Abstract
The cytosolic 70-kDa heat shock proteins (Hsp70s), Ssa and Ssb, of Saccharomyces cerevisiae are functionally distinct. Here we report that the ATPase activities of these two classes of Hsp70s exhibit different kinetic properties. The Ssa ATPase has properties similar to those of other Hsp70s studied, such as DnaK and Hsc70. Ssb, however, has an unusually low steady-state affinity for ATP but a higher maximal velocity. In addition, the ATPase activity of Hsp70s, like that of Ssa1, depends on the addition of K+ whereas Ssb activity does not. Suprisingly, the isolated 44-kDa ATPase domain of Ssb has a Km and Vmax for ATP hydrolysis similar to those of Ssa, rather than those of full length Ssb. Analysis of Ssa/Ssb fusion proteins demonstrates that the Ssb peptide-binding domain fused to the Ssa ATPase domain generates an ATPase of relatively high activity and low steady-state affinity for ATP similar to that of native Ssb. Therefore, at least some of the biochemical differences between the ATPases of these two classes of Hsp70s are not intrinsic to the ATPase domain itself. The differential influence of the peptide-binding domain on the ATPase domain may, in part, explain the functional uniqueness of these two classes of Hsp70s.
Molecular chaperones are ubiquitous proteins that bind to the nonnative conformations of proteins, thus facilitating cellular processes such as protein folding and translocation across membranes (1, 2). Seventy-kilodalton heat shock proteins (Hsp70s) are an abundant class of molecular chaperones. The highly conserved 44-kDa amino terminal domain of Hsp70 proteins has a low intrinsic ATPase activity in the native protein whereas the isolated ATPase domains have a much higher catalytic activity (3). The somewhat less conserved, adjacent, 18-kDa domain binds unfolded proteins and short extended peptides (4). The function of the C-terminal 10-kDa domain is, however, not yet defined well. Throughout this report, the 28-kDa C-terminal region containing the 18- and 10-kDa domains is collectively referred to as the “peptide-binding” domain unless explicitly stated otherwise.
It is believed that Hsp70 proteins have, like many GTP binding proteins, two distinct conformations. When an ADP molecule is bound to the ATP binding site, the Hsp70 exhibits stable peptide binding; when ATP is bound, binding of peptide is relatively unstable (5, 6). Therefore, ATP hydrolysis converts Hsp70 to the form that has a relatively stable interaction with unfolded proteins. Exchange of ADP for ATP results in rapid peptide binding and release.
Analogously, the peptide-binding domain affects the nucleotide state of the ATPase domain. The steady-state ATPase activity of several Hsp70s increases up to 5-fold on binding of peptide to the peptide-binding domain, indicating that the conformation of the C-terminal domain affects the ATPase activity of the N-terminal domain (7, 8, 9). Consistent with this idea, the 44-kDa ATPase domain when cleaved from the peptide-binding domain has severalfold higher ATPase activity than the intact protein (25). These results have led to the suggestion that, in the absence of bound peptide, the peptide-binding domain attenuates the intrinsic ATPase activity of the N-terminal domain.
Because the identity of the nucleotide bound to the ATPase domain profoundly affects the character of the interaction of Hsp70 with substrate proteins and the peptide-binding domain influences the ATPase activity, coordination between the peptide-binding activity and the ATPase activity must be fundamental for the chaperone activity of Hsp70 proteins. How this interdomain communication occurs is, at present, unknown. Based on the structure of the DnaK peptide-binding domain, Hendrickson and coworkers (10) have suggested that a helical domain serves as a “lid” over the peptide-binding domain. This lid opens and closes over the peptide via the breaking and forming of salt bridges. It is hypothesized that the opening and closing of the lid depends on the nucleotide state of the ATPase domain. However, the mechanism by which chemical energy in the ATPase domain is used to open and close this lid remains unresolved. Furthermore, it is unclear how the conformation of the peptide-binding domain is communicated to the ATPase domain.
The nucleotide bound to an Hsp70 also can be modulated by interacting proteins. For example, in both prokaryotes and eukaryotes, another type of chaperone, DnaJ, stimulates the ATPase activity of Hsp70s by accelerating the rate of ATP hydrolysis (11, 12, 13). In eukaryotes, a protein called Hip has been identified that stabilizes the ADP bound form by inhibiting the release of nucleotide (14). Alternatively, proteins have been found in both prokaryotes and eukaryotes—GrpE (11, 15) and Bag-1 (16), respectively—that act as nucleotide release factors stimulating exchange of ADP for ATP. By these means, the amount of time an Hsp70 remains in the ADP or ATP form is modulated in vivo.
Typically, eukaryotes have multiple Hsp70s distributed throughout several cellular compartments. The extensively studied Hsp70 family of Saccharomyces cerevisiae contains 14 members. The Ssa and Ssb Hsp70s are two cytosolic subfamilies (17). The Ssa subfamily has four members: Ssa1–4; the Ssb has two members: Ssb1 and Ssb2. The essential Ssa proteins, which are most closely related to the major cytosolic Hsp70s of mammalian cells, Hsp70/Hsc70, are involved in translocation of proteins across membranes and in the regulation of the heat shock response. On the other hand, Ssb1 and Ssb2, which differ from each other by only four amino acids and from the members of the Ssa family by ≈37%, seem to have a more specialized function. Ssb binds to translating ribosomes and can be crosslinked to the nascent chain (18, 19). This association, in addition to the fact that strains lacking Ssb are hypersensitive to certain inhibitors of protein synthesis, suggests that this class of Hsp70s may be involved in translation and/or very early folding events on the ribosome.
In addition to the antibiotic sensitivity, strains lacking Ssbs are cold-sensitive for growth. Genetic results using chimeric SSA:SSB genes have shown that these two phenotypes are separable (20). For example, rescue of the cold-sensitive phenotype requires the 44-kDa ATPase domain from Ssb. Any two of the three (44-, 18-, and 10-kDa) Ssb domains are sufficient for rescue of the antibiotic sensitivity and result in chimera association with ribosomes. For example, the expression of a chimera containing the Ssa1 ATPase domain and the 18-kDa and 10-kDa domains of Ssb1 allows for polysome association as well as growth in the presence of 70 μg/ml hygromycin B, a concentration that inhibits the growth of cells lacking Ssb.
Ssa1 has an ATPase activity very similar to that of other Hsp70s that have been analyzed, with a Km for ATP on the order of 0.2 μM and a kcat of 0.04 ATP⋅min−1 at physiological potassium concentrations (21). Although Ssb1 has been reported to have ATPase activity, its biochemical properties have not been studied extensively (22). Therefore, we began such an analysis. We confirm that Ssb has an intrinsic ATPase activity. However, this activity has several properties that differ from that of Ssa1 and other Hsp70s, including a higher Km for ATP and little dependence on added K+ for catalytic activity. These differences are, at least in part, caused by the influence of the Ssb peptide-binding domain on the ATPase domain.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions.
The yeast strains used in this study were BAS15: trp1–1 ura3–1 leu2–3,112 his3–11,15 ade2–1 can1–100 GAL2+ met2-Δ1 lys2-Δ2 ssa2∷TRP1 ssa3∷LYS2 ssa4∷ADE2 ssb1∷LEU2 ssb2∷HIS3 and TZ236: ssa1∷LEU2 his3 leu2 lys2 ura3. [YEp24-SSB1]. BAS15 was grown in 1% yeast extract, 2% peptone, 2% dextrose at 30°C. TZ236 was grown in minimal media (0.67% yeast nitrogen base without amino acids, 2% dextrose, supplemented with required amino acids as necessary) at 30°C. Cultures were grown to an OD600 of ≈3.
Protein Purification.
Ssa1, Ydj1, and Sis1. Ssa1 and Ydj1 were purified as described elsewhere (21, 22). Sis1 was expressed from a pET11a vector and was purified from Escherichia coli. Cells were lysed in buffer A (25 mM Mes, pH 6.1/5 mM MgCl2/5 mM 2-mercaptoethanol/10% glycerol), and 0.5 mM phenylmethylsulfonyl fluoride and 0.1 mg/ml Pepstatin A were added. Cleared lysate was applied to an S-Sepharose (Sigma) column equilibrated with buffer A and eluted with a gradient of 0–400 mM NaCl. Sis1-containing fractions were pooled and diluted 1:2 in 10 mM phosphate buffer and were applied to a CHT-II column (Bio-Rad) equilibrated with 10 mM phosphate buffer (pH 7.0) and eluted with a gradient of 10–500 mM phosphate buffer.
Ssb preparation.
Ssb was purified from TZ236. Cells were resuspended and lysed in buffer B (20 mM Hepes, pH 7.0/5 mM MgCl2/5 mM 2-mercaptoethanol/10% glycerol), and protease inhibitors were added (0.5 mM phenylmethylsulfonyl fluoride/2.5 mM EDTA/0.1 mg/ml Pepstatin A). After filtration to remove large particles, the lysate was applied to a DEAE-Sephacel (Pharmacia) column equilibrated with buffer B and eluted with a gradient containing 100–400 mM NaCl. Ssb-containing fractions were pooled together and were applied to an ATP-agarose column equilibrated with buffer B. After a 1 M NaCl wash step, Ssb was eluted with buffer B containing 2 mM ATP. Ssb containing fractions were pooled together, were diluted 1:1 with 5 mM sodium phosphate (pH 7.0), and were applied to a hydroxiapatite column (CHT-1, Bio-Rad). Ssb was eluted with a 5–300 mM sodium phosphate (pH 7.0) gradient. Fractions were collected, and protease inhibitors were added immediately, as above. Fractions not supplemented with protease inhibitors were degraded proteolytically in <24 hours at 4°C. Pooled fractions were dialyzed against buffer C (20 mM Tris⋅HCl, pH 7.5/20 mM NaCl/10 mM MgCl2/5 mM 2-mercapthoehanol/10% glycerol).
BAA and ABB.
BAA was purified from strain BAS15 carrying the BAA fusion on a plasmid essentially as Ssb except for the last chromatographic step. The elution from the hydroxiapatite column was performed with 50 mM sodium phosphate buffer and a subsequent 50–250 mM sodium phosphate buffer (pH 7.0) gradient. ABB was purified from strain BAS15 carrying the ABB fusion on a plasmid, as Ssb, but omitting the hydroxiapatite chromatography step.
ATPase fragments.
ATPase fragments of Ssb were obtained by either spontaneous hydrolysis maintaining a Ssb preparation for 24 hours at 4°C or by tryptic or chymotryptic hydrolysis. Digestions by trypsin and chymotrypsin (Sigma) were performed at 37°C for 10 minutes. The reactions were stopped by addition of phenylmethylsulfonyl fluoride up to a final concentration of 2 mM. Analytical preparations were denatured immediately and were analyzed by SDS/PAGE. Preparative reactions were immediately applied to a gel filtration column (Sephacryl S-200 HR, Sigma), equilibrated with buffer C. Protein concentrations for all proteins were calculated by using the Bradford assay (Bio-Rad) using ovalbumin as standard.
Western Blots.
Equivalent amounts of each protein were mixed with SDS-sample buffer and were subjected to SDS/PAGE. Gels then were stained with Coomassie brilliant blue or were transferred to nitrocellulose (Hybond-C, Amersham) and were immunoblotted by using polyclonal antibodies specific for Ssb (C-terminal 80 amino acids), Ssa (C-terminal 56 amino acids), or the conserved Hsp70 ATPase domain (peptide VGIDLGTTYSC). Detection was performed by using the ECL detection system (Amersham).
ATPase Assays.
ATPase assays were carried out essentially as described (21). Assays were carried out at 30°C in 50 mM Tris⋅HCl (pH 7.5), 2 mM MgCl2, 5 mM 2-mercapthoethanol, and variable ATP and/or potassium acetate concentrations. Activity was calculated by single-point assay in triplicate after assuring that the time point taken was in the linear range of the assay. Aliquots were removed and applied to polyethylenimine TLC plates. The fraction of ATP hydrolyzed to ADP in each reaction was quantitated by PhosphorImager analysis (Molecular Dynamics). Background hydrolysis was calculated and subtracted by using assays, also in triplicate, made in the absence of enzyme. To make the dependence measurements comparable in each assay, we used an ATP concentration near each protein’s respective Km value. Data were analyzed by using a nonlinear regression routine (Enzfitter, R. J. Leatherbarrow, Biosoft, Cambridge, U.K.). The statistical significance of stimulation of Hsp70 ATPase activity by peptides, carboxymethylated α-lactalbumin (CMLA), or DnaJ-homologues was analyzed by the Student’s t test (H. J. Motulsky, GraphPad, San Diego). Peptides A7 (RRLIEDAETAARG; catalog number A7433) and A5 (APRLRFTSL; catalog number A5308) and reduced CMLA used in the ATPase assays were obtained from Sigma and were used as 5 mg/ml and 10 mg/ml stock solutions, respectively. CMLA was boiled to remove contaminating ATPase activity. For each 40-μl ATPase assay, the following concentrations of each peptide, unfolded protein, or DnaJ-homologue were added: A5 (15 μg), A7 (15 μg), S32 (15 μg), CLMA (20 μg), Sis1 (12.9 μg), and Ydj1 (8.6 μg).
RESULTS
Kinetic Parameters of ATP Hydrolysis by Ssb.
To begin a kinetic characterization of Ssb, we compared the ATPase activity of Ssb to that of another yeast cytosolic Hsp70, Ssa1. By using a standard ATPase assay, the Km of Ssb for ATP was determined to be 147 μM, >1,000-fold higher that that of Ssa1 (Table 1) and other Hsp70s analyzed (23). However, the ability of Ssb to hydrolyze ATP is significantly higher than that of Ssa1, as demonstrated by its higher kcat (0.81 min−1 vs. 0.031 min−1).
Table 1.
Kinetic constants for Ssb, Ssa, B--, A--, BAA, and ABB at both 2.5 and 100 mM potassium acetate
| 2.5 mM KOAc
|
100 mM KOAc
|
|||||
|---|---|---|---|---|---|---|
| kcat, min−1 | Km, μM | kcat/Km | kcat, min−1 | Km, μM | kcat/Km | |
| BBB | 0.95 ± 0.13 | 270 ± 46 | 3.5 × 10−3 | 0.81 ± 0.13 | 147 ± 42 | 5.5 × 10−3 |
| B-- | 1.24 ± 0.10 | 14.0 ± 5.7 | 8.8 × 10−2 | 4.49 ± 0.98 | 0.37 ± 0.07 | 12.1 |
| BAA | 0.0143 ± 0.0011 | 20.1 ± 3.2 | 7.1 × 10−4 | 0.14 ± 0.02 | 1.69 ± 0.43 | 8.3 × 10−2 |
| ABB | 0.506 ± 0.014 | 205 ± 10 | 2.5 × 10−3 | 0.861 ± 0.064 | 98.6 ± 15.8 | 8.7 × 10−3 |
| AAA | 0.028 ± 0.003 | 4.2 ± 1 | 6.6 × 10−3 | 0.031 ± 0.004 | 0.11 ± 0.04 | 2.8 × 10−1 |
| A-- | 0.046 ± 0.07 | 45 ± 8.82 | 1.0 × 10−3 | 0.116 ± 0.021 | 0.49 ± 0.15 | 2.4 × 10−1 |
Ssb ATPase Activity Is not Stimulated by Peptides or DnaJs.
Because the ATPase activity of Hsp70s can be stimulated by peptide substrates or DnaJ proteins (4), we investigated the ability of Ssb to be stimulated by peptides and cochaperones. To determine stimulation by peptide substrates, standard ATPase assays were conducted in the presence of a panel of various peptides and CMLA. ATPase assays for several Hsp70s from S. cerevisiae were performed under the optimal concentrations of KOAc and ATP for each given Hsp70. As shown in Table 2, Ssb was not stimulated by CMLA or any of the peptides tested, which are clearly capable of stimulating one or more other Hsp70 subfamily members. Furthermore, Ssb ATPase activity was not stimulated by either yeast cytosolic DnaJ homolog, Ydj1 or Sis1, even when these proteins were added in excess to Ssb. However, both Ydj1 and Sis1 were able to stimulate two or more yeast Hsp70 subfamily members. These data suggest that purified Ssb ATPase activity is not affected by the addition of peptide or DnaJs and that, indeed, Ssb may differ from other Hsp70s in this respect. However, it is also possible that none of the peptides or DnaJs used in these assays interact with Ssb.
Table 2.
Stimulation factor of yeast Hsp70s ATPase activity by peptide, CMLA, and yeast DnaJ homologs
| Kar2 | Ssc1 | Ssa1 | Ssb | |
|---|---|---|---|---|
| A5 | NS | NS | 1.6 | NS |
| A7 | NS | 2.0 | 2.2 | NS |
| S32 | 2.8 | 1.9 | 2.7 | NS |
| CMLA | 2.7 | NS | 2.3 | NS |
| Ydj1 | NS | 4.0 | 10.2 | NS |
| Sis1 | 1.7 | 5.0 | 8.1 | NS |
NS, No significant stimulation; P < 0.05.
Ssb ATPase Activity Is Relatively Independent of Added Potassium.
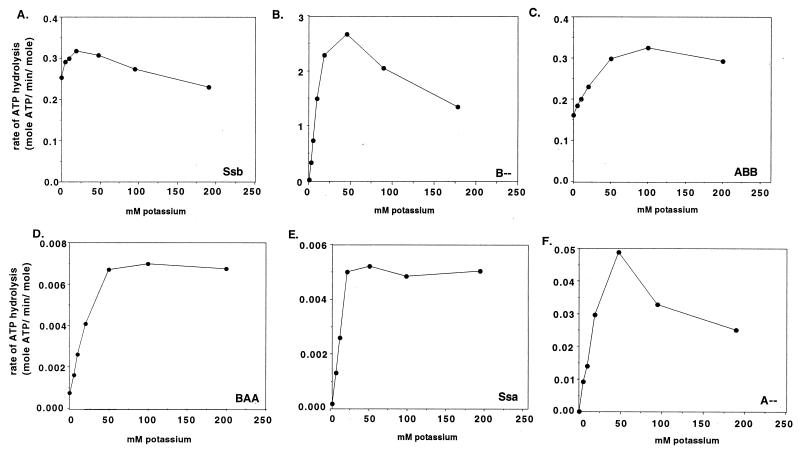
It has been shown that Ssa1 ATPase activity, like that of other Hsp70s analyzed, is highly K+-dependent (21). Ssa1 is nearly inactive at low concentrations of potassium, and its affinity for ATP increases ≈20-fold when the potassium concentration is raised from 2.5 to 200 mM. To compare Ssa1 and Ssb, we determined the K+ dependence of Ssb ATPase activity. There was little variation in ATPase activity of Ssb over a wide range of K+ concentrations (Fig. 1A), unlike that of Ssa1 (Fig. 1E). In addition, the Km of Ssb did not change substantially when the K+ concentration was raised from 2.5 to 100 mM (270 vs. 147 μM ATP) whereas Km of Ssa1 increased ≈38-fold (Table 1).
Figure 1.
Potassium dependence of ATPase activity. ATPase activities measured at 100 μM ATP for Ssb (A), 0.25 μM ATP for B-- (B), 100 μM ATP for ABB (C), 1 μM ATP for BAA (D), 0.25 ATP μM for Ssa (E), and 0.50 μM ATP for A-- (F) in the presence of increasing potassium concentrations.
ATPase Activity of the 44-kDa N-Terminal Fragment of Ssb.
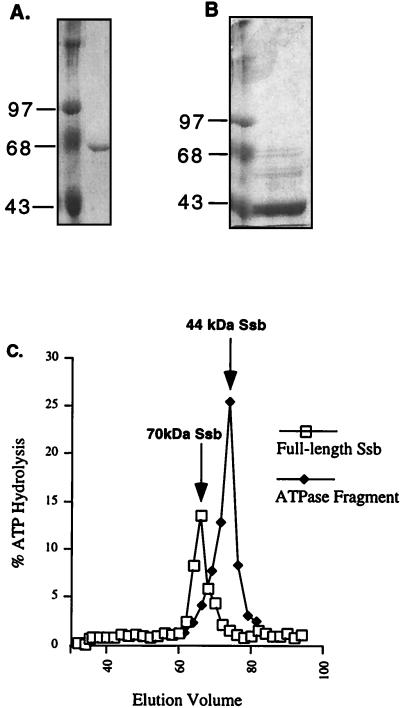
Although the Ssb preparations used in this analysis appeared homogeneous by SDS/PAGE analysis (Fig. 2A), the low ATPase activity of Hsp70 proteins typically makes the possibility of a contaminant protein with high ATPase activity a concern. This possibility was particularly an issue for us because of the unusually high Km and kcat, for an Hsp70, of our Ssb preparation and the fact that the Ssb ATPase activity was not stimulatable by peptides, CMLA, and DnaJ homologs. To address this concern, an Ssb sample (Fig. 2A) was passed through a Sephacryl S-200 gel filtration column. As expected, the ATPase activity coeluted with the Ssb protein peak (Fig. 2C) at an elution volume of 66 ml. This coelution suggested that the ATPase activity belonged to Ssb, although we cannot exclude the possibility that a contaminant ATPase of a molecular weight similar to that of Ssb was present in our preparation.
Figure 2.
Elution of Ssb ATPase domain from gel filtration column. Protein samples containing full length Ssb (A) or a spontaneously generated ATPase fragment of Ssb (B) were applied to a Sephacryl S-200 gel filtration column. In each case, the left lane is the molecular weight standard and the right lane is the protein sample. (C) Graphical representation of the coelution of full length Ssb with the ATPase activity at 66 ml (open squares) and an increased ATPase activity with the ATPase domain of Ssb at 76 ml (closed diamonds). An arrow marks the presence of each eluted protein as detected by SDS/PAGE.
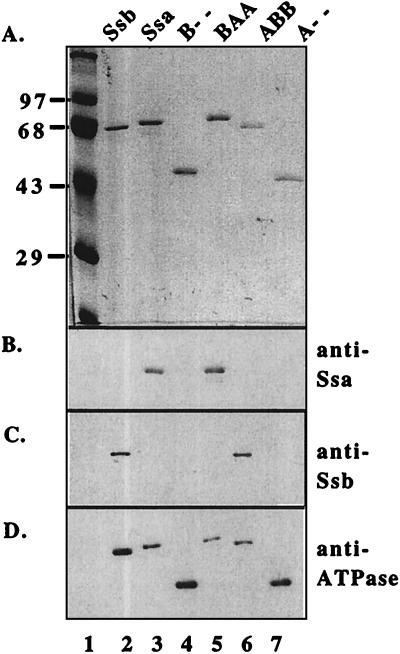
In a second experiment, an aliquot of the same sample, which had been left 24 hours at 4°C without protease inhibitors, was passed through the same column (Fig. 2B). Although a small amount of the 70-kDa Ssb band was still present, most had undergone proteolysis. Most of the ATPase activity coeluted at ≈76 ml together with the predominant fragment of ≈44-kDa, the expected size of the protease-resistant N-terminal ATPase domain. In fact, this fragment had an increased ATPase activity over that of full length Ssb. This separated domain was identified as the ATPase domain by Western blotting by using an antibody specific to the conserved Hsp70 ATPase domain (Fig. 3 A, lane 3, and D, lane 3). Coelution of the ATPase activity with the intact 70-kDa Ssb in the first sample and the 44-kDa Ssb ATPase domain in the sample that had been allowed to undergo partial proteolysis strongly suggested that the ATPase activity of the Ssb preparations was intrinsic to Ssb.
Figure 3.
Purifed Ssb, Ssa, ATPase domains, and Ssa/Ssb fusion proteins. Each protein [0.3 μg (A) or 0.15 μg (B–D)] was trichloroacetic acid precipitated and separated by SDS/PAGE. One gel (A) was stained with Coomassie blue. The second gel was transferred to nitrocellulose and was probed sequentially with antibodies against the C-terminal 56 amino acids of Ssa (B), the C-terminal 80 amino acids of Ssb (C), or the conserved ATPase domain that recognizes all Hsp70s (D).
The ATPase Domain of Ssb1 Shares Some Biochemical Properties with Ssa1.
Because of the differences between the biochemical properties of Ssb and Ssa1 we observed, the decision was made to characterize more thoroughly the 44-kDa ATPase fragment of Ssb1. It had been shown for several Hsp70s, including DnaK and Hsc70 (23), that the activity (kcat) of the 44-kDa fragment is severalfold higher than that of the intact protein but that the Km does not change substantially. Indeed, as for other Hsp70s, the kcat of the isolated Ssb ATPase domain (referred to here as B--), measured at 100 mM potassium, was ≈5-fold higher than the kcat of the intact molecule (4.49 min−1 vs. 0.81 min−1).
Surprisingly, unlike other Hsp70s tested, the Km of the 44-kDa fragment of Ssb (B--) was substantially different from that of the intact 70-kDa protein. The Km for B-- for ATP was calculated to be 0.37 μM (Table 1), only 0.25% of the intact Ssb. However this Km of B-- is very similar to that of Ssa (0.11 μM). As discussed above, the Km of Ssa for ATP is K+-dependent (21), being 20-fold higher at 2.5 mM K+ than at 200 mM K+ whereas the Km of intact Ssb is not. However, the Km of the 44-kDa Ssb fragment (B--) was 14 μM at 2.5 mM K+. This represents an ≈40-fold increase in the steady-state affinity for ATP when potassium is raised from 2.5 mM to nearly physiological concentrations (100–200 mM).
Similar to the ATPase activity of Ssa, the ATPase activity of B--, measured at 0.25 μM ATP, was highly dependent on K+ concentration (Fig. 1B); in the absence of added potassium, activity was barely detectable whereas maximal activity was attained at ≈50 mM K+. This potassium dependence was strikingly different from the activity profile of the intact protein. At 10 μM ATP, a concentration ≈30 times its Km, this profile did not change (data not shown). Therefore, in summary, the Km of B-- and the K+ dependence of activity is very different from that of the intact protein but very similar to Ssa. Yet, the kcat of B-- is >100-fold higher than that of Ssa.
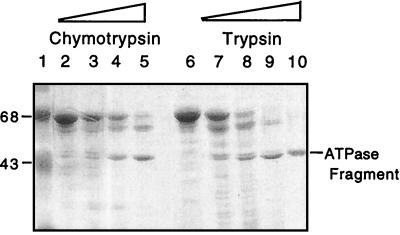
Tryptic and Chymotryptic Digestion of Ssb and Ssa1.
The ATPase domain of Ssb was generated under controlled conditions by using trypsin. Analytical digestions showed a slightly different pattern between trypsin and chymotrypsin hydrolysates. Trypsin hydrolysis yielded a slightly smaller ATPase fragment than chymotrypsin. Chymotryptic fragments had the same molecular weight, as judged by SDS/PAGE, as the ATPase fragment produced by spontaneous hydrolysis (Fig. 4). We therefore used chymotrypsin for a preparative hydrolysis of Ssb, and the ATPase domain so produced was separated by gel filtration chromatography, exactly as the spontaneously generated one. The potassium dependence for ATPase activity at 10 and 0.25 μM ATP and their Km at 100 mM potassium were calculated. The potassium dependence at both ATP concentrations was nearly identical to that of the spontaneously generated ATPase domain, and the Km for ATP at 100 μM was calculated to be 0.54 μM, very close to the Km for the spontaneously generated Ssb ATPase domain.
Figure 4.
Tryptic and chymotryptic generation of Ssb ATPase domain. SDS/PAGE separation of Ssb cleaved with increasing concentrations of chymotrypsin (lanes 2–5) or trypsin (lanes 6–9). The spontaneously generated ATPase fragment of Ssb is shown in lane 10.
To compare the effects of Ssb and Ssa peptide-binding domains on their respective ATPase domains, we purified Ssa1 and obtained its ATPase domain by chymotryptic hydrolysis and gel filtration chromatography under the same conditions as for Ssb (data not shown). As expected from analyses of other Hsp70s (3), at 100 mM potassium concentration, the kcat of this domain was ≈3-fold higher than that of intact Ssa1 (0.116 min−1 vs. 0.031 min−1), and its Km remained of the same order (0.49 vs. 0.11 μM). A-- potassium dependence was also similar to that of Ssa at both 0.5 μM (Fig. 1F) and 10 μM ATP (data not shown).
Biochemical Properties of Ssb ATPase Domain Under the Control of Ssa1 Peptide-Binding Domain in an Engineered Fusion Construct.
The ATPase domains of Ssb and Ssa share some biochemical properties, particularly Km and K+ dependence. The dramatic differences between the properties of full length Ssb and Ssa, however, suggest that the C terminus of each protein may affect the attached ATPase domain differently. To begin to test this idea, we purified a chimeric protein, BAA, containing the ATPase domain of Ssb and 18- and 10-kDa C-terminal domains of Ssa (Fig. 3 A, lane 5, and B, lane 5). If the ATPase domain of Ssb is similar to intact Ssa1, BAA having the Ssb ATPase domain under the control of Ssa1 peptide domain would yield a 70-kDa protein with an ATPase activity similar to that of Ssa1.
First, we carried out ATPase assays and calculated Km values for BAA at 2.5 and 100 mM potassium. The values obtained, 20 μM and 1.69 μM, respectively, were much closer to Ssa1 values of 4.2 and 0.11 μM than to Ssb values of 270 and 147 μM. (Table 1). The kcat values of BAA were strikingly similar to those of Ssa, particularly at 2.5 mM K+. The kcat of BAA and Ssa differed by only 2-fold (0.014 min−1 vs. 0.028 min−1) whereas the Ssb kcat was 0.95 min−1, 34-fold higher. In addition, the BAA ATPase activity was determined over a wide variety of potassium concentrations in the presence of 1 μM (Fig. 1D) and 10 μM ATP (data not shown). Like Ssa1 and the ATPase domains of Ssb and Ssa, the BAA chimera had very low activity at low K+ concentrations, and it reached maximal activity at ≈50 mM. Together, these results strongly suggest that the unique biochemical properties of the ATPase activity of Ssb are caused mostly by the effects of the peptide-binding domain on the ATPase domain and are not intrinsic to the ATPase domain itself.
Biochemical Properties of a Chimeric Ssa:Ssb Protein ABB.
We wanted to determine whether the peptide-binding domain of Ssb could influence the activity of other Hsp70 ATPase domains in the same way that it does its own. To test this idea, a chimeric Ssa1:Ssb protein in which the ATPase domain belonging to Ssa and the peptide-binding domain belonging to Ssb (ABB) was purified (Fig. 3 A, lane 6, and C, lane 6), and the potassium dependence and kinetic properties of its ATPase activity was measured. This particular fusion protein was difficult to purify. The best preparation was ≈90% pure, the major contaminants being Ssa1 and Kar2 (Fig. 3). Kinetic experiments demonstrated that the ATPase activity of ABB is more similar to Ssb than to Ssa. Km values were 205 and 99 μM at 2.5 and 100 mM potassium, respectively (Table 1). kcat values were 0.51 min−1 and 0.86 min−1 at 2.5 and 100 mM potassium, respectively, very close to those of Ssb (0.95 and 0.81 min−1). Accordingly, ABB showed little dependence on potassium for its ATPase activity; in the absence of added potassium, it retained more than half of its maximal activity whereas Ssa1 was virtually inactive under these conditions (Fig. 1C).
Stimulation of the ATPase Activity of Chimeric Proteins.
A general property of Hsp70 proteins is the ability of the ATPase activity to be stimulated by hydrophobic peptides and unfolded proteins (4). As reported here, the ATPase activity of Ssb could not be stimulated by peptides or unfolded proteins that were able to stimulate other Hsp70s of yeast. We also tested the ability of peptide A7 and CMLA, which stimulate the activity of Ssa 2-fold, respectively (Table 2), to stimulate the ATPase activity of the chimeric proteins BAA and ABB. A7 and CMLA stimulated the ATPase of BAA 2.4- and 2.5-fold, respectively, but had no effect on ABB ATPase activity (data not shown). Therefore, the ATPase domain of Ssb also shares the general property of Hsp70 proteins of being stimulatable by peptide within the context of the appropriate peptide-binding domain, but this ability is masked in the intact Ssb by the presence of the Ssb peptide-binding domain.
DISCUSSION
The two cytosolic hsp70 subfamilies Ssa1 and Ssb have distinct cellular functions even though they are in the same compartment of the cell (24). The results reported here uncover differences in the ATPase properties of these two classes of Hsp70s, which may play a role in determining their functional specificities. Although there are differences in the intrinsic properties between the ATPase domains of the two Hsp70s, more striking is the influence of the C-terminal regions. We demonstrate that the C-terminal regions not only influence the rate of ATP hydrolysis but also determine other properties of the ATPase activity. The differential influence of the C-terminal regions on the ATPase domains of Ssa1 and Ssb suggest that domain:domain interaction may in part determine functional specificity.
The 44-kDa ATPase domain of Ssa1 and Ssb—A-- and B--, respectively—have a similarly high steady-state affinity for ATP, as indicated by their similar Km values. However, the catalytic constant of B-- is ≈40-fold higher than that of A-- at physiological potassium concentrations. The activity of A-- is similar to that reported for the 44-kDa ATPase domain of Hsc70, as is the Km values for both yeast proteins (25). Therefore, B-- can be considered an Hsp70 ATPase of typical steady-state affinity but high activity.
It has been demonstrated previously that the ATPase activity of the N terminus of mammalian cytosolic Hsc70 is severalfold higher than that of the intact protein but is similar to the peptide-stimulated activity (25). This decrease in activity of the ATPase domain when in context of the intact protein suggests that the C terminus, in the absence of bound peptide, represses the activity of the N terminus. Similarly, both Ssa1 and Ssb have lower activity than their respective ATPase domains. However, the properties of Ssb ATPase domain are influenced more dramatically than that of Ssa in the context of the natural protein. The steady-state affinity for ATP is decreased greatly. The Km is 400-fold higher for the ATPase domain than for the full length protein. This modulatory effect of the Ssb C terminus is a property of the C terminus itself because it is capable of altering the ATPase domain of Ssa in a similar fashion. Fusion ABB has a 200-fold higher Km than A--. These data show that the Ssb peptide-binding domain can convert the high steady-state affinity ATPase domains of both Ssa1 and Ssb into low affinity ones.
The analysis of the K+ dependence of Ssa1 and Ssb provides another example of biochemical differences. K+ is required in the catalytic mechanism of ATP hydrolysis and, therefore, for the chaperone activity of Hsp70 proteins. Two potassium ions are bound to the catalytic site and are implicated in the correct positioning of the nucleotide and magnesium ions (26, 27). It is then somehow surprising that Ssb ATPase activity is nearly independent of the potassium concentration in the assay mixture. However, it is very improbable that Ssb has a different mechanism for ATP hydrolysis than the other hsp70s, especially because the residues implicated in the catalytic mechanism all are conserved in the different hsp70s, including Ssb (26). A more realistic explanation for the uncommon potassium dependence of Ssb ATPase activity is that the Ssb catalytic site has a higher affinity for potassium than the catalytic site of Ssa1 and that potassium remains bound to the catalytic site during the purification process. Following from this idea, Ssb peptide-binding domain would convert Ssb ATPase domain from an intrinsically potassium low affinity site to a potassium high affinity site. Furthermore, as demonstrated by the similar potassium profile of ABB and Ssb, the Ssb peptide-binding domain is also able to convert Ssa1 ATPase domain to a high affinity potassium binding site. However, regardless of the mechanism behind the K+ dependence, the alteration in potassium dependence as well as affinity for ATP by the presence of the C-terminal domain strongly argues for Hsp70-specific domain:domain interactions.
The reason(s) that any different Hsp70s of the same cellular compartment are unable to carry out the same cellular functions could be several: (i) differences in peptide-binding specificity, (ii) different ATPase characteristics and modifications of ATPase activity by the peptide-binding domain, the subject of this paper, and (iii) different interactions with other cellular proteins that function to modulation the chaperone activity of the Hsp70 or target it within the compartment. In the case of Ssb function, differences in the peptide-binding specificity between it and Ssa do not seem critical. The chimera BAB that contains the peptide-binding domain of Ssa, but is otherwise Ssb, rescues the two phenotypes of an ssb mutant strain, cold sensitivity and hypersensitivity to certain translation inhibiting drugs (20).
Here we show that there are both fundamental differences between the intrinsic ATPase activities of the Ssa and Ssb 44-kDa ATPase domains and the intrinsic ability of the two C-terminal domains to modulate the activity of an ATPase domain. However, whether these differences are critical for biological function will require more study because the results of the in vivo analysis carried out to date is complex. The fusion BAA rescues the cold-sensitive phenotype of a ssb1 ssb2 disruption strain. Here we demonstrate that the fusion BAA has biochemical properties more like Ssa1. This biochemical analysis is of particular interest because it suggests that it is not the B-like activity of the Ssb ATPase domain that confers rescue of the cold-sensitive phenotype. However, because wild-type Ssa1 cannot rescue Ssb function, there must be some feature of the Ssb ATPase domain that gives it Ssb-specific function. Perhaps this domain binds to a cochaperone or contains sequences that promote localization, giving BAA its Ssb-specific function.
On the other hand, ABB has biochemical characteristics like that of Ssb. It has a high Km for ATP and a high kcat, and its activity is not influenced by the addition of K+. ABB can rescue the drug sensitivity but not the cold sensitivity of the ssb1 ssb2 mutant. Of interest, the three fusions ABB, BAB, and BBA all rescue drug sensitivity and show ribosome association by our assays, raising the possibility that the ability to rescue is governed by proper localization on the ribosome. It is also possible that the biochemical characteristics of a particular fusion may determine its ability to rescue the drug sensitivity. It will be interesting to see whether these proteins have an ATPase activity like Ssb. If so, it would suggest that both the 18-kDa peptide-binding domains and 10-kDa domains can communicate with the ATPase domain to change its ability to bind ATP and hydrolyze it.
Perhaps the most surprising finding reported here is the difference between the ATPase characteristics of the two Hsp70s because, up to this time, such parameters have appeared to be similar amongst Hsp70s. But the different processes in which Hsp70 proteins are involved, such as protein translocation across membranes, protein refolding, de novo protein synthesis, etc., likely have different timetables and may require different ATPase binding and hydrolysis activities, which would be facilitated by different Hsp70 activities. Future studies should help us understand how the changes in biochemical properties of various hsp70s influence their abilities to function differently in the cell. The Ssa/Ssb Hsp70 subfamilies should provide a productive system for such analyses.
Acknowledgments
We thank P. Bertics, M. Krebs, K. Huchison, and P. James for helpful comments on this manuscript. This work was supported by a North Atlantic Treaty Organization Fellowship (to P.B.-L.), National Institutes of Health Training Grant T32-GM07215 (to C.P.), and National Institutes of Health Grant 5RO1 GM31107 (to E.A.C.).
ABBREVIATIONS
- hsp
heat shock protein
- CMLA
carboxymethylated α-lactalbumin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 2.Johnson J L, Craig E A. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 3.McKay D, Wilbanks S, Flaherty K, Ha J-H, O’Brien M, Shirvanee L. In: The Biology of Heat Shock Proteins & Molecular Chaperones. Morimoto R, Tissieres A, Georgopoulos C, editors. Plainview, NY: Coldo Spring Harbor Lab. Press; 1994. pp. 153–178. [Google Scholar]
- 4.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Schmid D, Baici A, Gehring H, Christen P. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 6.Theyssen H, Schuster H P, Packschies L, Bukau B, Reinstein J. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 7.Flynn G C, Chappell T G, Rothman J E. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 8.Ha J-H, McKay D. Biochemistry. 1994;33:14625–14635. doi: 10.1021/bi00252a031. [DOI] [PubMed] [Google Scholar]
- 9.Jordan R, McMacken R. J Biol Chem. 1995;270:4563–4569. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- 10.Zhu X, Zhao X, Burkholder W, Gragerov A, Ogata C, Gottesman M, Hendrickson W. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberek K, Marszalek J, Ang D, Georgopoulos C. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karzai A W, McMacken R. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 13.Miao B, Davis J E, Craig E A. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- 14.Hohfeld J, Minami Y, Hartl F-U. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 15.Packschies L, Theyssen H, Buchberger A, Bukau B, Goody R, Reinstein J. Biochemistry. 1997;36:3417–3422. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- 16.Hohfeld J, Jentsch S. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig E A, Baxter B K, Becker J, Halladay J, Ziegelhoffer T. In: The Biology of Heat Shock Proteins & Molecular Chaperones. Morimoto R I, Tissieres A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 31–52. [Google Scholar]
- 18.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 19.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke B A, Lopez-Buesa P, Walter W A, Wiedmann M, Craig E A. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James P, Pfund C, Craig E. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 21.Ziegelhoffer T, Lopez-Buesa P, Craig E A. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]
- 22.Cyr D M, Douglas M G. J Biol Chem. 1994;269(1):9798–9804. [PubMed] [Google Scholar]
- 23.McKay D B. Adv Protein Chem. 1993;44:67–98. doi: 10.1016/s0065-3233(08)60564-1. [DOI] [PubMed] [Google Scholar]
- 24.Craig E A, Ziegelhoffer T, Nelson J, Laloray S, Halladay J. Cold Spring Harbor Symposia on Quantitative Biology: Protein Kinesis: The Dynamics of Protein Trafficking & Stability. XV. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 441–449. [Google Scholar]
- 25.O’Brien M, McKay D. J Biol Chem. 1993;268:24323–24329. [PubMed] [Google Scholar]
- 26.Flaherty K M, DeLuca-Flaherty C, McKay D B. Nature (London) 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 27.Palleros D R, Reid K L, Shi L, Welch W, Fink A L. Nature (London) 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]