Abstract
Rhox5, the founding member of the reproductive homeobox on the X chromosome (Rhox) gene cluster, encodes a homeodomain-containing transcription factor that is selectively expressed in Sertoli cells, where it promotes the survival of male germ cells. To identify Rhox5-regulated genes, we generated 15P-1 Sertoli cell clones expressing physiological levels of Rhox5 from a stably transfected expression vector. Microarray analysis identified many genes altered in expression in response to Rhox5, including those encoding proteins controlling cell cycle regulation, apoptosis, metabolism, and cell-cell interactions. Fifteen of these Rhox5-regulated genes were chosen for further analysis. Analysis of Rhox5-null male mice indicated that at least nine of these are Rhox5-regulated in the testes in vivo. Many of them have distinct postnatal expression patterns and are regulated by Rhox5 at different postnatal time points. Most of them are expressed in Sertoli cells, indicating that they are candidates to be directly regulated by Rhox5. Transfection analysis with expression vectors encoding different mouse and human Rhox family members revealed that the regulatory response of a subset of these Rhox5-regulated genes is both conserved and redundant. Given that Rhox5 depends on androgen receptor (AR) for expression in Sertoli cells, we examined whether some Rhox5-regulated genes are also regulated by AR. We provide several lines of evidence that this is the case, leading us to propose that RHOX5 serves as a key intermediate transcription factor that directs some of the actions of AR in the testes.
Using expression profiling, we identify androgen receptor (AR)-regulated genes whose expression is controlled by the androgen/AR-regulated Rhox5 homeobox gene in Sertoli cells.
Homeobox genes encode transcription factors harboring a 60-amino acid DNA-binding motif called a homeodomain. These transcription factors govern key embryonic developmental processes, including body-axis formation, organogenesis, and limb development (1). Whereas the roles of homeobox genes in embryonic development have been studied intensely for over two decades, their functions in controlling postembryonic developmental events have only begun to be examined. Homeobox genes are good candidates to control postembryonic events, because many homeobox genes are expressed during postnatal development and in adult tissues. Indeed, mouse knockout studies have shown that the HoxA9, Hoxc13, and Pdx1 homeobox genes are crucial for hematopoiesis, hair growth, and gut homeostasis, respectively (2,3,4).
Another postembryonic process likely to be controlled by homeobox genes is spermatogenesis. This idea is supported by the fact that over 50 of the approximately 200 known mouse homeobox genes are expressed in the testis and other male reproductive organs (5,6,7,8,9,10,11,12). Although this suggests a role for homeobox transcription factors in male reproduction, the functions of most homeobox transcription factors in the postnatal and adult male reproductive tract are not yet clear, because efforts to elucidate their roles by use of knockout mice have been clouded by putative functional redundancies and embryonic lethality (6). These efforts have identified some homeobox genes that control the formation of the male reproductive tract during embryogenesis, but the identity of homeobox genes that have roles in the male reproductive tract after birth have remained largely obscure (5,12,13,14).
To our knowledge, the only mammalian homeobox gene demonstrated to have a role in spermatogenesis is Rhox5 (previously known as Pem), the founding member of the reproductive homoeobox genes on the X chromosome (Rhox) gene family (7). All the members of the Rhox gene family form a tandem array on the X chromosome in mammals. The mouse Rhox gene cluster contains 33 members, and thus is the largest homeobox gene cluster known in any species (7,8,9,10,11,15). Virtually all of the Rhox genes are selectively expressed in reproductive tissues and the placenta, suggesting that they encode transcription factors devoted to regulating gametogenesis and embryogenesis (7).
Rhox5 was first identified in a search for genes differentially expressed between two related T-lymphoma cell clones that exhibited marked differences in developmental status and tumorigenicity (16). Rhox5 was initially of interest because it was found to be selectively expressed in T-cell lymphomas, not normal T cells, and also in a wide variety of other tumor cells of diverse developmental and tissue origins (17). Later investigations revealed that Rhox5 encodes a tumor antigen (18), interacts with protein involved in tumorigenesis (19,20), and its expression is induced by the proto-oncogene Ras (21,22). Although these studies have suggested the possibility that Rhox5 has a role in tumorigenesis, its precise role in malignancy has not yet been determined. Other studies have focused on Rhox5’s normal expression pattern and biological roles. Rhox5 is normally expressed in the early mouse embryo in a developmentally regulated manner (23), and is later expressed in placenta (24,25) and in both male and female germ cells in developing fetal gonads (26,27). In neonatal and adult mice and rats, Rhox5 expression is primarily confined to somatic cells in the testis, epididymis, and ovary (28,29). In the testis, its expression is restricted to Sertoli cells, where it is expressed in a stage-specific manner during the seminiferous epithelial cycle by an androgen- and androgen receptor (AR)-dependent mechanism (30,31).
The expression of Rhox5 in somatic cells in the male reproductive tract is crucial for normal spermatogenesis and sperm maturation. Targeted mutation of Rhox5 in male mice leads to subfertility, marked by increased germ-cell apoptosis, reduced sperm number, and reduced sperm motility (7). Rhox5-null mice exhibit an increased frequency of apoptotic germ cells in seminiferous epithelial cycle stages that normally contain dying germ cells (stages I–IV and stage XII) and they also have apoptotic germ cells in stages that normally lack dying germ cells (stages V–XI). Rhox5 is unlikely to act directly on germ cells to control germ cell survival and motility, because it is not detectably expressed in germ cells, but instead is expressed in Sertoli cells, the nurse cells in intimate contact with germ cells (29,30). This suggests a model in which Rhox5 functions in spermatogenesis by virtue of its ability to regulate genes encoding Sertoli cell secreted and cell surface proteins that, in turn, control key events in germ cells, including their differentiation and survivial (32,33,34).
To begin to address the molecular mechanisms by which Rhox5 helps drive spermatogenesis, we used transcriptional profiling to identify genes regulated by physiological levels of Rhox5 in 15P-1 cells, a well-established Sertoli cell line (35,36,37). This genome-wide analysis led to the discovery of many Rhox5-regulated genes, a large proportion of which encode cell surface proteins. Many of these were expressed in Sertoli cells, which was of interest because such genes could mediate Rhox5-induced signals that impinge on germ cells. Also identified were Rhox5-regulated genes encoding other kinds of proteins, including secreted factors, transcription factors, and metabolic enzymes. We focused on a subset of these genes and found that most are also Rhox5-regulated in the testes in vivo. We determined the testicular cell types and developmental stages in which the regulation of each gene occurs. We also identified other Rhox family members that regulate their expression, and provided evidence that the Rhox-dependent regulatory circuitry controlling these genes is conserved. Finally, we addressed whether any of the Rhox5 gene targets are secondary androgen-response (SAR) genes. These are long-sought-after genes that are not directly regulated by androgen and AR, but rather are controlled by androgen-dependent regulators, such as androgen-induced transcription factors (38,39). Because Rhox5 is an AR- and androgen-inducible gene in Sertoli cells, it is a good candidate to regulate SAR genes. Indeed, we obtained many lines of evidence that several Rhox5-regulated genes are SAR genes. We also showed that other androgen-inducible Rhox genes (besides Rhox5) are capable of regulating these candidate SAR genes. The discovery of genes coregulated by AR and RHOX transcription factors has the potential to begin illuminating the molecular mechanisms by which androgens drive spermatogenesis.
Results
Establishment of Sertoli cell clones expressing physiological levels of Rhox5
To identify Rhox5 target genes by a gain-of-function approach, we screened Sertoli cell lines for lines that express low endogenous Rhox5 mRNA levels. Our rationale was that a low Rhox5-expressing line would provide an optimal baseline for identifying genes regulated by physiological levels of Rhox5 expressed from a Rhox5-expression vector. We predicted that most Sertoli cell lines would express low levels of Rhox5 because we previously found that primary mouse Sertoli cells dramatically down-regulate Rhox5 mRNA expression after only 1 d in culture (30). Indeed, we found that all three Sertoli cell lines that we tested—15P-1, MSC-1, and Tm4— expressed very low levels of Rhox5 mRNA, as judged by quantitative (q) PCR analysis (supplemental Fig. S1, which is published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). The two cell lines with the lowest expression, 15P-1 and Tm4, expressed approximately 300-fold less Rhox5 mRNA than adult testes. This is a conservative estimate of the difference given that Rhox5 mRNA is only expressed in Sertoli cells in the adult testis (26,29). Indeed, RHOX5 protein was undetectable in 15P-1 cells, as judged by Western blot analysis (supplemental Fig. S2). We chose 15P-1 over Tm4 for our gain-of-function study because we found that 15P-1 cells could be transfected more efficiently than Tm4 cells (using Lipofectamin; data not shown). We also chose the 15P-1 cell line because it exhibits many normal Sertoli cell characteristics, including the expression of genes normally expressed by Sertoli cells (e.g. WT1 and Steel) and the ability to support the transit of male germ cells through meiosis (35,40,41).
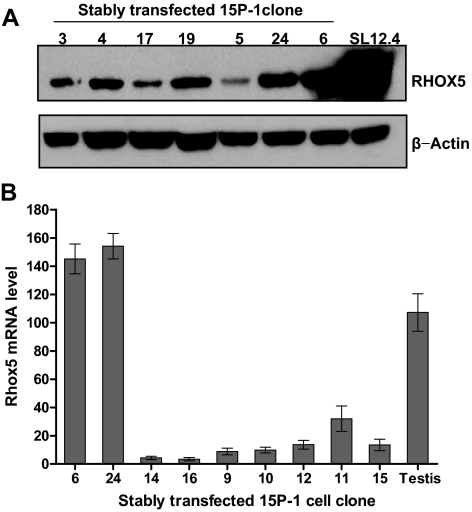
To express Rhox5 in 15P-1 cells, a murine Rhox5 full-length cDNA was cloned into an expression vector under the control of the CMV immediate early promoter. This construct was then stably transfected into 15P-1 Sertoli cells and several independent G418-resistant subclones were obtained. Northern blot analysis demonstrated that many of the cell clones expressed Rhox5 mRNA at levels comparable to that in postnatal and adult testes (data not shown). To confirm that Rhox5 was expressed, we used Western blot analysis. This showed that the stably transfected cell clones also expressed RHOX5 protein (Fig. 1A). To obtain a quantitative assessment of Rhox5 mRNA levels, we used qPCR analysis (Fig. 1B). This demonstrated that clones 6 and 24 expressed the highest level of Rhox5 mRNA and thus these two clones were chosen as the Rhox5-positive clones for microarray analysis. Clones 14 and 16, which had the lowest level of Rhox5 mRNA (comparable with the level in parental 15P-1 cells; data not shown), were chosen as the Rhox5-negative clones for microarray analysis.
Figure 1.
Generation of 15P-1 Sertoli cell clones expressing physiological levels of Rhox5. A, Western blot analysis of RHOX5 protein expression in 15P-1 stable clones stably transfected with a RHOX5-expression vector. SL12.4 cells served as positive control for RHOX5 expression. After probing with RHOX5 antiserum, the blot was stripped and reprobed with a β-actin antiserum as an internal control for loading. B, qPCR analysis of total cellular RNA from the 15P-1 Sertoli cell clones. Rhox5 mRNA expression levels in the 15P-1 cell clones relative to parental 15P-1 cells (which was arbitrarily given a value of 1), all normalized against ribosomal protein L19 transcript levels. Adult testis served as a positive control. Values denote the mean fold change ± sem.
Identification of genes regulated by Rhox5 in Sertoli cells
Total cellular RNA was isolated from clones 6, 24, 14, and 16 and analyzed by microarray analysis. The digitally captured data was analyzed by use of GeneSpring software to determine the genes that were differentially expressed between the four samples in all pairwise combinations. This analysis revealed that 182 genes were either up-regulated or down-regulated by 2-fold or more in the Rhox5-positive cell clones compared with the Rhox5-negative cell clones (supplemental Tables 1 and 2). The Rhox5-regulated genes that we identified by this approach encode proteins encompassing a broad range of functions. We used OntoExpress software to classify them and put them into categories based on gene ontology classification (42). Supplemental Fig. 3 provides a graphical representation of the molecular function analysis, biological process analysis, and cellular component analysis for the proteins encoded by all 182 genes. Supplemental Figs. 4 and 5 provide the same type of information for only the Rhox5 up-regulated and down-regulated genes, respectively. Compared with the whole mouse genome, the Rhox5-regulated genes were not statistically overrepresented in any one category, but instead were involved in a wide array of molecular and cellular functions.
In vitro regulation and cellular site of expression in vivo
To verify the regulation observed by microarray analysis, we performed qPCR analysis on 15 of the 182 genes (Table 1). Nine of them encode membrane-bound proteins, which we chose because such proteins could mediate Rhox5-dependent signaling between Sertoli cells and germ cells. The other six genes were selected for various reasons. Carboxypeptidase X1 (Cpxm1) and ganglioside-induced differentiation-associated protein 1 (Gdap1) were chosen because they are dramatically regulated by Rhox5, according to our microarray analysis (Table 1). BTB and CNC Homology 2 (Bach2) and kruppel-like factor 9 (Klf9) encode transcription factors with well-established functions; the latter is known to have a role in female reproduction and neuronal development (43,44). Last, isocitrate dehydrogenase 3 (Idh3a) and 2,3-bisphosphoglycerate mutase (Bpgm) were selected because they encode proteins directly involved in metabolism. This was of interest because we have found that Rhox5 regulates many other genes involved in metabolism (determined by use of an independent approach to the one we took here; MacLean, J., Z. Hu, and M. F. Wilkinson, unpublished observations).
Table 1.
Rhox5-regulated genes
| Genesymbol | Gene name | Array | qPCR | Molecularfunction | Cellularcomponent |
|---|---|---|---|---|---|
| Tmem47 | Transmembrane protein 47 | 253 | 519 | Signal transduction | Membrane |
| Cd24a | CD24a antigen | 16.6 | 15.6 | Apoptosis | Membrane |
| Gdap1 | Ganglioside-induced differentiation associatedprotein 1 | 10.5 | 4.0 | Signal transduction | Cytoplasm |
| Tfrc | Transferrin receptor | 8.5 | 2.8 | Transport | Membrane |
| Bpgm | 2,3-bisphosphoglycerate mutase | 3.0 | 2.7 | Metabolism | Unknown |
| Idh3a | Isocitrate dehydrogenase 3 | 2.6 | 5.0 | Metabolism | Mitochondrion |
| Fzd2 | Frizzled homolog 2 | −2.1 | −2.7 | Signal transduction | Membrane |
| Tmem176b | Transmembrane protein 176B | −2.1 | −1.6 | Signal transduction | Membrane |
| Tmem176a | Transmembrane protein 176A | −2.2 | −2.2 | Unknown | Membrane |
| Bach2 | BTB and CNC homology 2 | −2.6 | −6.6 | Transcription factor | Nucleus |
| Pltp | Phospholipid transfer protein | −3.2 | −5.0 | Transport | Membrane |
| Klf9 | Kruppel-like factor 9 | −3.2 | −2.3 | Signal transduction | Nucleus |
| Ifnar2 | Interferon (α and β) receptor 2 | −3.5 | −3.2 | Signal transduction | Membrane |
| Unc5c | Unc5 homolog C | −3.5 | −9.5 | Signal transduction | Membrane |
| Cpxm1 | Carboxypeptidase ×1 | −8.1 | −10.3 | Transport | Extracellular |
The “Array” column indicates the average change in mRNA level in the Rhox5-positive expressing 15P-1 Sertoli cell clones (nos. 6 and 24) relative to the Rhox5-negative cell clones (nos. 14 and 16), as judged by microarray analysis. The “qPCR” column lists the average mRNA levels of the same genes, as judged by use of qPCR analysis, normalized against the level of L19 mRNA (see Fig. 1B). Positive and negative values denote gene expression that was up-regulated and down-regulated, respectively, in response to Rhox5. The genes were classified into “molecular function” and “cellular component” by use of OntoExpress software.
To verify the regulation of these 15 genes, qPCR was performed on total cellular RNA from the four original 15P-1 clones used for microarray hybridization (6, 24, 14, and 16), and other 15P-1 stable clones expressing different levels of Rhox5 mRNA (clones 1, 4, 11, 19, and 26; data not shown). This analysis demonstrated that all 15 of the transcripts that we tested were regulated in precisely the same manner as indicated by microarray analysis (Table 1). Furthermore, the magnitude of the regulation for most of the transcripts was similar when measured by either qPCR analysis or microarray analysis.
To assess which of these 15 genes are candidates to be direct targets of Rhox5, we assessed whether they are expressed in Sertoli cells, the site of Rhox5 expression (29). To do this, we generated an enriched Sertoli cell fraction from adult testes by use of a modified protocol involving gravity sedimentation and enzymatic treatments (see Materials and Methods). This procedure also generates purified interstitial (mainly Leydig) cells, which allowed us to determine whether any of these Rhox5-regulated genes are also expressed in this somatic cell subset. Hypotonic shock was used to lyse most of the contaminating germ cells in both Sertoli- and interstitial-enriched fractions (45). By comparing gene expression in Sertoli and interstitial cell fractions prepared with or without hypotonic shock, we were able to assess the contribution of germ cells. Although it is not possible to generate entirely pure populations of these cell types, our protocol yielded fractions that were substantially enriched, based on their relative expression of Sertoli (Gata1 and Rhox5) (29,30,31,46) and Leydig cell markers (LH receptor [Lhr]) (47). To assess the effectiveness of hypotonic shock in removing germ cells, we assessed the levels of two germ cell-specific genes: testicular orphan receptor-2 (Tr2) and Deleted in Azoospermia-like (Dazl) (48,49).
Table 2 shows the expression of the 15 Rhox5-regulated genes in the different cell subsets, as assessed by qPCR analysis. Five of the genes were predominantly expressed in Sertoli cells (group 1), based on two criteria. First, they were more highly expressed in the Sertoli cell fraction that underwent hypotonic shock and than the one that did not (under the headings Sertoli cells and Sertoli no-shock, respectively in Table 2). Second, they were more highly expressed in the purified Sertoli cell fraction than in total testes (the latter of which was given a value of 1). Of note, however, we found that the magnitude of the increased expression in the purified Sertoli cell fraction (relative to total testes) varied considerably between genes. This may, in part, be the result of some of the genes being expressed in other cell types in the testes. Another possible contributing factor is that our procedure may not equally purify Sertoli cells in all stages of the seminiferous epithelial cycle. This would lead to different levels of enrichment, depending on the pattern of expression of a given gene during the seminiferous epithelial cycle. For example, the positive control, Gata1, has been shown to be expressed exclusively by Sertoli cells, but because it exhibits stage-specific expression (50), this could explain why we found its expression was increased by less in the purified Sertoli cell fraction (∼4-fold) than one of the Rhox5-regulated genes, interferon receptor-2 (Ifnar2), which exhibited approximately 8-fold increased expression. As another example, the Rhox5-regulated genes, Tfc, which encodes the transferrin receptor, was previously shown to be specifically expressed only by Sertoli cells in the testes (51), but we found it was only expressed at approximately 2-fold higher levels in the purified Sertoli cell fraction than in total testes. We suggest that this may be explained by its stage-specific expression (52). We did find that Tfc expression was relatively Sertoli cell-specific, as Tfc mRNA was at approximately 10-fold higher levels in the purified Sertoli cell fraction than in the purified Leydig cell fraction.
Table 2.
Cell types expressing Rhox5-regulated genes
| Gene | Sertoli cells | Interstitial cells | Sertoli no-shock | Interstitial no-shock | Primary source |
|---|---|---|---|---|---|
| Group 1 | |||||
| Gdap1 | 2.2 | 1.0 | 1.2 | 1.6 | Sertoli |
| Tfrc | 1.9 | 0.2 | 0.7 | 0.5 | Sertoli |
| Tmem176b | 2.7 | 0.4 | 0.2 | 0.3 | Sertoli |
| Tmem176a | 2.8 | 0.8 | 0.3 | 0.3 | Sertoli |
| Ifnar2 | 8.3 | 0.9 | 0.4 | 1.4 | Sertoli |
| Group 2 | |||||
| Tmem47 | 8.9 | 2.2 | 2.7 | 0.1 | Sertoli + interstitial |
| Fzd2 | 8.3 | 6.3 | 1.7 | 0.5 | Sertoli + interstitial |
| Pltp | 5.5 | 4.3 | 0.9 | 1.6 | Sertoli + interstitial |
| Unc5c | 1.9 | 2.0 | 1.1 | 1.4 | Sertoli + interstitial |
| Group 3 | |||||
| Cd24a | 0.7 | 7.7 | 0.7 | 1.3 | Interstitial |
| Group 4 | |||||
| Bpgm | 0.4 | 0.7 | 0.1 | 0.4 | Non-Sertoli/interstitial |
| Idh3a | 0.9 | 0.1 | 1.1 | 1.1 | Non-Sertoli/interstitial |
| Bach2 | 0.5 | 0.4 | 0.8 | 1.1 | Non-Sertoli/interstitial |
| Klf9 | 0.8 | 0.7 | 1.1 | 0.9 | Non-Sertoli/interstitial |
| Cpxm1 | 0.2 | 0.04 | 0.4 | 0.9 | Non-Sertoli/interstitial |
| Markers | |||||
| Gata1 | 4.2 | 0.1 | 3.8 | 0.1 | Sertoli |
| Rhox5 | 5.3 | 0.9 | 1.5 | 0.2 | Sertoli |
| Lhr | 0.6 | 19.9 | 0.2 | 1.0 | Interstitial |
| Tr2 | 0.7 | 0.2 | 1.0 | 1.9 | Germ |
| Dazl | 0.5 | 0.1 | 2.2 | 1.0 | Germ |
qPCR analysis of total cellular RNA from purified Sertoli and interstitial cells obtained from adult testes. The “no shock” columns refer to the semipurified Sertoli and interstitial cell fractions that had not undergone hypotonic shock to remove most germ cells. The numbers are mRNA level (normalized against L19 mRNA; see Fig. 1B) in the cell fractions relative to total testes, the latter of which was arbitrarily given a value of “1.’
We divided the other 9 Rhox5-regulated genes into three categories: 1) those expressed in both the Sertoli- and interstitial-cell fractions (group 2), 2) those expressed predominantly in the interstitial-cell fraction (group 3), and 3) those predominantly expressed in testicular cell types other than Sertoli cells or interstitial cells (group 4). The group 2 genes are candidates for directly regulation by Rhox5, whereas those in groups 3 and 4 probably are indirectly regulated through paracrine signaling from Sertoli cells. We note, however, that Bpgm, which is in group 4 because it was less expressed in the Sertoli and interstitial fractions than in total testes, exhibited significantly increased expression in purified Sertoli cells than in nonshocked Sertoli cells, which suggests that it is expressed in at least a subset of Sertoli cells. Together, these results provide evidence that approximately 60% of the Rhox5-regulated genes that we selected for in-depth analysis are expressed in Sertoli cells in the adult testes in vivo.
In vivo regulation
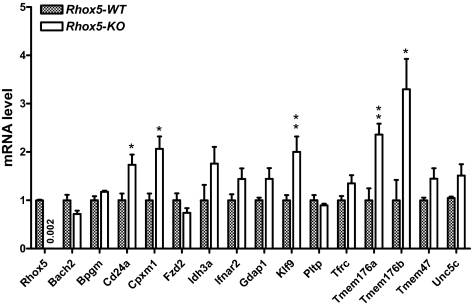
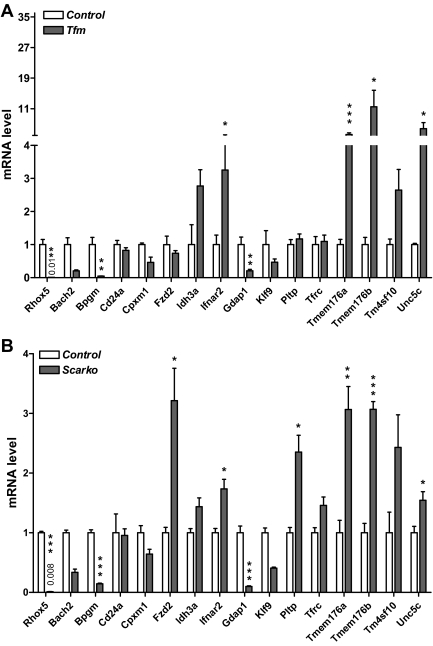
Although the 15P-1 cell line has many Sertoli cell characteristics (35,37), it is an immortalized cell line adapted to tissue culture, and thus it does not completely mimic normal Sertoli cells. Hence, it was crucial to determine whether the genes regulated by Rhox5 in 15P-1 cells were also regulated by Rhox5 in vivo. We took three approaches to address this question. First, we compared their expression in adult testes from Rhox5-null and control mice. Second, we compared their expression in purified somatic cell fractions from Rhox5-null and control adult testes. Third, we compared their postnatal pattern of expression in Rhox5-null and control mice. The first approach revealed that 5 of the 15 genes were significantly regulated by Rhox5 in the adult testis: cluster-w4 antigen (Cd24a), transmembrane protein (Tmem) 176a, Tmem176b, Klf9, and Cpxm1 (Fig. 2). All five of these genes were up-regulated in the Rhox5-null adult testes, indicating that they are negatively regulated by Rhox5 in vivo. Consistent with this finding, four of these genes were repressed by Rhox5 in 15P-1 cells (Table 1). The lone exception, Cd24a, was strongly up-regulated by Rhox5 in 15P-1 cells, whereas it was modestly down-regulated by Rhox5 in the adult testes. Three of the genes—Tmem176a, Tmem176b, and Klf9—were down-regulated by similar magnitudes in 15P-1 cells and the adult testis.
Figure 2.
Genes regulated by Rhox5 in the adult testis. qPCR analysis of total cellular RNA from adult Rhox5-null (KO) and wild-type (WT) littermate testes. mRNA levels were quantified as described in Fig. 1B. Statistical analysis was performed by use the two-tailed Student’s t test: *, P < 0.05; **, P < 0.01.
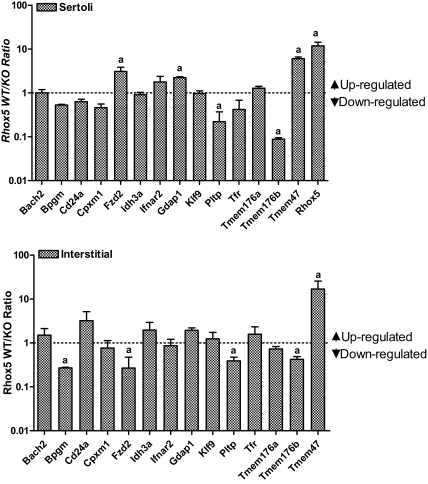
Because molecular analysis of the whole testes will often not reveal genes regulated by Rhox5 in only a specific subset of testicular cells, we also examined Rhox5-mediated regulation in adult testicular cell subsets. Because we found that most of the Rhox5-regulated genes are expressed in the enriched Sertoli cell and interstitial cell fractions, we examined these two cell subsets. Figure 3 shows the ratio of gene expression in these two fractions prepared from Rhox5-null and control mice (note that the ratio is the average of two independent experiments and is presented on a log scale). The data indicate that several genes are regulated by Rhox5 in the Sertoli cell fraction, the interstitial cell fraction, or both. Of the five genes up- or down-regulated by 2-fold or more in Sertoli cells (indicated with an a in Fig. 3, upper panel), four were regulated in the same manner in response to Rhox5 in 15P-1 cells (Table 1). The lone exception, Frizzled homolog 2 (Fzd2), was up-regulated by Rhox5 in the Sertoli cell fraction in vivo and modestly down-regulated by Rhox5 in 15P-1 cells. However, Fzd2 expression was down-regulated by Rhox5 in the interstitial fraction, indicating that it is regulated in the same manner by Rhox5 in interstitial cells in vivo as it is in 15P-1 cells. Three of the other four genes regulated by 2-fold or more in the interstitial cell fraction (indicated with an a in Fig. 3, lower panel) were also regulated in the same manner in vitro (Table 1). Together, these data indicated that four additional genes (beyond those identified in total adult testes) are Rhox5 regulated in vivo: Fzd2, Gdap1, Pltp, and Tmem47 (also known as Tm4sf10). All four of these genes are expressed in purified Sertoli cells (Table 2) and regulated by Rhox5 in Sertoli cells (Fig. 3), indicating that they are all candidates to be direct targets of RHOX5.
Figure 3.
Cell subset expression pattern of Rhox5-regulated genes. qPCR analysis of total cellular RNA from purified Sertoli and interstitial cells (prepared by use of hypotonic shock to remove germ cells) from adult Rhox5-null (KO) and wild-type (WT) littermate mice testes. The value shown for each gene is the average WT/KO mRNA ratio from two independent cell preparations, quantified as described in Fig. 1B. a, mRNA ratios of either ≥2 or ≤0.5 (i.e. >2-fold change).
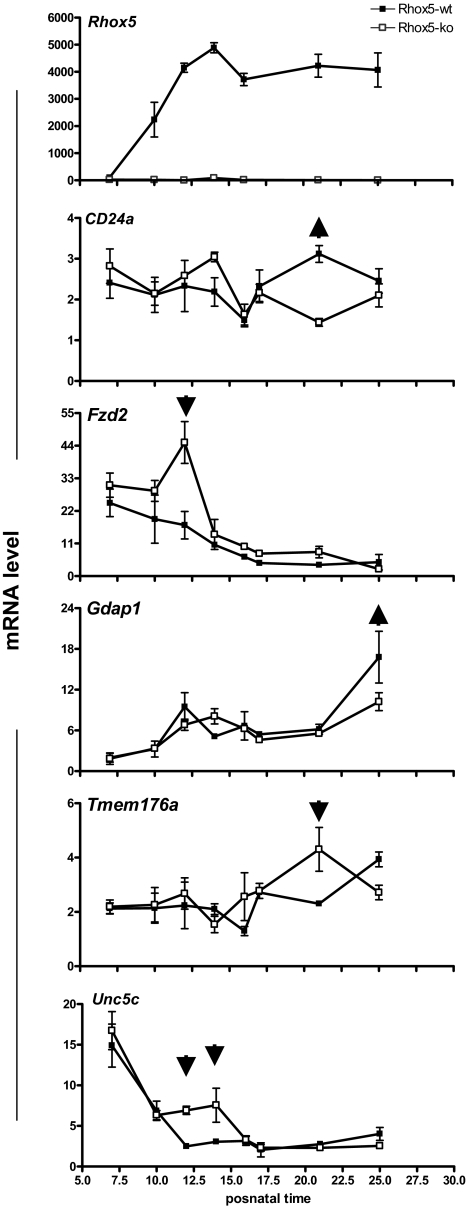
As a final approach to assess the regulation of these 15 genes by Rhox5 in vivo, we examined their expression pattern during the first wave of spermatogenesis in Rhox5-null and control mice. This approach provided an independent assessment of Rhox5-mediated regulation and it had the potential to pinpoint the stage(s) of spermatogenesis at which Rhox5 acts on its target genes. This analysis demonstrated that 5 of the 15 genes were significantly regulated by Rhox5 at one or more time points during the first wave of spermatogenesis (Fig. 4 and supplemental Fig. S6). Although some of these genes were regulated by Rhox5 only transiently during the first wave of spermatogenesis (i.e. at very specific postnatal time points), the regulation was reproducible. Furthermore, all five of these genes exhibited the same regulation in vitro. Thus, three of the genes (Fzd2, Unc5c, and Tmem176a) were down-regulated by Rhox5 both in vivo and in 15P-1 cells, whereas the other two genes (Cd24a and Gdap1) were up-regulated by Rhox5 both in vivo and in 15P-1 cells (Fig. 4 and Table 1). Three of the genes (Fzd2, Gdap1, and Tmem176a) were also regulated in adult testes in the same manner as they were in postnatal testes (Figs. 2 and 3). Four of the genes were expressed and/or regulated in purified Sertoli cells (Table 2 and Fig. 3), indicating that they are candidates to be direct targets of Rhox5.
Figure 4.
Developmental expression pattern of Rhox5-regulated genes. qPCR analysis of testes RNA from Rhox5-null (KO) and wild-type (WT) littermate mice of the indicated postnatal ages (six mice per time point). All values were quantified as described in Fig. 1B. Postnatal time points at which Rhox5 regulates the indicated genes are denoted by arrows (upward and downward arrows indicate genes that are up-regulated and down-regulated, respectively). A value of 1 was arbitrarily given to the lowest expression level among the time points examined for a given gene. Error bars indicate standard error.
Together, the data derived from these three different approaches indicated that 10 of the 15 genes regulated by Rhox5 in the 15P-1 Sertoli cell line are also regulated by Rhox5 in the testis in vivo. Seven of these 10 genes are expressed in the enriched Sertoli cell fraction and thus may be directly regulated by Rhox5 (Table 2).
Rhox5 targets Ar-regulated genes
The Rhox5 gene depends on the nuclear hormone receptor AR and androgen for expression in Sertoli cells (see the introductory section and Discussion). This led us to hypothesize that a subset of Rhox5-regulated genes are also regulated by AR and androgen. In other words, we hypothesized that a subset of Rhox5-regulated genes are SAR genes that are androgen-regulated through the action of Rhox5. As a first test of this hypothesis, we determined whether any of the 15 Rhox5-regulated genes described above are regulated by AR in vivo. To do this, we examined their expression in Sertoli cell AR-knockout (Scarko) mice, which have Ar mutated conditionally in Sertoli cells (53), and testicular feminized (Tfm) mice, which have a naturally occurring mutation in the Ar gene that inactivates AR function (54). qPCR analysis of adult testes from these mutant mice revealed that 11 of the 15 Rhox5-regulated genes had significantly altered expression compared with control testes (Fig. 5). Eight of these genes were regulated in the same manner by Ar and Rhox5 (either positively or negatively in response to both), indicating that these are candidate SAR genes. Six of them had significantly altered expression in both Scarko and Tfm mice, providing strong evidence that these genes are bona fide AR-regulated genes. The other two genes, Fzd2 and Pltp, were only regulated in Scarko mice, not Tfm mice. Although many possible explanations for this exist, a likely one is that because Tfm mice are cryptorchid (55), the abnormally high temperature of the testes in these mice prevents normal AR-mediated regulation of these two genes.
Figure 5.
Identification of Ar- and Rhox5-regulated genes. A, qPCR analysis of testes RNA from adult Tfm and littermate control mice. B, qPCR analysis of testes RNA from adult Scarko and littermate control mice. All values were quantified as described in Fig. 1B. Control mRNA levels were arbitrarily given a value of 1. Statistical analysis was performed by use the two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The signature feature of SAR genes is that they do not directly respond to AR (39,56). To determine whether the eight candidate SAR genes have the potential to respond to AR, we examined their 5′- and 3′-flanking regions for androgen-response elements (AREs) by use of the Gemomatix program. Four of these genes had no consensus AREs, whereas the other four had either one or two consensus AREs (supplemental Table 3). To empirically determine whether the eight candidate SAR genes do not directly respond to AR, we examined whether their expression was altered in response to AR and androgen under conditions when Rhox5 is not induced. This was accomplished by use of 15P-1 cells, which do not exhibit increased Rhox5 mRNA levels in response to the testosterone analog R1881 and transfection with an AR expression plasmid (supplemental Fig. S6). We found that AR and R1881 did not significantly affect the expression of any of the eight candidate SAR genes in the 15P-1 cell line (supplemental Fig. S7). In contrast, AR and R1881 did increase the expression of the AR-regulated gene Fabp (data not shown).
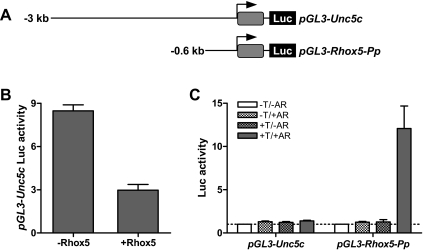
As another test of AR/androgen responsiveness, we generated a luciferase reporter construct containing 3 kb of 5′-flanking sequence from the candidate SAR gene Unc5c (Fig. 6A). We chose Unc5c because it harbors no consensus AREs in this region (Table 3 and data not shown). When cotransfected into the MSC1 Sertoli cell line with an AR expression plasmid and incubated with R1881, this Unc5c promoter construct did not exhibit altered luciferase expression (Fig. 6B). In contrast, the positive control, a luciferase reporter construct containing the Rhox5 proximal promoter (Pp) (Fig. 6A), expressed greatly elevated levels of luciferase in response to AR and R1881 (Fig. 6B). Note that, whereas the Rhox5 Pp reporter was greatly increased in expression in response to AR and R1881, the endogenous Rhox5 gene was not significantly elevated in expression (as judged by qPCR analysis; data not shown), probably because it is in a repressed state because of DNA methylation (17,57). The ability of Unc5c to respond to AR only under conditions in which Rhox5 expression is also induced (Figs. 5 and 6), coupled with the ability of Unc5c to be regulated by Rhox5 both in vitro and in vivo (Table 1 and Fig. 4, respectively), provides strong evidence that Unc5c is regulated by AR through the action of Rhox5.
Figure 6.
The candidate SAR gene Unc5c responds to Rhox5 but not Ar and androgen in vitro. A, Schematic diagram of Unc5c promoter and Rhox5 proximal promoter (Pp) constructs harboring the firefly luciferase (Luc) reporter. B, Luc expression from MSC1 cells transiently cotransfected with pGL3-Unc5c (100 ng), a Renilla luciferase vector (pRL-Tk) (25 ng) for an internal control, and either a Rhox5-expression vector or the corresponding empty expression vector (300 ng). C, Luc expression from MSC1 cells transiently transfected with the constructs shown in panel A (100 ng) and pRL-Tk (25 ng). Where indicated with +AR, the cells were cotransfected with an AR expression vector (100 ng). Where indicated with +T, the cells were incubated with the synthetic androgen R1881. The relative Luc activities shown are relative to nontreated cells (which were arbitrarily given a value of 1) from three independent transfection experiments. Error bars indicate standard error. Note that AR + T did not induce endogenous Rhox5 expression in these cells (data not shown), explaining why Unc5c expression was not regulated.
Identification of common targets of AR-regulated Rhox genes
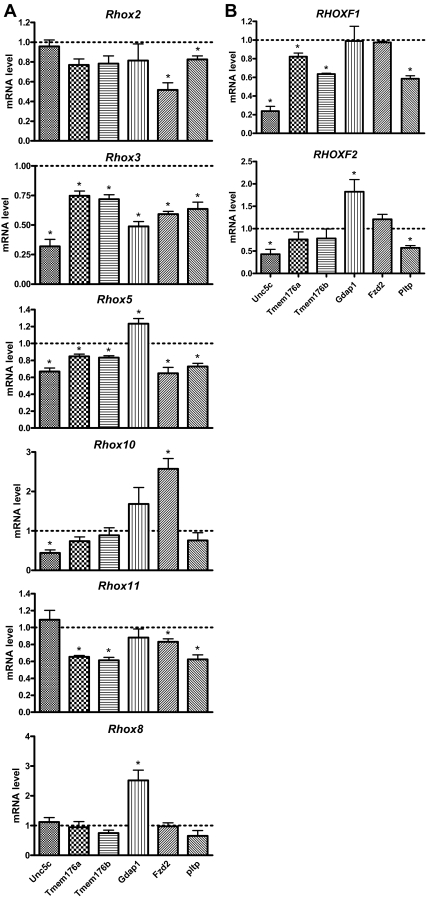
In the previous section, we provided evidence that we had identified SAR genes that are regulated by AR through the action of the AR-inducible gene Rhox5. As another means to address whether we identified bona fide SAR genes, we determined whether they are regulated by other AR-inducible Rhox genes. We previously demonstrated that Rhox2, Rhox3, Rhox10, and Rhox11 are all strongly up-regulated in expression in response to AR and androgen in Sertoli cells (7). Thus, we examined whether any of these other AR-inducible Rhox genes were capable of regulating the putative SAR genes we had identified. The putative SAR genes that we selected for this analysis were those that fulfilled three criteria: 1) regulated by Rhox5 in vivo, as judged by our three-prong analysis (Figs. 2–4); 2) regulated by AR in vivo (Fig. 5); and 3) significantly expressed in the purified Sertoli cell fraction (Table 2, category 1 or 2). The six genes that fulfilled these criteria were Gdap1, Tmem176a, Tmem176b, Fzd2, Pltp, and Unc5c. To determine whether these six genes are regulated by AR-regulated Rhox genes, we transiently transfected expression vectors encoding RHOX2, RHOX3, RHOX5, RHOX10, and RHOX11 into 15P-1 cells (we previously showed that all these RHOX proteins are all expressed at similar levels from these expression vectors) (32). qPCR analysis revealed that each of the six SAR genes was significantly regulated in response to at least two of the AR-inducible Rhox genes (Fig. 7A). For example, Unc5c was negatively regulated by Rhox3, Rhox5, and Rhox10. Tmem176a and Tmem176b were also regulated by Rhox3 and Rhox5, but instead of being regulated by Rhox10, these two genes responded to Rhox11. Pltp had the same pattern of expression as Tmem176a and Tmem176b, except that Rhox2 also negatively regulated its expression. Fzd2 had an unusual pattern of expression in that it was negatively regulated by Rhox2, Rhox3, Rhox5, and Rhox11, but was positively regulated by Rhox10. To assess specificity, we also evaluated the effect of Rhox8, which is not AR regulated, either as assessed in a Sertoli cell line (7) or in postnatal and adult testes in vivo (MacLean, J., K. de Gendt, G. Verhoeven, and M. F. Wilkinson, unpublished observations). We found that forced expression of Rhox8 did not significantly affect the expression of any of the SAR genes except for Gdap1. Rhox8 positively regulated Gdap1, as did Rhox5 and Rhox10. It is noteworthy that we found that transiently transfected Rhox5 only modestly regulated the SAR genes (Fig. 7A), which contrasted with its more dramatic effects on some of the SAR genes in cells stably transfected with Rhox5 (Table 1; i.e. Gdap1, Pltp, and Unc5c). Regardless of this quantitative difference, Rhox5 had the same qualitative effect on the six SAR genes in transiently and stably transfected cells (i.e. Rhox5 negatively regulated all the SAR genes except for Gdap1 under both conditions).
Figure 7.
Regulation of candidate SAR genes by Ar-inducible Rhox genes. A, qPCR analysis of total cellular RNA from 15P-1 Sertoli cells transiently transfected with expression vectors encoding the indicated mouse Rhox genes or the empty expression vector (800 ng). A value of 1 was arbitrarily given to the expression of the indicated genes in cells transfected with the empty expression vector. All values were quantified as described in Fig. 1B. B, Same as panel A, except the cells were transiently transfected with expression vectors encoding the human RHOXF1 and RHOXF2 proteins. Statistical analysis was performed by using the two-tailed student’s t-test (*, P < 0.05).
To determine whether the regulation of the putative SAR genes is a conserved response, we examined the effect of forced expression of the human RHOX genes RHOXF1 and RHOXF2. These are the only human RHOX genes that have been characterized; both are selectively expressed in human testes (58,59). RHOXF1 is an androgen-induced gene, whereas RHOXF2 has not been characterized in this regard (58). We found that transient transfection of expression vectors encoding either RHOXF1 or RHOXF2 triggered the down-regulation of Unc5c and Pltp (Fig. 7B). RHOXF2, but not RHOXF1, up-regulated Gdap1 expression, whereas RHOXF1, but not RHOXF2, modestly decreased the expression of both Tmem176 and Tmem176b. The only putative SAR gene that did not significantly respond to either RHOXF1 or RHOXF2 was Fzd2. We conclude that human RHOXF1 and RHOXF2 share with androgen-regulated mouse Rhox family members the ability to regulate most of the SAR genes, suggesting that this is a conserved response.
Discussion
Homeobox genes were originally identified over 25 yr ago and have since been shown to control a wide variety of developmental and cellular events from yeast to mammals, yet surprisingly little is known about the gene networks downstream of homeobox genes. The discovery of the Rhox homeobox gene cluster provides an opportunity to identify gene networks important for male and female gametogenesis. In this report, we focused on identifying genes downstream of Rhox5, the founding member of the Rhox gene cluster. This Sertoli cell-expressed gene promotes the survival of mouse male germ cells in all stages of the seminiferous epithelial cycle (7). Rhox5 is also required to generate normal levels of motile spermatozoa and for normal male fertility (7). To identify physiologically relevant genes downstream of Rhox5 that might mediate these functions, we took a two-prong approach in which we first identified Rhox5-regulated genes by use of genome-wide expression profiling in a Sertoli cell line, and then followed that up with in vivo analysis. We favored the use of a Sertoli cell line for our initial screen of Rhox5-regulated genes for several reasons. First, this allowed us to identify regulated genes in a relatively homogenous set of cells. By contrast, analysis of whole testes and testicular cell subsets has potential problems with cellular heterogeneity and cellular contamination, respectively. Second, the Sertoli cell line we chose for our analysis, 15P-1, expresses little or no Rhox5 mRNA or RHOX5 protein (supplemental Figs. 1 and 2, respectively), which allowed us to take a simple gain-of-function approach to identify Rhox5-regulated genes. Third, we carefully selected stably transfected 15P-1 cell clones that expressed normal levels of Rhox5 to allow us to screen for physiologically relevant Rhox5-regulated genes. Finally, 15P-1 is an ideal cell line to perform molecular studies on the molecular mechanisms by which RHOX5 regulates its target genes because these cells can be efficiently transfected and they exhibit many normal Sertoli cell characteristics, including many Sertoli cell markers and the ability to support the differentiation of germ cells (35,36,37,40,41).
Using this approach, we identified many genes regulated by Rhox5 (Table 1), including those encoding proteins involved in a wide variety of functions (supplemental Figs. 3–5). We focused on 15 of these genes first by verifying that they are regulated by Rhox5 in 15P-1 cells (Table 1) and then by conducting an in-depth analysis of their expression pattern and regulation in response to Rhox5 in vivo (Figs. 2–4). By analyzing 1) adult testes, 2) purified cell fractions from adult testes, and 3) postnatal testes from many time points during the first wave of spermatogenesis from both Rhox5-null and control littermate mice, we found that at least 11 of these 15 genes are regulated by Rhox5 in vivo (Table 1, Figs. 2–4). Seven of these 11 in vivo Rhox5-regulated genes are expressed in Sertoli cells, based on our cell-subset fractionation analysis (Table 2), and thus are candidates to be directly regulated by Rhox5. Direct analysis of whether they are direct targets is complicated by the need for a ChIP-grade RHOX5 antisera and by the fact that the cis elements responsible for regulation could be far from their promoter regions. We showed that many of these Sertoli cell-expressed genes are androgen and/or AR regulated (Fig. 5), and thus they may mediate some of the actions of androgen in the testes, as discussed in more detail below. Six of these seven genes encode cell surface proteins and thus they may be involved in cell-cell signaling between Sertoli cells and/or between Sertoli and developing germ cells.
Although some Rhox5-regulated genes appeared to be expressed and regulated exclusively in Sertoli cells, many Rhox5-regulated genes were expressed and/or regulated in other testicular cell types. For example, we found that a number of the Rhox5-regulated genes were expressed in both the Sertoli and interstitial cell fractions (Table 2, category 2); some of these were regulated by Rhox5 in the interstitial cell fraction (Fig. 3). We also identified a gene exclusively expressed in the interstitial cell fraction—Cd24a (Table 2, category 3)—that exhibited modestly altered expression in Rhox5-null mice (Figs. 3 and 4), suggesting that Cd24a is regulated by Rhox5 in a paracrine manner. Other Rhox5-regulated genes were expressed at very low levels in both the Sertoli and interstitial cell fractions (Table 2, category 4) and thus these genes may be regulated by Rhox5 in other testicular cell types, such as germ cells or myoid cells. The overall picture that emerges from this analysis of 15 Rhox5-regulated genes is that, even though RHOX5 protein is restricted to Sertoli cells (30,31), its web of regulation extends to various cell types in the testes.
Because Rhox5 is an androgen- and AR-induced homeobox gene (29,30,31), it is a candidate to encode a transcription factor that mediates some of the actions of androgen in the testes. This predicts that a subset of RHOX5 targets are SAR genes; i.e. genes indirectly regulated by AR through the action of intermediate factors (60). Although several SAR genes have been defined in the prostate (39,61), to our knowledge none have been defined in the testes. It is crucial to define SAR genes in the testis, as such genes are likely to be mediators of androgen action in this male reproductive organ. Progress on this front has been stymied because very few androgen-regulated transcription factors have been identified in the Sertoli cell, the only cell type definitively known to mediate androgen action in the testes (6). To our knowledge, the only androgen-induced transcription factor genes that have been defined in Sertoli cells are Rhox5 and c-myc. Rhox5 is androgen- and AR- induced in both cultured Sertoli cells and Sertoli cells in vivo (29,30,31,46,53,62,63,64), whereas c-myc is androgen-inducible in cultured Sertoli cells (65), but it appears not to be AR-regulated in vivo (53,55). In this paper, we provide several lines of evidence that we have identified eight SAR genes that are regulated by AR through the action of Rhox5 in the testes. First, all eight of these genes are regulated by Rhox5 in 15P-1 Sertoli cells (Table 1), and all but one of them (Ifnar2) are regulated by Rhox5 in the testes in vivo (Figs. 2–4). Second, our analysis of Ar-deficient mice indicated that all eight of these genes are AR-regulated in adult testes in vivo (Fig. 5). As confirmation of this, we found that five of these genes (Bpgm, Cpxm1, Ifnar2, Tmem176a, and Tmem176b) were independently defined as AR-regulated by a study that used microarray analysis to identify genes differentially expressed in postnatal Tfm Ar-deficient and control mice testes (55). Third, our analysis of the putative regulatory regions in the candidate SAR genes showed that four of them had no consensus AREs and the other four had few consensus AREs (Table 3). Because, by definition, SAR genes are regulated indirectly by AR, this paucity of AREs is consistent with these genes being bona fide SAR genes. We should note, however, that ARE consensus sites are often not functional, and functional AREs often differ considerably from the consensus sequence (66). Fourth, none of the eight genes were regulated by AR and androgen under conditions in which Rhox5 was not induced (supplemental Fig. 7). Fifth, a reporter construct harboring the promoter region of one of the candidate SAR genes (Unc5c) was responsive to Rhox5, but was not responsive to AR and androgen under conditions in which Rhox5 was not induced (Fig. 6). Sixth, several androgen/AR-inducible mouse Rhox genes besides Rhox5 regulated many of the candidate SAR genes (Fig. 7A). Finally, RHOXF1, an androgen-inducible human RHOX gene (58), was also capable of regulating some of the candidate SAR genes (Fig. 7B), providing evidence that this is a conserved response. Human RHOXF2 also regulated many of these genes (Fig. 7B), but whether it is an androgen-inducible gene has not yet been determined.
Consistent with the often noted connection between the testis and the brain (67), we found that a remarkably large proportion of the candidate SAR genes encode proteins with known functions in the nervous system. Unc5c encodes a transmembrane receptor of the immunoglobulin superfamily that binds to netrin-1, a protein important for many developmental events in the brain, including governing axon migration by triggering chemorepulsion (68). UN5C also has a death domain, which engenders it with proapoptotic activity that may also be important for its roles in brain development (69). We recently showed that UNC5C functions similarly in the testes by promoting the death of male germ cells (32). This germ cell death-promoting property, coupled with the fact that Unc5c gene expression is negatively regulated by Rhox5, makes Unc5c a good candidate to have a role in Rhox5’s ability to promote the survival of germ cells (7). Another candidate SAR gene, Gdap1, is up-regulated during neuron differentiation (70) and, when mutated in humans, is responsible for many cases of Charcot-Marie-Tooth disease, the most common inherited form of peripheral neuropathy. GDAP1 is a member of the glutathione-S-transferase family, and thus, its neurological protective role may derive from its antioxidant properties. Tmem176b (also known as Clast1, Lr8, and TORID) encodes a ubiquitously expressed four-transmembrane cell surface protein that is crucial for normal brain development and function. Targeted deletion of Tmem176b in mice causes defects in the cerebellum and severe ataxia (71). It has not been reported whether Tmem176b-mutant mice have defects in male fertility, but our finding that both Ar and Rhox5 regulate its expression suggests it may have some role in androgen-mediated events in the testes. A related four-transmembrane protein-encoding gene, Tmem176a (also known as GS188 and HCA112), is another putative SAR gene. At present, there is no information on either the biochemical or biological function of Tmem176a. Another candidate SAR gene, Pltp, encodes a protein that acts both inside and outside cells to mediate the transfer of lipids. Although PLTP is fairly widely expressed and therefore probably acts in many cell types, it appears to be particularly important for brain function, in part, by virtue of its being secreted by neurons and glial cells. PLTP is likely to have roles in neurodegenerative and inflammatory brain diseases, including Alzheimer’s disease, because PLTP levels are altered in many of these disease states. PLTP also alters the phosphorylation state of TAU, a neuron-specific protein that undergoes shifts in phosphorylation status in many neurological diseases (72).
The remaining three candidate SAR genes, Fzd2, Ifnr2, and Bpgm, are likely to have roles in many cell types. Fzd2, encodes one of the 10 known frizzled receptors in mice; these are cell surface receptors that bind to the WNT family of secreted glycoproteins. On binding of WNT proteins, frizzled receptors elicit signal transduction events crucial for conserved developmental steps in many organisms. Because frizzled receptors tend to act redundantly, it has been difficult to assign precise functions to them, including FZD2 (73). Ifnar2 encodes the cell surface receptor for antiviral cytokines called interferons. On interferon binding, IFNAR2 activates the STAT signaling pathway, which in turn elicits an antiviral response, including rapid mRNA decay and translational repression (74). Bpgm encodes a multifunctional enzyme best known for its role in controlling the metabolism of 2,3-diphosphoglycerate in erythrocytes (75). It will be interesting to know what specific roles BPGM has in the testes.
In addition to the candidate SAR genes described above, we identified four other genes regulated by Rhox5 in the testes in vivo: Cd24a, Klf9, Cpxm1, and Tmem47 (Figs. 2–4). Two of these, Cd24a and Klf9, have known roles in the nervous system. Cd24a encodes a glycosylphosphatidylinositol-anchored glycoprotein that was originally identified based on its expression in immune cells, but it was subsequently shown to also be expressed and have a role in neurons. In the adult mouse central nervous system, CD24 expression is restricted to immature proliferating neurons in regions of the brain undergoing neurogenesis. Cd24a-knockout mice exhibit increased numbers of proliferating neurons in these zones, indicating that CD-24 serves to dampen the generation of adult neurons (76). Kfl9 (also known as Bteb1) encodes a zinc-finger transcription factor that when knocked out in mice leads to cerebellar Purkinje cell defects and behavioral defects consistent with its having roles in the cerebellum, amygdala, and hippocampus (77). Whereas exactly how KLF9 functions in the brain is not known, in vitro experiments have suggested it has a role in neurite extension and branching in vitro (44). Tmem47 encodes a four-transmembrane protein that is a member of a different subfamily than Tmem176a and Tmem176b, the other four-transmembrane protein genes regulated by Rhox5. The functional role of TMEM47 in mammals is not known, but its ortholog in Caenorhabditis elegans, VAB-9, is crucial for the formation of adherens junctions in epithelial cells (78). Thus, it is tempting to speculate that TMEM47 might function in the formation and/or maintenance of the testes-specific adherens junctions that form between Sertoli cells and germ cells: the ectoplasmic specialization.
Future studies will be required to precisely define the gene networks downstream of AR and the AR-regulated Rhox genes and to identify their roles in spermatogenesis. One essential question will be to delineate which functions of AR require the action of androgen-regulated Rhox genes. Studies in AR-deficient mice harboring either a nonfunctional Ar allele in Sertoli cells or a hypomorphic Ar allele in all cells have revealed that AR has at least three functions: 1) it promotes the progression of meiotic spermatocytes, 2) it promotes the maturation and/or survival of round spermatids, allowing the generation of elongating spermatids, and 3) it promotes the final maturation and release of elongated spermatids into the lumen of the seminiferous tubules (62,64,79,80). Thus, androgen-inducible Rhox genes may be involved in any or all of these events. Another question for the future will be to determine which Rhox5-regulated genes in Sertoli cells are direct targets of RHOX5. This will be a challenge to answer, because a RHOX5-binding consensus sequence has yet to be defined and RHOX5 may act on enhancers far upstream or downstream of its target genes. Finally, it will be intriguing to understand how Rhox5 regulates genes in cell types in addition to the Sertoli cell, the only cell type that significantly expresses Rhox5 (26,29). Given that a large proportion of Rhox5-regulated genes encode cell surface and secreted proteins (supplemental Fig. S3), an obvious mechanism by which it could act is to influence cell-cell interactions and signaling events occurring between Sertoli cells and other cell types in the testis. By defining and characterizing Rhox-regulated genes in vitro and in vivo, we believe that the work presented in this paper provides a foundation to address these many future issues.
Materials and Methods
Cell culture and transfection
The 15P-1 and MSC1 cell lines were maintained in DMEM supplemented with 10% fetal bovine serum and 50 mg/ml of both penicillin and streptomycin. The cells were grown at 37 C in 5% CO2 atmosphere and split when approximately 80% confluent.
To generate stable Rhox5 expression plasmid-transfected clones, 15P-1 cells cultured to approximately 60% confluence in 100-mm plates were transfected with 2 μg of Rhox5 expression plasmid by use of lipofectamine (Invitrogen, Carlsbad, CA) and clones resistant to antibiotic (700 μg/ml G418) were selected.
The concentration of plasmids used for transfection was independently determined by use of both a fluorometer and analytical gel electrophoresis. For transient transfection analysis, 15P-1 cells were plated in 60-mm dishes the day before transfection at a density of approximately 2 × 106 cells per dishes in 4 ml of growing medium. On the day of transfection, 800 ng of plasmid DNA was diluted with 150 μl of serum-free medium and mixed with 150 μl of serum-free media containing 3 μl of Lipofectamine (Invitrogen, Carlsbad, CA) and incubated at room temperature for 30 min; then, the DNA-lipid complex was added to the wells and incubated at 37 C in a CO2 incubator for 5 h. Cells were harvested 36 h post transfection for RNA analysis. MSC1 cells were transfected by use of Fugene 6 (Roche Diagnostics, GmbH, Mannheim, Germany) according to the manufacturer’s instructions. Cells were plated on 12-well culture dishes and then cotransfected with 100 ng of the Rhox5/Unc5c-containing plasmids and 100 ng of either the AR expression plasmid or empty vector. After a 6-h incubation in DMEM supplemented with 10% charcoal-stripped bovine serum (HyClone Laboratories, Inc., Logan, UT), R1881 was added at final concentration of 10 nm. Cells were harvested 36 h post transfection for luciferase assay by use of the Dual Luciferase Assay System (Promega, Madison, WI), according to the manufacturer’s instructions. The relative luciferase activity was calculated by normalizing against the cotransfected internal control pRL-null. The results shown are the mean ± sem of three independent transfection experiments.
Plasmids
The Unc5c-P (G734), Rhox5-Pp (Pem250), and Rhox expression plasmids were generated previously (32,46). The human AR pcDNA 3.1 plasmid (G541) was kindly provided by Zhengxin Wang (The University of Texas M.D. Anderson Cancer Center, Houston, TX).
RNA and protein analysis
Total cellular RNA was prepared from cell lines and tissues by guanidinium isothiocyanate lysis and centrifugation over a 5.7 m CsCl cushion as described previously (81). The Rhox5 cDNA probe used for Northern blot analysis was prepared as described previously (24). Real-time RT-PCR analysis was performed as described previously (7) with primers as described in Table 1. Microarray analysis with 15P-1 stable cell clones was performed at the University of Iowa DNA core facility on total cellular RNA from clones 6, 24, 14, and 16, using an Affymetrix mouse genome GeneChip 430 2.0 array. This gene chip independently samples approximately 45,101 probe sets, which corresponds to approximately 39,000 genes.
For Western blot analysis, 20 μg of total cell lysates were electrophoresed in a 10% SDS-polyacrylamide, transferred to Hybond ECL nitrocellulose (Amersham, Piscataway, NJ), and probed with antibodies against RHOX5 or β-actin (Sigma Chemical Co., St. Louis, MO). The membranes were given three 10-min washes with 0.1% Tween 20 in PBS at room temperature and then incubated for 45 min at room temperature with the secondary antibody (ECL kit antirabbit or antimouse from Amersham) at a dilution of 1:5000. The filter was developed by using the ECL-Plus reagent according to the manufacturer’s protocol (Amersham).
Testis cell fractionation
Sertoli and interstitial cells were purified from testes as previously described (7,82). In brief, testes were decapsulated and the seminiferous tubules were allowed to settle in PBS, followed by incubation in collagenase (C2674; Sigma). After another round of settling, the supernatant, which was enriched for interstitial cells, was subjected to hypotonic shock (by incubation in 1:7 diluted PBS for 3 min) to remove germ cells, and the surviving cells were pelleted and the RNA extracted. The pellet obtained after collagenase treatment, which was enriched for Sertoli cells, was incubated in a solution containing a mixture of enzymes [0.1% collagenase, 0.2% hyaluronidase (H6254; Sigma), 0.04% DNase I (D5025; Sigma), and 0.03% trypsin inhibitor (T6522; Sigma) in 1× PBS, pH 7.4] at 30 C for 40 min. The cells were pelleted and subjected to the same hypotonic shock as the interstitial cells, followed by centrifugation and RNA extraction of the pellet.
Supplementary Material
Acknowledgments
We thank Miriam Buttigieg for mouse tissue collection, and Drs. James MacLean and Wai-kin Chan for helpful advice throughout the course of this project.
Footnotes
This work was supported by NIH Grants HD-45595.
Present address for D.D.: Fox Chase Cancer Center, Philadelphia, Pennsylvania 19111.
Disclosure Summary: The authors have nothing to disclose.
First Published Online November 9, 2009
Abbreviations: AR, Androgen receptor; ARE, androgen-response element; qPCR, quantitative PCR; SAR, secondary androgen-response.
References
- Ingham PW, Martinez Arias A 1992 Boundaries and fields in early embryos. Cell 68:221–235 [DOI] [PubMed] [Google Scholar]
- Chiba S 1998 Homeobox genes in normal hematopoiesis and leukemogenesis. Int J Hematol 68:343–353 [DOI] [PubMed] [Google Scholar]
- Awgulewitsch A 2003 Hox in hair growth and development. Naturwissenschaften 90:193–211 [DOI] [PubMed] [Google Scholar]
- Lee CS, Kaestner KH 2004 Clinical endocrinology and metabolism. Development of gut endocrine cells. Best Pract Res Clin Endocrinol Metab 18:453–462 [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P 2007 Sex determination and gonadal development in mammals. Physiol Rev 87:1–28 [DOI] [PubMed] [Google Scholar]
- Maclean 2nd JA, Wilkinson MF 2005 Gene regulation in spermatogenesis. Curr Top Dev Biol 71:131–197 [DOI] [PubMed] [Google Scholar]
- MacLean JA, Chen MA, Wayne CM, Bruce SR, Rao MK, Meistrich ML, MacLeod CL, Wilkinson MF 2005 Rhox: a new homeobox gene cluster. Cell 120:369–382 [DOI] [PubMed] [Google Scholar]
- Jackson M, Watt AJ, Gautier P, Gilchrist D, Driehaus J, Graham GJ, Keebler J, Prugnolle F, Awadalla P, Forrester LM 2006 A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genomics 7:212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean 2nd JA, Lorenzetti D, Hu Z, Salerno WJ, Miller J, Wilkinson MF 2006 Rhox homeobox gene cluster: recent duplication of three family members. Genesis 44:122–129 [DOI] [PubMed] [Google Scholar]
- Morris L, Gordon J, Blackburn CC 2006 Identification of a tandem duplicated array in the Rhox alpha locus on mouse chromosome X. Mamm Genome 17:178–187 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J 2006 Remarkable expansions of an X-linked reproductive homeobox gene cluster in rodent evolution. Genomics 88:34–43 [DOI] [PubMed] [Google Scholar]
- Rao M, Wilkinson MF 2002 Homeobox genes and the male reproductive system. In: Robaire B, Hinton BT, eds. The epididymis: From molecules to clinical practice. New York: Kluwer Academic/Plenum Publishers [Google Scholar]
- Lindsey S, Wilkinson MF 1996 Homeobox genes and male reproductive development. J Assist Reprod Genet 13:182–192 [DOI] [PubMed] [Google Scholar]
- MacLaughlin DT, Donahoe PK 2004 Sex determination and differentiation. N Engl J Med 350:367–378 [DOI] [PubMed] [Google Scholar]
- Geyer CB, Eddy EM 2008 Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene 423:194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod CL, Fong AM, Seal BS, Walls L, Wilkinson MF 1990 Isolation of novel complementary DNA clones from T lymphoma cells: one encodes a putative multiple membrane-spanning protein. Cell Growth Differ 1:271–279 [PubMed] [Google Scholar]
- Sasaki AW, Doskow J, MacLeod CL, Rogers MB, Gudas LJ, Wilkinson MF 1991 The oncofetal gene Pem encodes a homeodomain and is regulated in primordial and pre-muscle stem cells. Mech Dev 34:155–164 [DOI] [PubMed] [Google Scholar]
- Ono T, Sato S, Kimura N, Tanaka M, Shibuya A, Old LJ, Nakayama E 2000 Serological analysis of BALB/C methylcholanthrene sarcoma Meth A by SEREX: identification of a cancer/testis antigen. Int J Cancer 88:845–851 [DOI] [PubMed] [Google Scholar]
- Agarwal SK, Lee Burns A, Sukhodolets KE, Kennedy PA, Obungu VH, Hickman AB, Mullendore ME, Whitten I, Skarulis MC, Simonds WF, Mateo C, Crabtree JS, Scacheri PC, Ji Y, Novotny EA, Garrett-Beal L, Ward JM, Libutti SK, Richard Alexander H, Cerrato A, Parisi MJ, Santa Anna-A S, Oliver B, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ 2004 Molecular pathology of the MEN1 gene. Ann NY Acad Sci 1014:189–198 [DOI] [PubMed] [Google Scholar]
- Guo F, Huang X, Li S, Sun L, Li Y, Li H, Zhou Y, Chu Y, Zhou T 2007 Identification of prosaposin as a novel interaction partner for Rhox5. J Genet Genomics 34:392–399 [DOI] [PubMed] [Google Scholar]
- Rao MK, Maiti S, Ananthaswamy HN, Wilkinson MF 2002 A highly active homeobox gene promoter regulated by Ets and Sp1 family members in normal granulosa cells and diverse tumor cell types. J Biol Chem 277:26036–26045 [DOI] [PubMed] [Google Scholar]
- MacLean 2nd JA, Rao MK, Doyle KM, Richards JS, Wilkinson MF 2005 Regulation of the Rhox5 homeobox gene in primary granulosa cells: preovulatory expression and dependence on SP1/SP3 and GABP. Biol Reprod 73:1126–1134 [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS 2004 Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6:117–131 [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Kleeman J, Richards J, MacLeod CL 1990 A novel oncofetal gene is expressed in a stage-specific manner in murine embryonic development. Dev Biol 141:451–455 [DOI] [PubMed] [Google Scholar]
- Lin TP, Labosky PA, Grabel LB, Kozak CA, Pitman JL, Kleeman J, MacLeod CL 1994 The Pem homeobox gene is X-linked and exclusively expressed in extraembryonic tissues during early murine development. Dev Biol 166:170–179 [DOI] [PubMed] [Google Scholar]
- Pitman JL, Lin TP, Kleeman JE, Erickson GF, MacLeod CL 1998 Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Dev Biol 202:196–214 [DOI] [PubMed] [Google Scholar]
- Daggag H, Svingen T, Western PS, van den Bergen JA, McClive PJ, Harley VR, Koopman P, Sinclair AH 2008 The rhox homeobox gene family shows sexually dimorphic and dynamic expression during mouse embryonic gonad development. Biol Reprod 79:468–474 [DOI] [PubMed] [Google Scholar]
- Maiti S, Doskow J, Sutton K, Nhim RP, Lawlor DA, Levan K, Lindsey JS, Wilkinson MF 1996 The Pem homeobox gene: rapid evolution of the homeodomain, X chromosomal localization, and expression in reproductive tissue. Genomics 34:304–316 [DOI] [PubMed] [Google Scholar]
- Lindsey JS, Wilkinson MF 1996 Pem: a testosterone- and LH-regulated homeobox gene expressed in mouse Sertoli cells and epididymis. Dev Biol 179:471–484 [DOI] [PubMed] [Google Scholar]
- Sutton KA, Maiti S, Tribley WA, Lindsey JS, Meistrich ML, Bucana CD, Sanborn BM, Joseph DR, Griswold MD, Cornwall GA, Wilkinson MF 1998 Androgen regulation of the Pem homeodomain gene in mice and rat Sertoli and epididymal cells. J Androl 19:21–30 [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Meistrich ML, Wilkinson MF 2003 Pem homeobox gene promoter sequences that direct transcription in a Sertoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. Mol Endocrinol 17:223–233 [DOI] [PubMed] [Google Scholar]
- Hu Z, Shanker S, MacLean 2nd JA, Ackerman SL, Wilkinson MF 2008 The RHOX5 homeodomain protein mediates transcriptional repression of the netrin-1 receptor gene Unc5c. J Biol Chem 283:3866–3876 [DOI] [PubMed] [Google Scholar]
- Hu Z, MacLean JA, Bhardwaj A, Wilkinson MF 2007 Regulation and function of the Rhox5 homeobox gene. Ann NY Acad Sci 1120:72–83 [DOI] [PubMed] [Google Scholar]
- Shanker S, Hu Z, Wilkinson MF 2008 Epigenetic regulation and downstream targets of the Rhox5 homeobox gene. Int J Androl 31:462–470 [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Paquis-Flucklinger V, Bertino B, Sage J, Jasin M, Miyagawa K, van Heyningen V, Besmer P, Cuzin F 1993 Transmeiotic differentiation of male germ cells in culture. Cell 75:997–1006 [DOI] [PubMed] [Google Scholar]
- Grandjean V, Sage J, Ranc F, Cuzin F, Rassoulzadegan M 1997 Stage-specific signals in germ line differentiation: control of Sertoli cell phagocytic activity by spermatogenic cells. Dev Biol 184:165–174 [DOI] [PubMed] [Google Scholar]
- Jabado N, Canonne-Hergaux F, Gruenheid S, Picard V, Gros P 2002 Iron transporter Nramp2/DMT-1 is associated with the membrane of phagosomes in macrophages and Sertoli cells. Blood 100:2617–2622 [DOI] [PubMed] [Google Scholar]
- Payne AH 1990 Hormonal regulation of cytochrome P450 enzymes, cholesterol side-chain cleavage and 17 alpha-hydroxylase/C17–20 lyase in Leydig cells. Biol Reprod 42:399–404 [DOI] [PubMed] [Google Scholar]
- Verhoeven G, Swinnen JV 1999 Indirect mechanisms and cascades of androgen action. Mol Cell Endocrinol 151:205–212 [DOI] [PubMed] [Google Scholar]
- Del Rio-Tsonis K, Covarrubias L, Kent J, Hastie ND, Tsonis PA 1996 Regulation of the Wilms’ tumor gene during spermatogenesis. Dev Dyn 207:372–381 [DOI] [PubMed] [Google Scholar]
- Rossi P, Albanesi C, Grimaldi P, Geremia R 1991 Expression of the mRNA for the ligand of c-kit in mouse Sertoli cells. Biochem Biophys Res Commun 176:910–914 [DOI] [PubMed] [Google Scholar]
- Rogers MF, Ben-Hur A 2009 The use of gene ontology evidence codes in preventing classifier assessment bias. Bioinformatics (Oxf) 25:1173–1177 [DOI] [PubMed] [Google Scholar]
- Velarde MC, Geng Y, Eason RR, Simmen FA, Simmen RC 2005 Null mutation of Kruppel-like factor9/basic transcription element binding protein-1 alters peri-implantation uterine development in mice. Biol Reprod 73:472–481 [DOI] [PubMed] [Google Scholar]
- Bonett RM, Hu F, Bagamasbad P, Denver RJ 2009 Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: implications for neural development and plasticity. Endocrinology 150:1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle JR, Heindel JJ, Steinberger A, Sanborn BM 1986 Effect of hypotonic treatment on Sertoli cell purity and function in culture. In Vitro Cell Dev Biol 22:325–331 [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Rao MK, Kaur R, Buttigieg MR, Wilkinson MF 2008 GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Mol cell Biol 28:2138–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuopio T, Pelliniemi LJ, Huhtaniemi I 1989 Rapid Leydig cell proliferation and luteinizing hormone receptor replenishment in the neonatal rat testis after a single injection of human chorionic gonadotropin. Biol Reprod 40:135–143 [DOI] [PubMed] [Google Scholar]
- Zhang H, Denhard LA, Zhou H, Liu LH, Lan ZJ 2008 0610009K11Rik, a testis-specific and germ cell nuclear receptor-interacting protein. Biochem Biophys Res Commun 366:898–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz-Mercer B, Elliott DJ, Issakov J, Leider-Trejo L, Schreiber L, Misonzhnik F, Eisenthal A, Maymon BB 2002 Localization of a specific germ cell marker, DAZL1, in testicular germ cell neoplasias. Virchows Arch 440:387–391 [DOI] [PubMed] [Google Scholar]
- Yomogida K, Ohtani H, Harigae H, Ito E, Nishimune Y, Engel JD, Yamamoto M 1994 Developmental stage- and spermatogenic cycle-specific expression of transcription factor GATA-1 in mouse Sertoli cells. Development (Camb) 120:1759–1766 [DOI] [PubMed] [Google Scholar]
- Roberts KP, Griswold MD 1990 Characterization of rat transferrin receptor cDNA: the regulation of transferrin receptor mRNA in testes and in Sertoli cells in culture. Mol Endocrinol 4:531–542 [DOI] [PubMed] [Google Scholar]
- Linder CC, Heckert LL, Roberts KP, Kim KH, Griswold MD 1991 Expression of receptors during the cycle of the seminiferous epithelium. Ann NY Acad Sci 637:313–321 [DOI] [PubMed] [Google Scholar]
- Denolet E, De Gendt K, Allemeersch J, Engelen K, Marchal K, Van Hummelen P, Tan KA, Sharpe RM, Saunders PT, Swinnen JV, Verhoeven G 2006 The effect of a Sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Mol Endocrinol 20:321–334 [DOI] [PubMed] [Google Scholar]
- He WW, Kumar MV, Tindall DJ 1991 A frame-shift mutation in the androgen receptor gene causes complete androgen insensitivity in the testicular-feminized mouse. Nucleic Acids Res 19:2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Abel M, Charlton HM, Hu B, Johnston H, Baker PJ 2007 Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive tfm mouse testis. Endocrinology 148:2914–2924 [DOI] [PubMed] [Google Scholar]
- Heemers HV, Verhoeven G, Swinnen JV 2006 Androgen activation of the sterol regulatory element-binding protein pathway: current insights. Mol Endocrinol 20:2265–2277 [DOI] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E, Okano M 2006 DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev 20:3382–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geserick C, Weiss B, Schleuning WD, Haendler B 2002 OTEX, an androgen-regulated human member of the paired-like class of homeobox genes. Biochem J 366:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne CM, MacLean JA, Cornwall G, Wilkinson MF 2002 Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene 301:1–11 [DOI] [PubMed] [Google Scholar]
- Dean DM, Sanders MM 1996 Ten years after: reclassification of steroid-responsive genes. Mol Endocrinol 10:1489–1495 [DOI] [PubMed] [Google Scholar]
- Collett GP, Betts AM, Johnson MI, Pulimood AB, Cook S, Neal DE, Robson CN 2000 Peroxisome proliferator-activated receptor alpha is an androgen-responsive gene in human prostate and is highly expressed in prostatic adenocarcinoma. Clin Cancer Res 6:3241–3248 [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lécureuil C, Heyns W, Carmeliet P, Guillou F, Sharpe RM, Verhoeven G 2004 A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci USA 101:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon SF, Walther N, Saunders PT 2005 Expression of androgen and estrogen receptors in Sertoli cells: studies using the mouse SK11 cell line. Endocrinology 146:5304–5312 [DOI] [PubMed] [Google Scholar]
- Tan KA, De Gendt K, Atanassova N, Walker M, Sharpe RM, Saunders PT, Denolet E, Verhoeven G 2005 The role of androgens in Sertoli cell proliferation and functional maturation: studies in mice with total or Sertoli cell-selective ablation of the androgen receptor. Endocrinology 146:2674–2683 [DOI] [PubMed] [Google Scholar]
- Lim K, Yoo JH, Kim KY, Kweon GR, Kwak ST, Hwang BD 1994 Testosterone regulation of proto-oncogene c-myc expression in primary Sertoli cell cultures from prepubertal rats. J Androl 15:543–550 [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M 2007 A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhu P, Wu C, Yu L, Zhao S, Gu X 2003 In silico analysis indicates a similar gene expression pattern between human brain and testis. Cytogenet Genome Res 103:58–62 [DOI] [PubMed] [Google Scholar]
- Round J, Stein E 2007 Netrin signaling leading to directed growth cone steering. Curr Opin Neurobiol 17:15–21 [DOI] [PubMed] [Google Scholar]
- Mehlen P, Llambi F 2005 Role of netrin-1 and netrin-1 dependence receptors in colorectal cancers. Br J Cancer 93:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Nakagawa T, Kanematsu T, Uchida T, Tsuji S 1999 Isolation of 10 differentially expressed cDNAs in differentiated Neuro2a cells induced through controlled expression of the GD3 synthase gene. J Neurochem 72:1781–1790 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Fujimura L, O-Wang J, Hatano M, Sakamoto A, Arima M, Ebara M, Ino H, Yamashita T, Saisho H, Tokuhisa T 2006 Role of Clast1 in development of cerebellar granule cells. Brain Res 1104:18–26 [DOI] [PubMed] [Google Scholar]
- Dong W, Albers JJ, Vuletic S 2009 Phospholipid transfer protein reduces phosphorylation of tau in human neuronal cells. J Neurosci Res 87:3176–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Guimera J, Wurst W, Prakash N 2007 Distinct but redundant expression of the Frizzled Wnt receptor genes at signaling centers of the developing mouse brain. Neuroscience 147:693–711 [DOI] [PubMed] [Google Scholar]
- Domanski P, Fish E, Nadeau OW, Witte M, Platanias LC, Yan H, Krolewski J, Pitha P, Colamonici OR 1997 A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with Jak1 and is sufficient to activate the Jak-Stat pathway and induce an antiviral state. J Biol Chem 272:26388–26393 [DOI] [PubMed] [Google Scholar]
- Joulin V, Peduzzi J, Roméo PH, Rosa R, Valentin C, Dubart A, Lapeyre B, Blouquit Y, Garel MC, Goossens M, Rosa J, Cohen-Solal M 1986 Molecular cloning and sequencing of the human erythrocyte 2,3-bisphosphoglycerate mutase cDNA: revised amino acid sequence. EMBO J 5:2275–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvindrah R, Rougon G, Chazal G 2002 Increased neurogenesis in adult mCD24-deficient mice. J Neurosci 22:3594–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Kobayashi A, Yamashita T, Shimanuki T, Nakajima O, Takahashi S, Ikegami S, Inokuchi K, Yamashita K, Yamamoto M, Fujii-Kuriyama Y 2003 Functional analysis of basic transcription element binding protein by gene targeting technology. Mol Cell Biol 23:2489–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS, Köppen M, Sims P, Hodgkin J, Yonkof A, Hardin J 2003 The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol 5:619–625 [DOI] [PubMed] [Google Scholar]
- Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, Guillou F, Lardy H, Yeh S 2004 Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc Natl Acad Sci USA 101:6876–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel MH, Baker PJ, Charlton HM, Monteiro A, Verhoeven G, De Gendt K, Guillou F, O'Shaughnessy PJ 2008 Spermatogenesis and Sertoli cell activity in mice lacking Sertoli cell receptors for follicle-stimulating hormone and androgen. Endocrinology 149:3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M 2000 Purification of RNA. New York: Oxford University Press [Google Scholar]
- Anway MD, Folmer J, Wright WW, Zirkin BR 2003 Isolation of Sertoli cells from adult rat testes: an approach to ex vivo studies of Sertoli cell function. Biol Reprod 68:996–1002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.