The cloning, expression, purification and crystallization of the mouse Elf3 C-terminal DNA-binding domain in complex with mouse type II TGF-β receptor promoter DNA are reported. The crystals were characterized and an X-ray diffraction data set was collected to a resolution of 2.2 Å.
Keywords: transcription factors, Elf3, Ets, cancer
Abstract
Ets proteins are transcription factors that activate or repress the expression of genes that are involved in various biological processes, including cellular proliferation, differentiation, development, transformation and apoptosis. Like other Ets-family members, Elf3 functions as a sequence-specific DNA-binding transcriptional factor. A mouse Elf3 C-terminal fragment (amino-acid residues 269–371) containing the DNA-binding domain has been crystallized in complex with mouse type II TGF-β receptor promoter (TβR-II) DNA. The crystals belonged to space group P212121, with unit-cell parameters a = 42.66, b = 52, c = 99.78 Å, and diffracted to a resolution of 2.2 Å.
1. Introduction
Elf3 is a member of the Ets transcription-factor family, the members of which play a role in normal growth and development as well as in a variety of cancers and diseases (Wasylyk et al., 1993 ▶; Hromas & Klemsz, 1994 ▶; Dittmer & Nordheim, 1998 ▶). Currently, 29 human Ets-family members have been recognized based on the presence of a highly conserved DNA-binding Ets domain (Gallant & Gilkeson, 2006 ▶). Ets-family members are defined by the Ets domain, which is a highly conserved winged helix–loop–helix-type DNA-binding domain (DBD) that recognizes a core motif 5′-GGA(A/T)-3′, which is often referred to as the Ets-binding site (EBS; Nye et al., 1992 ▶; Donaldson et al., 1996 ▶).
Elf3 (E74-like factor 3), also known as ESE1, ERT, jen and ESX, was first described in human mammary-cancer cells (Jobling et al., 2002 ▶). Mouse Elf3 (mElf3) is translated as a 371-amino-acid protein that displays 89% identity and 93% similarity to the human Elf3 amino-acid sequence (Tymms et al., 1997 ▶). Elf3 is composed of five defined domains: a pointed domain (amino-acid residues 60–128), a transactivation domain (TAD; amino acids 129–159), a serine- and aspartic acid-rich (SAR) domain (amino acids 189–229), an AT-hook domain (amino acids 238–259) and an Ets DNA-binding domain (DBD; amino acids 274–354) (Kopp et al., 2007 ▶).
Elf3 has been identified in a wide range of epithelial carcinoma cells and is aberrantly expressed in cancers of the lung and breast (Chang et al., 1997 ▶). Studies in several cell-culture model systems have shown that the promoter of the type II TGF β receptor (TβRII) gene is transactivated by Elf3. Expression of the mouse TβRII (mTβRII) gene behaves as a tumor suppressor and it is expressed in nearly all cell types. mElf3 binds to EBS and strongly stimulates the expression of the mTβRII promoter in differentiated cells derived from mouse F9 embryonal carcinoma cells (Choi et al., 1998 ▶; Kim et al., 2002 ▶; Kopp et al., 2004 ▶). Other studies using Hs578t breast-cancer cells revealed that the expression of Elf3 dramatically elevates the expression of TβRII and decreases the tumorigenicity of these cells (Chang et al., 2000 ▶).
The Ets domain is highly conserved within the Ets family of transcription factors and has been structurally characterized for several family members. Interestingly, Elf3 (but not Ets2) was found to activate the TβRII gene in F9 differentiated cells (Kim et al., 2002 ▶). In order to determine the unique structural determinants of TβRII gene recognition by Elf3, we initiated structural and functional studies of Elf3–DNA complexes. Here, we report the preliminary X-ray crystallographic analysis of mElf3 C-terminal residues 269–371 (mElf3269–371), which contain an Ets domain, in complex with mTβRII dsDNA.
2. Materials and methods
2.1. Cloning, expression and purification
The plasmid mElf3-pET-103 (residues 269–371) of the mElf3 gene was PCR-amplified using primers 5′-AGATCTGGTCTCCCATGGCACCAAGAGGTACTC-3′ and 5′-AGATGGATCCTCAATTCCGACTCTCTCCAACC-3′. The PCR-amplified product was digested with NcoI and BamHI restriction enzymes and inserted into similar restriction sites of pET-28b vector (Novagen).
The protein mElf3269–371 was overproduced in Escherichia coli BL21 (DE3) cells grown at 295 K for 16 h in LB medium supplemented with 0.05 mg ml−1 kanamycin. The harvested cell pellet was resuspended in a solution containing 50 mM Tris–HCl pH 8.0, 800 mM NaCl, 10% glycerol, 2 mM DTT, 2 mM PMSF and 1 mM EDTA, which was sonicated and centrifuged to remove cell debris. The supernatant was treated with 1.4% streptomycin sulfate at 277 K for 30 min and centrifuged. The clear lysate was diluted with two volumes of 50 mM Tris–HCl pH 8.0 and applied onto an SP-Sepharose Cation Exchanger resin column (GE Healthcare Life Sciences) and subsequently onto an SP Hi-Trap column (GE Healthcare Life Sciences) pre-equilibrated with 25 mM Tris–HCl pH 8.0, 150 mM NaCl, 5% glycerol, 1 mM DTT, 1 mM PMSF and 1 mM EDTA (buffer A). The protein was eluted with buffer A plus 0.8 M NaCl. The collected fractions were pooled together and dialyzed against 25 mM Tris–HCl pH 8.0, 2.5% glycerol and 1 mM DTT (buffer B). This preparation was applied onto a Heparin Hi-Trap column (GE Healthcare Life Sciences) pre-equilibrated with buffer B. The column was washed and eluted with buffer B plus 1 M NaCl. The fractions containing Elf3 protein were pooled, dialyzed against 25 mM MES pH 6.2, 150 mM NaCl and 5 mM DTT and concentrated to 9.4 mg ml−1. The sample purity and molecular weight were determined at all stages by SDS–PAGE using 15% gels stained with Coomassie Blue R-250.
2.2. Preparation of dsDNA and DNA-binding assay
A double-stranded (ds) oligonucleotide corresponding to the mTβRII gene (Kim et al., 2002 ▶) was prepared by annealing the synthetic oligonucleotides 5′-GAGGAGTTTCCTGTTT-3′ and 5′-CAAACAGGAAACTCCT-3′. The oligonucleotides were dissolved in 10 mM Tris–HCl pH 7.5, 1 mM EDTA and 1 M NaCl and then annealed together by heating to 368 K for 5 min and gradually cooling to room temperature over a 3 h period using a PCR thermal cycler. The dsDNA solution was desalted, dried and dissolved in 10 mM Tris–HCl pH 8.0 and stored at 253 K.
For the DNA-binding experiments, a reaction mixture consisting of mElf3269–371 and mTβRII dsDNA was prepared in 25 mM MES buffer pH 6.2, 150 mM NaCl and 5 mM DTT and incubated at room temperature for 20 min, mixed with 1 µl glycerol and applied onto 6% polyacrylamide gel. Electrophoresis was performed in 0.5× TBE buffer at 150 V for 35 min on ice. DNA and protein bands were visualized by subsequent ethidium bromide and Coomassie Blue staining, respectively.
2.3. Crystallization
The mElf3269–371–mTβRII dsDNA complex was prepared by mixing the components in a 1:1.05 ratio and was concentrated to 10 mg ml−1 in 25 mM MES pH 6.2, 150 mM NaCl and 5 mM DTT. Complex formation was confirmed by 6% polyacrylamide gel electrophoresis. Initial crystallization screening was performed using Natrix Screen (Hampton Research) with the sitting-drop vapour-diffusion method at 295 K by mixing 1 µl protein–DNA complex solution with 1 µl reservoir solution. Small aggregated crystals appeared in condition No. 27 of Natrix Screen. Optimization of the conditions was performed by variation of the polyethylene glycol (PEG) 8000 concentration. Diffraction-quality crystals were finally obtained at 295 K in 0.2 M ammonium acetate, 0.01 M magnesium acetate tetrahydrate, 0.05 M sodium cacodylate trihydrate pH 6.5 and 25%(w/v) PEG 8000.
2.4. X-ray diffraction data collection and processing
The crystal was soaked in cryoprotectant for a few seconds, scooped up in a nylon-fiber loop and flash-cooled in a dry nitrogen stream at 100 K. The cryoprotectant solution consisted of 0.2 M ammonium acetate, 0.01 M magnesium acetate tetrahydrate, 0.05 M sodium cacodylate trihydrate pH 6.5, 33%(w/v) PEG 8000 and 15% glycerol. Preliminary X-ray examinations of crystals were carried out at cryo-temperature using a Rigaku R-AXIS IV imaging plate with Osmic VariMax HR mirror-focused Cu Kα radiation from a Rigaku FR-E rotating-anode generator operated at 45 kV and 45 mA. The final data set was collected on Argonne National Laboratory Advanced Photon Source beamline 24ID-C using an ADSC Q315 detector. All intensity data were indexed, integrated and scaled with DENZO and SCALEPACK from the HKL-2000 program package. The crystal diffracted to a resolution of 2.05 Å. However, careful observation of reflections revealed crystal twinning, especially at resolutions above 2.2 Å. For this reason, we limited the current data processing to a resolution of 2.2 Å. The crystal parameters and data-processing statistics are summarized in Table 1 ▶.
Table 1. Crystal parameters and data-collection statistics.
Values in parentheses are for the last shell.
| Crystal parameters | |
| Unit-cell parameters (Å) | |
| a | 42.66 |
| b | 52.00 |
| c | 99.78 |
| Space group | P212121 |
| Data collection | |
| Temperature (K) | 100 |
| Resolution (Å) | 50.0–2.2 (2.24–2.2) |
| Unique reflections | 11575 |
| Redundancy | 5.8 (6.0) |
| Completeness (%) | 97.6 (97.5) |
| Rmerge† (%) | 6.4 (39.0) |
| 〈I/σ(I)〉 | 53.5 (9.9) |
| Mosaicity (°) | 0.66–1.01 |
R
merge = 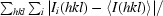
 , where I
i(hkl) and 〈I(hkl)〉 are the intensity of measurement i and the mean intensity of the reflection with indices hkl, respectively.
, where I
i(hkl) and 〈I(hkl)〉 are the intensity of measurement i and the mean intensity of the reflection with indices hkl, respectively.
3. Results and discussion
Purified mElf3269–371 has a tendency to aggregate at concentrations of 6 mg ml−1 and higher. Protein aggregation was monitored by the dynamic light-scattering (DLS) technique and was reduced considerably using the solubility screening described by Jancarik et al. (2004 ▶). After dialysis against a solution containing 150 mM NaCl, 5 mM DTT and 25 mM MES buffer pH 6.2 and binding with mTβRII dsDNA, the protein–DNA complex was successfully concentrated to 10 mg ml−1 without aggregation.
The crystallization of protein–DNA complexes strongly depends on the DNA construct selected. Our previous crystallization experiments revealed that certain DNA constructs increase the chance of crystallization of protein–DNA complexes (Tahirov et al., 2001 ▶). Crystals often grew when the DNA duplexes were paired with overhanging bases and formed continuous double-stranded B-DNA pseudo-helices that contained a whole number of helical turns plus a half-turn. The DNA fragment used for the crystallization of the mElf3269–371–mTβRII dsDNA complex was selected in a similar way. It contained one-and-a-half helical turns with overhanging G and C bases:
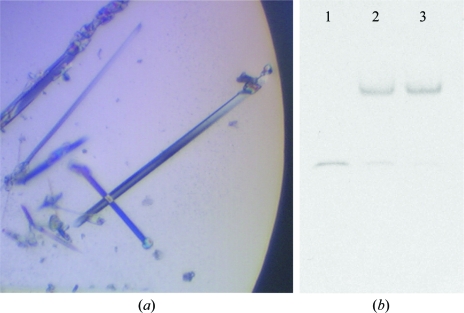
mElf3269–371 bound to the DNA fragment described above produced small needle-like crystals with Natrix screen condition No. 27. Further optimization of the precipitant concentration improved the size and shape of the crystal, which diffracted to 2.05 Å resolution (Fig. 1 ▶ a). The crystal was orthorhombic and belonged to space group P212121. A solvent-content calculation (Matthews, 1968 ▶) suggested that one molecule of mElf3269–371–mTβRII dsDNA was located in the asymmetric unit, with a solvent content of the crystal of 50.6%. An electrophoretic mobility shift assay (EMSA) of a dissolved crystal sample confirmed the presence of both protein and DNA components (Fig. 1 ▶ b). The structure was solved by the molecular-replacement method and will be reported elsewhere.
Figure 1.
(a) A photomicrograph of the crystals of mElf3269–371–mTβRII dsDNA. (b) EMSA. Lane 1, DNA. Lane 2, initial sample. Lane 3, washed and dissolved crystals.
Acknowledgments
We thank J. Lovelace and G. E. Borgstahl for maintenance and management of the Eppley Institute’s X-ray Crystallography facility. The primers were synthesized in the Eppley Institute’s Molecular Biology Core facility. Both facilities are supported by Cancer Center Support Grant P30CA036727. This work is supported by the UNMC Eppley Cancer Center Pilot Project LB595 and in part by NIGMS grant R01GM082923 to THT and also in part by Nebraska Department of Health and Human Services grant LB506 to AR. This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357.
References
- Chang, C. H., Scott, G. K., Kuo, W. L., Xiong, X., Suzdaltseva, Y., Park, J. W., Sayre, P., Erny, K., Collins, C., Gray, J. W. & Benz, C. C. (1997). Oncogene, 14, 1617–1622. [DOI] [PubMed]
- Chang, J., Lee, C., Hahm, K. B., Yi, Y., Choi, S. G. & Kim, S. J. (2000). Oncogene, 19, 151–154. [DOI] [PubMed]
- Choi, S. G., Yi, Y., Kim, Y. S., Kato, M., Chang, J., Chung, H. W., Hahm, K. B., Yang, H. K., Rhee, H. H., Bang, Y. J. & Kim, S. J. (1998). J. Biol. Chem.273, 110–117. [DOI] [PubMed]
- Dittmer, J. & Nordheim, A. (1998). Biochim. Biophys. Acta, 1377, F1–F11. [DOI] [PubMed]
- Donaldson, L. W., Petersen, J. M., Graves, B. J. & McIntosh, L. P. (1996). EMBO J.15, 125–134. [PMC free article] [PubMed]
- Gallant, S. & Gilkeson, G. (2006). Arch. Immunol. Ther. Exp. (Warsz), 54, 149–163. [DOI] [PubMed]
- Hromas, R. & Klemsz, M. (1994). Int. J. Hematol.59, 257–265. [PubMed]
- Jancarik, J., Pufan, R., Hong, C., Kim, S.-H. & Kim, R. (2004). Acta Cryst. D60, 1670–1673. [DOI] [PubMed]
- Jobling, A. I., Fang, Z., Koleski, D. & Tymms, M. J. (2002). Invest. Ophthalmol. Vis. Sci.43, 3530–3537. [PubMed]
- Kim, J. H., Wilder, P. J., Hou, J., Nowling, T. & Rizzino, A. (2002). J. Biol. Chem.277, 17520–17530. [DOI] [PubMed]
- Kopp, J. L., Wilder, P. J., Desler, M., Kim, J. H., Hou, J., Nowling, T. & Rizzino, A. (2004). J. Biol. Chem.279, 19407–19420. [DOI] [PubMed]
- Kopp, J. L., Wilder, P. J., Desler, M., Kinarsky, L. & Rizzino, A. (2007). J. Biol. Chem.282, 3027–3041. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Nye, J. A., Petersen, J. M., Gunther, C. V., Jonsen, M. D. & Graves, B. J. (1992). Genes Dev.6, 975–990. [DOI] [PubMed]
- Tahirov, T. H., Inoue-Bungo, T., Sasaki, M., Fujikawa, A., Kimura, K., Sato, K., Adachi, S., Kamiya, N. & Ogata, K. (2001). Acta Cryst. D57, 854–856. [DOI] [PubMed]
- Tymms, M. J., Ng, A. Y., Thomas, R. S., Schutte, B. C., Zhou, J., Eyre, H. J., Sutherland, G. R., Seth, A., Rosenberg, M., Papas, T., Debouck, C. & Kola, I. (1997). Oncogene, 15, 2449–2462. [DOI] [PubMed]
- Wasylyk, B., Hahn, S. L. & Giovane, A. (1993). Eur. J. Biochem.211, 7–18. [DOI] [PubMed]