Abstract
Background and Aims
Polyploids, organisms that have multiple sets of chromosomes, are common in certain plant and animal taxa. However, there are only a few reports of intraspecific ploidy variation within the genus Quercus. The aim of the study was to investigate the suspected ploidy level of two oaks that have unusual microsatellite banding patterns.
Methods
Polyploidy was investigated by using microsatellite analysis, stomata length measurements and nuclear DNA content estimation by flow cytometry.
Key Results
Each putative triploid tree has patterns of microsatellite variation unexpected for diploid genomes, with up to three alleles at some loci, significantly longer stomata and 1·5 times more DNA per nucleus compared with diploids.
Conclusions
To our knowledge, this report contains the first evidence for triploidy in Q. petraea and confirmation of this phenomenon in Q. robur. Regardless of the positive or negative aspects of the presence of triploid oaks in forest stands, it is of value to be able to screen for them. This study demonstrates that nuclear microsatellites and estimation of DNA content by flow cytometry can readily be used for this purpose.
Key words: Quercus robur, Quercus petraea, microsatellites, stomata, flow cytometry, DNA amounts, triploidy
INTRODUCTION
The heritable condition of possessing more than two complete sets of chromosomes (polyploidy) is a prominent and significant force in evolution and speciation in plants and, to a lesser extent, in animals (Schultz, 1980; Levin, 1983; Soltis and Soltis, 1995). Well-documented examples of genome doubling exist in various groups of plants, insects, yeasts, amphibians, reptiles and fishes (Otto and Whitton, 2000; Wendel, 2000). Among trees, polyploids including triploids are common in angiosperms, where they represent one-third of all species, but they are rare in gymnosperms (Wright, 1962).
The genus Quercus includes about 300 species and belongs to the family Fagaceae, which represents the most important source of timber among broad-leaved species of the northern hemisphere. Oak wood is well recognized for its strength, durability and all-round usefulness (Ohri and Ahuja, 1990). However, despite the great economical value of polyploidy in crop and forest tree species, no thorough studies on polyploidy have been carried out in oaks. There have been a few, isolated reports on spontaneous triploidy and artificially induced polyploidy in Q. robur (Johnsson, 1946; Burda and Shchepotiev, 1973; Butorina, 1993; Naujoks et al., 1995; Lefort et al., 1998, 2000; Lefort and Douglas, 1999), but these have focused mainly on detection methods, providing evidence of triploidy rather than any indication of its frequency in nature (Lefort and Douglas, 1999).
There are several ways to detect polyploidy. In trees, cytological techniques are standard, but they are difficult to perform because of the small size of tree chromosomes (D'Emerico et al., 1995) and the presence of extra chromosomes in some individuals (Ohri and Ahuja, 1990; Zaldoš et al., 1998). The measurement of stomata length (Burda and Shchepotiev, 1973; Naujoks et al., 1995; Lefort and Douglas, 1999) and stomata area (Burda and Shchepotiev, 1973) may also enable the detection of triploids. However, detecting triploid trees seems to be much easier with genetic markers, such as isozymes (Naujoks et al., 1995) or microsatellites (Lefort et al., 1998, 2000; Lefort and Douglas, 1999). Among the various techniques available for estimating genome size, flow cytometry is convenient, rapid, precise and accurate for detecting small differences in DNA content (Zaldoš et al., 1998).
The purpose of this study was to explain the unusual microsatellite banding patterns observed in two mature trees during a genotyping study in a mixed oak stand of Q. robur and Q. petraea. Suspecting that these trees were triploid, their polyploidy was investigated by using conventional and molecular methods.
MATERIALS AND METHODS
Plant material
The oak trees were in a study site located in the Forest District Jamy, in the northern part of Poland near Grudziadz. This site is a 120-year-old mixed oak stand of Q. petraea and Q. robur. Within approx. 5 ha all adult oak trees were mapped and classified according to a set of morphological traits, including leaf characters and acorn morphology. Among 421 trees, 266 individuals (63 %) were assigned as typical Q. petraea and 145 individuals (34 %) were classified as Q. robur. Ten individuals (3 %) revealed characters of intermediate type and were classified as putative hybrids of the two oak species.
Vicia villosa ‘Minikowska’ was used as an internal standard for flow cytometry. Its DNA content (3·32 pg/2C) was estimated by using male human leucocytes (2C = 7·0 pg; Tiersch et al., 1989).
Microsatellite analysis
All mapped trees were sampled for leaves and genotyped using a set of six microsatellite markers — ssrQpZAG9, ssrQpZAG110 (Steinkellner et al., 1997), ssrQrZAG7, ssrQrZAG20 (Kampfer et al., 1998), MSQ4 and MSQ13 (Dow et al., 1995) — by the PCR multiplex protocol described by Dzialuk et al. (2005). PCR products were analysed using an ABI 310 automatic sequencer (Applied Biosystems, California, USA). DNA fragment analyses for two trees, namely no. 275 classified as Q. robur and no. 354 classified as Q. petraea, revealed patterns of microsatellite variation unexpected for diploid genomes, with up to three alleles at some loci.
For the two putative triploid trees sampling of leaf material was repeated using leaves collected at two crown levels: from the lowest branch by cutting with pruning scissors, and from within the crown by shooting down branches with a shotgun. In total, leaf samples were collected three times during the vegetation period. The leaves were used for microscopic stomata length measurements and for nuclear DNA content estimation by flow cytometry. In addition, wood samples were collected for simple sequence repeat (SSR) analyses.
In addition, leaves of two diploid individuals of Q. robur (nos. 89 and 356) and two of Q. petraea (nos. 187 and 220) were sampled as a reference for stomata length measurements and DNA content estimation to confirm the polyploid nature of the putative triploid trees.
Stomata length measurements
Stomata length was measured by using a light microscope at 400 × magnification, coupled with a camera and appropriate computer software (LUCIA-version G). Microscope slides (approx. 1·5 cm2 of fresh leaves) were prepared immediately prior to observation. For each tree, the lengths of 80 stomata were measured; in total, 480 stomata were analysed.
Nuclear DNA content estimation by flow cytometry
Plant material was prepared for flow cytometric analysis according to Galbraith et al. (1983), with some modifications necessary for species containing staining inhibitors in their cytosol (Zaldoš et al., 1998). Quercus and Vicia (internal standard) leaf blade fragments of about 0·5–1 cm2 were chopped simultaneously with a sharp razor blade in a plastic Petri dish with 1 ml of nucleus-isolation buffer (45 mm MgCl2.6H2O, 30 mm sodium citrate, 20 mm 3-[N-morpholino]propane sulfonic acid, 0·5 % v/v Triton X-100, pH 7·0), supplemented with two reductants [combination and concentrations established in a preliminary experiment, in which the two antioxidants, PVP and β-mercaptoethanol, were added to the buffer separately (PVP at 1 and 2 %, β-mercaptoethanol at 10 and 15 mm) or together, using different combinations of these concentrations], polyvinylpyrrolidone (PVP; 2 % v/v) and β-mercaptoethanol (15 mm), as well as with RNse A (50 µg mL–1). After chopping, the suspension was passed through a 50-μm mesh nylon filter and incubated for 10 min at room temperature, then propidium iodide (PI; 50 µg mL–1) was added and the sample was incubated for another 5 min at room temperature. For each sample, about 7000–10 000 nuclei were analysed using a Partec CCA (Münster, Germany) flow cytometer. Analyses were replicated seven times (for seven different leaves) for each tree. Histograms were analysed using a DPAC v.2·2 computer program. Nuclear DNA content was calculated according to the linear relationship between the ratio of the 2C peak positions of Quercus/Vicia on histograms of fluorescence intensities.
RESULTS
Microsatellite analysis
Among 421 trees, two trees, namely no. 275 classified as Q. robur and no. 354 classified as Q. petraea, revealed patterns of microsatellite variation unexpected for diploid genomes, with up to three alleles at some loci.
Multiplex PCR analyses of microsatellite variation repeated for the total material sampled from the two putative triploids demonstrated the same pattern, regardless of DNA source tissue (leaves or wood), crown level and sampling time. Thus, standard single microsatellite locus PCR reactions were performed. The results were identical to those obtained in multiplex reactions, confirming the presence of three peaks in some loci. Genotypes of both putative triploid trees are shown in Table 1. In both cases four of six marker loci revealed a three-peak pattern, which is expected for a triploid genome. The diploid-like pattern (i.e. two PCR products) was observed in the MSQ 4 locus for both putative hybrid trees. However, no homozygous genotype was found in either of the two genotyped trees.
Table 1.
Genotypes of two putative triploid trees at six microsatellite loci
| Tree | AG 7 | AG 9 | AG 20 | AG 110 | MSQ 4 | MSQ 13 |
|---|---|---|---|---|---|---|
| 275 | 130/132 | 191/195/197 | 157/161/175 | 204/210/212 | 208/214 | 218/224/230 |
| 354 | 140/146/148 | 193/195/197 | 161/163/189 | 208/240 | 202/204 | 190/220/224 |
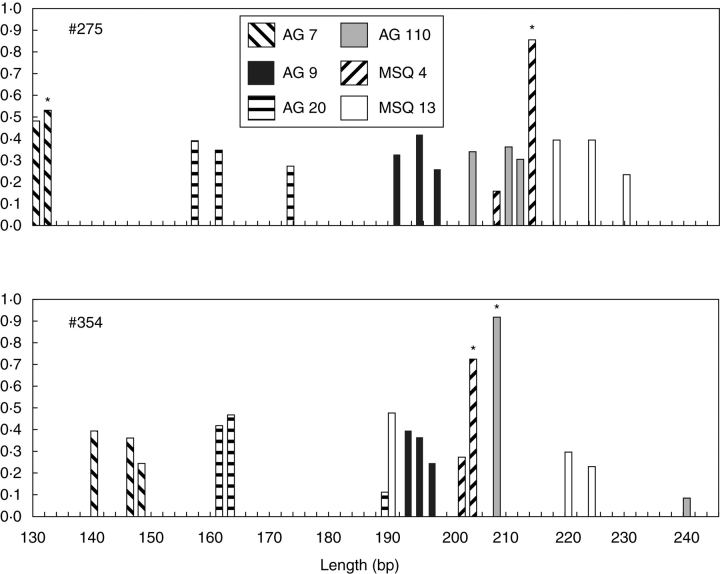
In the diploid-like genotypes, unusual differences were observed in the amount of PCR product between allelic products (scored as peak height), which may be related to the presence of the same allele in two out of three chromosomes. Figure 1 shows the relative amount (i.e. percentage) of the allelic products at all six loci. In the diploid-like MSQ 4 locus, the larger amount of product was scored for longer DNA fragments. The same pattern, but much less evident, was also observed for the AG 7 locus in tree no. 275. The opposite pattern was detected for the products of the AG 110 locus for tree no. 354. However, in this case the amount of PCR product was variable, as the shorter allele took over 80 % of the total PCR product of the locus.
Fig. 1.
Relative amounts of allelic PCR products scored for particular loci for two putative triploid trees. Abscissa: allele length in base pairs (bp). Ordinate: relative amounts. Peaks suspected to be a result of overlaps in allele fragment size are marked with asterisks.
Stomata length measurements
The stomata length of the two putative triploid trees ranged from 22·1 to 40·0 µm (mean 30·47 µm), whereas for the control sample it varied from 20·8 to 33·9 µm (mean 27·72 µm for Q. robur and 25·86 µm for Q. petraea) (Table 2). Each putative triploid tree had significantly longer stomata compared with control trees, as confirmed by Tukey's test (α < 0·05).
Table 2.
Stomata length and DNA content of diploid and putative triploid (*) Q. petraea and Q. robur trees
| Species | Tree no. | Stomata length (μm) |
Nuclear DNA content (pg, mean ± SD) |
Ploidy level | ||
|---|---|---|---|---|---|---|
| Range | Mean (s.e.) | 2C | 1Cx | |||
| Q. robur | 89 | 21·22–32·11 | 26·67 (0·353) | 1·62 ± 0·02 | 0·81 ± 0·01 | 2x |
| 356 | 21·96–33·91 | 28·77 (0·343) | 1·59 ± 0·01 | 0·79 ± 0·01 | 2x | |
| 275* | 23·73–40·00 | 30·58 (0·325)a | 2·44 ± 0·02 | 0·81 ± 0·01 | 3x | |
| Q. petraea | 187 | 22·11–31·91 | 26·23 (0·407) | 1·64 ± 0·02 | 0·82 ± 0·01 | 2x |
| 220 | 20·80–29·46 | 25·49 (0·166) | 1·62 ± 0·03 | 0·81 ± 0·01 | 2x | |
| 354* | 22·13–38·85 | 30·36 (0·189)b | 2·45 ± 0·03 | 0·82 ± 0·01 | 3x | |
a,bValues significantly different compared with diploid trees at α < 0·05 (Tukey's test).
DNA content estimation by flow cytometry
The flow cytometric histograms of nuclei isolated from Quercus leaves, regardless of the ploidy level of the tree, contained only a peak corresponding to the G0/G1 phase of the cell cycle, which facilitated the use of Vicia villosa (2C = 3·32 pg) as an internal standard. Modification of Galbraith's buffer (Galbraith et al., 1983) prevented any reduction of the fluorescence of the internal standard nuclei and improved histogram quality; the coefficient of variation (CV) for the Quercus peak ranged from 3·90 to 7·0 %, values regarded as acceptable for species containing staining inhibitors in their cytosol. 2C DNA values ranged from 1·59 to 2·45 pg (Table 2). The putative triploids contained 1·5-fold more DNA per nucleus (3Cx) than the diploids (2Cx). The monoploid genome size (Cx) of all trees was about 0·8 pg.
DISCUSSION
Plants can possess different ploidy values, even at the species level. Spontaneous intraspecific ploidy variation has been observed in several tree species including Acacia dealbata (Blakesley et al., 2002), Ulmus americana (Sherald et al., 1994) and Fraxinus americana (Schaefer and Miksche, 1977; Armstrong, 1982). Within the genus Quercus there are only few reports of spontaneous intraspecific ploidy variation in Q. robur. Triploidy has been confirmed in only 11 naturally occurring individuals, although most of these were found by coincidence during other studies (Table 3). Triploid oaks appear to occur very rarely. This study provides evidence for triploidy in a mixed stand of Q. petraea and Q. robur at a frequency of 0·48 %. Other authors have observed similar frequencies in Q. robur (Table 3). However, much higher frequencies (five triploids among 16 individuals analysed) were observed among selected trees of Q. robur (Lefort et al., 2000). To our knowledge, this report contains the first evidence of triploidy in Q. petraea. Triploid acorns are more frequent in oak trees producing twin seedlings at a high frequency (Johnsson, 1946). The origin of triploids is probably from gametes, which fail to undergo meiotic reduction. Among diploid oaks, several individuals produce 2n pollen grains comprising 5–10 % of the microspores (Butorina, 1993).
Table 3.
Frequency of triploidy and stomata length of triploid and diploid Q. robur trees
| Triploids | Diploids | Source | ||
|---|---|---|---|---|
| No. of individuals | Frequency | Mean stomata length (µm) | Stomata length (µm) | |
| 3 | 0·41 % | N/A | N/A | Johnsson (1946) |
| 2 | 0·57 % | 26·3 | Mean 20·2 | Burda and Shchepotiev (1973), Butorina (1993) |
| 1 | 0·25 % | 24·5 | 18·2–20·1 | Naujoks et al. (1995) |
| 5 | N/A | 28·5 | Mean 22·4–26·4 | Lefort et al. (1998, 2000), Lefort and Douglas (1999) |
This present study was motivated by the presence of an unusual tri-allelic pattern at some microsatellite loci in two oak individuals (one Q. robur and one Q. petraea). Unusual tri-allelic patterns for some nuclear microsatellite loci were reported for Q. robur earlier by Lefort and co-workers (Lefort et al., 1998, 2000; Lefort and Douglas, 1999). This tri-allelic pattern at some loci could result from aneuploidy of one or more chromosomes, or a duplication of one or several regions of the genome. Indeed, hyperaneuploidy and hypoaneuploidy were reported for oak in a cytological study previously (Butorina, 1993). However, the loci used were each located on a different chromosome (Barreneche et al., 1998, 2004); therefore, the two individuals investigated could have been examples of euploidy. Unusual banding patterns observed in only a few of several loci analysed seem to be typical for triploid trees. In oaks, Naujoks et al. (1995) observed additional alleles for two of 11 isozyme loci analysed in one individual; Lefort and Douglas (1999) found tri-allelic profiles at 2–6 microsatellite loci in six triploid oaks. Based on the amounts of amplification products, the diploid-like patterns can be explained to be a result of the overlapping of two allelic fragments of identical size. We note that tri- or multi-allelic profiles at microsatellite markers are also frequently observed in diploids due to localized duplications of chromosomal segments and/or past genome duplication events (Lynch and Conery, 2000). It is thus important to confirm differences in ploidy level using additional data.
Ploidy level was confirmed by measuring stomata length. According to Burda and Shchepotiev (1973), the stomata of triploid oaks are always significantly longer (sometimes up to 1·3 times) than in diploids. A similar tendency was observed in this study (Table 2) and by Naujoks et al. (1995) and by Lefort and Douglas (1999) (Table 3). Burda and Shchepotiev (1973) observed also a significantly larger area of leaf stomata (455·5 µm2) than in diploids (269·3 µm2).
The ploidy level was also confirmed by flow cytometric analysis of nuclear DNA content. The diploids of both Quercus species contained about 1·6 pg DNA. Thus, assuming that the monoploid genome size is 0·8 pg, the trees possessing 2·4 pg/2C clearly are triploids. The 2C values obtained here are slightly lower then those reported by Favre and Brown (1996) and Zoldoš et al. (1998; over 1·8 pg), probably because another internal standard, isolation buffer and/or fluorescent dye was used. However, the values were very close to those estimated using Feulgen microdensitometry (Ohri and Ahuja, 1990; Bennett and Smith, 1991).
Polyploid plants have larger cells but also individual plants are often larger. This has led to deliberate creation of polyploid varieties of such plants as watermelons, marigolds and snapdragons. In general, polyploids are considered to possess a greater ability to colonize a wider range of habitats and survive better in harsh unstable environments compared with their diploid progenitors, probably due to increased heterozygosity and genic and biochemical flexibility provided by the presence of additional alleles (Stebbins, 1985; Song et al., 1995; Matzke et al., 1999). In addition, compared with their diploid progenitors, triploids seem to be more tolerant to drought, and more resistant to pathogens and pests due to enhanced production of various secondary plant metabolites (Levin, 1983). Triploid oaks have the potential to be highly productive. They differ from other individuals of similar age because they are unusually large, having all of the characteristics of elite trees (Butorina, 1993; Naujoks et al., 1995; Lefort et al., 1998). The observed giant characters have fostered optimistic expectations regarding breeding of polyploid forest trees (Naujoks et al., 1995; Lefort and Douglas, 1999). Their natural occurrence presents an opportunity for studying the fertility and vigour of triploid oaks in the wild, and the possibility of testing the hypothesis that they can be used in forest tree breeding programmes. Although triploid oaks are capable of producing acorns, they have rather low fertility (Butorina, 1993). This fertility (the occurrence of flowering and fruit set) offers opportunities for further research concerning allele segregation and gene flow. Meiosis in microsporogenesis of these trees is highly disturbed and, as a consequence, pollen with unbalanced chromosome numbers is produced. The number of cells showing meiotic disturbances may vary in different years, reaching percentages of abnormal divisions of up to 98 % of the total number of dividing microspocytes (Butorina, 1993).
The presence of triploid oaks in forest stands where seeds are collected for reforestation purposes may have further implications. Triploid oaks that are unusually large may be selected as elite trees (Lefort and Douglas, 1999) utilized for harvesting seeds or cuttings with the aim of establishing seed orchards. Such sampling may unintentionally increase the proportion of triploids in further generations, probably reducing their fertility. Regardless of the positive or negative aspects of the presence of triploid oaks in forest stands it is of value to be able to screen for them. This study demonstrated that nuclear microsatellites and estimation of DNA content by flow cytometry can readily be used for this purpose.
ACKNOWLEDGEMENTS
We wish to thank Dr Roman Gout (National Forestry University of Ukraine) and Professor J. Derek Bewley (University of Guelph, Canada) for comments and suggestions on the manuscript. This research was partly supported by research grant 3 P06L 034 23 obtained from the Ministry of Science and Higher Education, Poland.
LITERATURE CITED
- Armstrong JE. Polyploidy and wood anatomy of mature white ash, Fraxinus americana. Wood and Fiber Science. 1982;14:331–339. [Google Scholar]
- Barreneche T, Bodenes C, Lexer C, Trontin JF, Fluch S, Streiff R, et al. A genetic linkage map of Quercus robur L. (pedunculate oak) based on RAPD, SCAR, microsatellite, minisatellite, isozyme, and 5S rDNA markers. Theoretical and Applied Genetics. 1998;97:1090–1103. [Google Scholar]
- Barreneche T, Casasoli M, Russel K, Akkak A, Meddour H, Plomion C, Villani F, Kremer A. Comparative mapping between Quercus and Castanea using simple sequence repeats (SSRs) Theoretical and Applied Genetics. 2004;108:558–556. doi: 10.1007/s00122-003-1462-2. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. Nuclear DNA amount in angiosperms. Philosophical Transactions of the Royal Society of London B. 1991;334:309–345. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Blakesley D, Allen A, Pellny TK, Roberts AV. Natural and induced polyploidy in Acacia dealbata Link. and Acacia mangium Wild. Annals of Botany. 2002;90:391–398. doi: 10.1093/aob/mcf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda RI, Shchepotiev FL. Spontaneous polyploidy in seedlings of multi-seeded acorns of Quercus robur L. Cytology and Genetics. 1973;7:140–143. (in Russian) [Google Scholar]
- Butorina AK. Cytogenetic study of diploid and spontaneous triploid oaks, Quercus robur L. Annales des Sciences Forestieres. 1993;50:144–150. [Google Scholar]
- D'Emerico S, Bianco P, Medagli P, Schirone B. Karyotype analysis in Quercus spp. (Fagaceae) Silvae Genetica. 1995;44:66–70. [Google Scholar]
- Dow BD, Ashley MV, Howe HF. Characterization of highly variable (GA/CT)n microsatellites in the bur oak, Quercus macrocarpa. Theoretical and Applied Genetics. 1995;91:137–141. doi: 10.1007/BF00220870. [DOI] [PubMed] [Google Scholar]
- Dzialuk A, Chybicki I, Burczyk J. PCR-multiplexing of nuclear SSR loci in Quercus sp. Plant Molecular Biology Reporter. 2005;23:121–128. [Google Scholar]
- Favre JM, Brown S. A flow cytometric evaluation of the nuclear DNA content and GC percent in genomes of European oak species. Annales des Sciences Forestieres. 1996;53:915–917. [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JR, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Johnsson H. Chromosome numbers of twin plants of Quercus robur and. Fagus sylvatica. Hereditas. 1946;32:469–472. doi: 10.1111/j.1601-5223.1946.tb02787.x. [DOI] [PubMed] [Google Scholar]
- Kampfer S, Lexer Ch, Glössl J, Steinkellner H. Characterization of (GA)n microsatellite loci from Quercus robur. Hereditas. 1998;129:183–186. [Google Scholar]
- Lefort F, Douglas GC. Occurrence and detection of triploids by microsatellite analysis. In: Douglas GC, editor. Strategies for improvement of forest tree species.; Proceedings of the Teagasc/TDC Symposium on Forest Genetics; COFORD, Dublin, Ireland. 1999. pp. 19–35. [Google Scholar]
- Lefort F, Lally M, Thompson D, Douglas GC. Morphological traits, microsatellite fingerprinting and genetic relatedness of a stand of elite oaks (Q. robur L.) at Tullynally, Ireland. Silvae Genetica. 1998;47:257–262. [Google Scholar]
- Lefort F, Douglas GC, Thompson D. Microsatellite DNA profililng of phenotypically selected clones of Irish oak (Quercus spp.) and ash (Fraxinus exxelsior L.) Silvae Genetica. 2000;49:21–28. [Google Scholar]
- Levin DA. Polyploidy and novelty in flowering plants. American Naturalist. 1983;122:1–24. [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Scheid OM, Matzke AJM. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. Bioessays. 1999;21:761–767. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Naujoks G, Hertel H, Ewald D. Characterization and propagation of an adult triploid pedunculate oak (Quercus robur L.) Silvae Genetica. 1995;44:282–286. [Google Scholar]
- Ohri D, Ahuja MR. Giemsa C-banded karyotype in Quercus L. (oak) Silvae Genetica. 1990;39:216–219. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Sherald JL, Santamour FS, Hajela RK, Jr, Hajela N, Sticklen MB. A Dutch elm disease resistant triploid elm. Canadian Journal of Forest Research. 1994;24:647–653. [Google Scholar]
- Schultz RJ. Role of polyploidy in the evolution of fishes. In: Lewis WH, editor. Polyploidy: biological relevance. New York: Plenum Press; 1980. pp. 313–340. [Google Scholar]
- Schaefer VG, Miksche JP. Microspectrophotometric determination of DNA per cell and polyploidy in. Fraxinus americana. Silvae Genetica. 1977;26:184–192. [Google Scholar]
- Soltis DE, Soltis PS. The dynamic nature of polyploid genomes. Proceedings of the National Academy of Sciences of the USA; 1995. pp. 8089–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proceedings of the National Academy of Sciences of the USA; 1995. pp. 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL., Jr Polyploidy, hybridization and invasion of new habitats. Annals of the Missouri Botanical Garden. 1985;72:824–832. [Google Scholar]
- Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A, et al. Identification and characterization of (GA/CT)n microsatellite loci from. Quercus petraea. Plant Molecular Biology. 1997;33 doi: 10.1023/a:1005736722794. [DOI] [PubMed] [Google Scholar]
- Tiersch TR, Chandler RW, Wachtel SSM, Ellias S. Reference standards for flow cytometry and application in comparative studies of nuclear DNA content. Cytometry. 1989;10:706–710. doi: 10.1002/cyto.990100606. [DOI] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Wright JW. Genetics of forest tree improvement. Rome: FAO; 1962. FAO Forestry and Forest Products Studies, No. 16. [Google Scholar]
- Zaldoš V, Papeš D, Brown SC, Panaus O, Šiljak-Yakovlev S. Genome size and base composition of seven Quercus species: inter- and intra-population variation. Genome. 1998;41:162–168. [Google Scholar]