI] [PubMed]
- Leiros, I., Wang, E., Rasmussen, T., Oksanen, E., Repo, H., Petersen, S. B., Heikinheimo, P. & Hough, E. (2006). Acta Cryst. F62, 1185–1190. [DOI] [PMC free article] [PubMed]
- Li, S. J., Umena, Y., Yorita, K., Matsuoka, T., Kita, A., Fukui, K. & Morimoto, Y. (2007). Biochem. Biophys. Res. Commun. 358, 1002–1007. [DOI] [PubMed]
- Lindqvist, Y. (1989). J. Mol. Biol. 209, 151–166. [DOI] [PubMed]
- Lindqvist, Y. (1992). Chemistry and Biochemistry of Flavoenzymes, edited by F. Müller, Vol. 3, pp. 367–387. Boca Raton: CRC Press.
- Lindqvist, Y., Brändén, C. I., Mathews, F. S. & Lederer, F. (1991). J. Biol. Chem. 266, 3198–3207. [PubMed]
- Lohkamp, B. & Emsley, P. (2005). CCP4 Newsl. 42, contribution 7.
- Macheroux, P., Kieweg, V., Massey, V., Soderlind, E., Stenberg, K. & Lindqvist, Y. (1993). Eur. J. Biochem. 213, 1047–1054. [DOI] [PubMed]
- Macheroux, P., Massey, V., Thiele, D. J. & Volokita, M. (1991). Biochemistry, 30, 4612–4619. [DOI] [PubMed]
- Macheroux, P., Mulrooney, S. B., Williams, C. H. Jr & Massey, V. (1992). Biochim. Biophys. Acta, 1132, 11–16. [DOI] [PubMed]
- Maeda-Yorita, K., Aki, K., Sagai, H., Misaki, H. & Massey, V. (1995). Biochimie, 77, 631–642. [DOI] [PubMed]
- Miziorko, H. M. & Lorimer, G. H. (1983). Annu. Rev. Biochem. 52, 507–535. [DOI] [PubMed]
- Murray, M. S., Holmes, R. P. & Lowther, W. T. (2008). Biochemistry, 47, 2439–2449. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Randall, W. C., Streeter, K. B., Cresson, E. L., Schwam, H., Michelson, S. R., Anderson, P. S., Cragoe, E. J. Jr, Williams, H. W. R., Eichler, E. & Rooney, C. S. (1979). J. Med. Chem. 22, 608–614. [DOI] [PubMed]
- Rao, K. S. & Lederer, F. (1998). Protein Sci. 7, 1531–1537. [DOI] [PMC free article] [PubMed]
- Rooney, C. S., Randall, W. C., Streeter, K. B., Ziegler, C., Cragoe, E. J. Jr, Schwam, H., Michelson, S. R., Williams, H. W., Eichler, E., Duggan, D. E., Ulm, E. H. & Noll, R. M. (1983). J. Med. Chem. 26, 700–714. [DOI] [PubMed]
- Schuman, M. & Massey, V. (1971a). Biochim. Biophys. Acta, 227, 500–520. [DOI] [PubMed]
- Schuman, M. & Massey, V. (1971b). Biochim. Biophys. Acta, 227, 521–537. [DOI] [PubMed]
- Schwam, H., Michelson, S., Randall, W. C., Sondey, J. M. & Hirschmann, R. (1979). Biochemistry, 18, 2828–2833. [DOI] [PubMed]
- Sobrado, P., Daubner, S. C. & Fitzpatrick, P. F. (2001). Biochemistry, 40, 994–1001. [DOI] [PubMed]
- Stenberg, K., Clausen, T., Lindqvist, Y. & Macheroux, P. (1995). Eur. J. Biochem. 228, 408–416. [PubMed]
- Stenberg, K. & Lindqvist, Y. (1996). Protein Expr. Purif. 8, 295–298. [DOI] [PubMed]
- Stenberg, K. & Lindqvist, Y. (1997). Protein Sci. 6, 1009–1015. [DOI] [PMC free article] [PubMed]
- Sukumar, N., Dewanti, A. R., Mitra, B. & Mathews, F. S. (2004). J. Biol. Chem. 279, 3749–3757. [DOI] [PubMed]
- Umena, Y., Yorita, K., Matsuoka, T., Kita, A., Fukui, K. & Morimoto, Y. (2006). Biochem. Biophys. Res. Commun. 350, 249–256. [DOI] [PubMed]
- Vignaud, C., Pietrancosta, N., Williams, E. L., Rumsby, G. & Lederer, F. (2007). Arch. Biochem. Biophys. 465, 410–416. [DOI] [PubMed]
- Williams, E., Cregeen, D. & Rumsby, G. (2000). Biochim. Biophys. Acta, 1493, 246–248. [DOI] [PubMed]
- Williams, H. W., Eichler, E., Randall, W. C., Rooney, C. S., Cragoe, E. J. Jr, Streeter, K. B., Schwam, H., Michelson, S. R., Patchett, A. A. & Taub, D. (1983). J. Med. Chem. 26, 1196–1200. [DOI] [PubMed]
- Xia, Z. X. & Mathews, F. S. (1990). J. Mol. Biol. 212, 837–863. [DOI] [PubMed]
- Xu, Y. & Mitra, B. (1999). Biochemistry, 38, 12367–12376. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: glycolate oxidase–4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole complex, 2w0u, r2w0usf
Enhanced figure: interactive version of Fig. 1