Abstract
Background and Aims
The recent assembly of the complete sequence of the plastid genome of the model taxon Physcomitrella patens (Funariaceae, Bryophyta) revealed that a 71-kb fragment, encompassing much of the large single copy region, is inverted. This inversion of 57% of the genome is the largest rearrangement detected in the plastid genomes of plants to date. Although initially considered diagnostic of Physcomitrella patens, the inversion was recently shown to characterize the plastid genome of two species from related genera within Funariaceae, but was lacking in another member of Funariidae. The phylogenetic significance of the inversion has remained ambiguous.
Methods
Exemplars of all families included in Funariidae were surveyed. DNA sequences spanning the inversion break ends were amplified, using primers that anneal to genes on either side of the putative end points of the inversion. Primer combinations were designed to yield a product for either the inverted or the non-inverted architecture.
Key Results
The survey reveals that exemplars of eight genera of Funariaceae, the sole species of Disceliaceae and three generic representatives of Encalyptales all share the 71-kb inversion in the large single copy of the plastid genome. By contrast, the plastid genome of Gigaspermaceae (Funariales) is characterized by a gene order congruent with that described for other mosses, liverworts and hornworts, and hence it does not possess this inversion.
Conclusions
The phylogenetic distribution of the inversion in the gene order supports a hypothesis only weakly supported by inferences from sequence data whereby Funariales are paraphyletic, with Funariaceae and Disceliaceae sharing a common ancestor with Encalyptales, and Gigaspermaceae sister to this combined clade. To reflect these relationships, Gigaspermaceae are excluded from Funariales and accommodated in their own order, Gigaspermales order nov., within Funariideae.
Key words: Plastid genome, Physcomitrella patens, Funariaceae, Funariales, Bryophyta, inversion, Gigaspermaceae
INTRODUCTION
The structure and size of the plastid genome has been greatly altered since its endosymbiotic origin (Dyall et al., 2004; Hackett et al., 2004; Timmis et al., 2004). Many more genes have been transferred to the nucleus (Martin et al., 1998, 2002; Korpelainen, 2004) or more rarely to the mitochondrion (Nakazano and Hirai, 1993; Zheng et al., 1997) than gained (Goremykin et al., 2003). Most of the size reduction in the genome occurred prior to the diversification of land plants, and hence the size and structure of the plastid genome of embryophytes is fairly well conserved (Raubeson and Jansen, 2005). Phylogenetic reconstructions in plants have relied extensively on plastid DNA as a source of characters, either for raw nucleotide sequences (Soltis and Soltis, 1998) or for genomic rearrangements (Olmstead and Palmer, 1994; Raubeson and Jansen, 2005). Changes in the composition of the plastid genome have been reported for several plants and their green algal ancestors (e.g. Turmel et al., 1999; Martin et al., 2002; Hackett et al., 2004). Some gene transfers to the nucleus are rare and hence phylogenetically highly informative (Kelch et al., 2004), whereas others have occurred repeatedly and hence carry a more complex phylogenetic signal (e.g. Lavin et al., 1990; Doyle et al., 1995; Millen et al., 2001).
Other structural rearrangements relate to the size of the inverted repeat, a duplicated region that separates the large and small single copy units (e.g. Plunkett and Downie, 2000), and to the actual order of genes (e.g. Raubeson and Jansen, 1992). Unlike gene losses and changes in the composition of the inverted repeat, inversions of segments comprising several genes appear to be rather rare events (Soltis and Soltis, 1998) and hence compose a class of powerful phylogenetic markers (Rokas and Holland, 2000). For example, a 22·8-kb and a 3·3-kb inversion mark the split between Barnadesioideae and the remainder of Asteraceae, which all share these genomic changes (Kim et al., 2005). Similarly the phylogenetic distribution of a 50-kb inversion in the plastid genomes in Fabaceae (Saski et al., 2005) supports the paraphyly of certain suprageneric taxa and suggests that this extensive rearrangement occurred only once during the evolutionary history of Fabaceae (Doyle et al., 1996).
Less is known about the potential systematic significance of genome structural changes in mosses, but early indications suggest that this is a fruitful avenue of research. The plastid genome of Physcomitrella patens (Hedw.) Bruch and Schimp. (Sugiura et al., 2003) differs from typical plant genomes in the loss of the DNA-directed RNA polymerase alpha chain gene (rpoA) to the nuclear genome and from genomes of other early land plants such as liverworts and hornworts by a 71-kb inversion in the large single copy (LSC). Although the gene loss was initially considered diagnostic of mosses (Sugiura et al., 2003), Sugita et al. (2004) and Goffinet et al. (2005) demonstrated that the gene was present in the plastid genome of early diverging lineages, such as the peatmosses (Sphagnopsida), and lost twice during the evolutionary history of Bryophyta. The 71-kb long inverted fragment comprises 57% of the genome and is the largest inversion reported to date in the plastid genome of plants. Sugiura et al. (2003) considered the inversion to be diagnostic of Physcomitrella since a survey of several arthrodontous mosses (i.e. Bryopsida) revealed that the order of genes in their plastid genomes followed a sequence similar to that found in liverworts (Ohyama et al., 1986) and hornworts (Kugita et al., 2003).
Physcomitrella belongs to Funariaceae, a family of terricolous mosses defined by rather small gametophytes bearing unicostate leaves composed of smooth, lax rectangular cells, and a unique peristomial architecture (Vitt, 1982; Fife, 1985; Shaw et al., 1989). Funariaceae comprise 16 genera (Goffinet and Buck, 2004; Werner et al., 2007), of which three accommodate approx. 90% of the species diversity (Crosby et al., 1999). The family is considered to be closely allied to Disceliaceae (Goffinet and Cox, 2000), which accommodates a single species, the gametophore of which is highly reduced and arises from a persistent protonema (Vitt, 1982). Gigaspermaceae, a family of highly specialized mosses, with short foliate branches developing from underground rhizomes and immersed capsules holding large spores, have traditionally been associated with the former families within Funariales (e.g. Vitt, 1982, 1984; Thouvenot, 2000). Phylogenetic inferences suggest that Funariales are most closely related to Encalyptales, although their shared ancestry is only weakly to moderately supported (e.g. Goffinet and Cox, 2000; Cox et al., 2004). The phylogenetic affinities within this clade, and in particular those of Gigaspermaceae, are also ambiguous. Indeed, inferences from plastid and nuclear data, resolved with low bootstrap support, Gigaspermaceae as the sister-group to the remainder of Funariales and Encalyptales (Goffinet and Cox, 2000; Goffinet et al., 2001), rather than a member of Funariales. Timmiaceae, which comprise a single genus, Timmia Hedw., may share a unique common ancestor with Funariales and Encalyptales (Cox et al., 2004), but their inclusion in Funariidae (Goffinet and Buck, 2004) also remains unsettled.
Goffinet et al. (2005) reported that the 71-kb inversion characterized not only the genome of Physcomitrella but also that of two other taxa within Funariaceae [i.e. Funaria hygrometrica Hedw. and Entosthodon laevis (Mitt.) Fife] and Encalypta ciliata Hedw. (Encalyptaceae). By contrast, the plastid genome of Timmia lacked the rearrangement. Whether the inversion characterizes all remaining members of Funariidae (i.e. all Funariales and Encalyptales) remained ambiguous. Here, DNA sequences spanning the putative end points of the inversion in the LSC unit are surveyed in various members of Funariidae to assess the distribution and the phylogenetic significance of the inversion in this lineage of mosses.
MATERIALS AND METHODS
Taxon sampling
Exemplars of 13 genera of Funariales (eight Funariaceae, one Disceliaceae and four Gigaspermaceae) and all three genera of Encalyptales (sensu Goffinet and Buck, 2004) were studied for the organization of their plastid genome. The species sampled were: Discelium nudum (Smith 47503, NYSM-Disceliaceae), Aphanorrhegma serrata (Goffinet s.n.), Entosthodon bonplandii (Goffinet 6326), E. laevis (Goffinet 5601), E. serratum (Bowers 13109), Funaria flavicans (Goffinet 4798), F. hygrometrica 1 (Goffinet 5576), F. hygrometrica 2 (Goffinet 9278), Funariella curviseta (Ros and Werner 15/1/2006), Goniomitrium acuminatum (Curnow 6532), G. seroi (Puche 26/1/2004), Physcomitrella patens (Culture WT-CH, University of Geneva, Switzerland), Physcomitrium lorentzii (Goffinet 5348), P. pyriforme 1 (Goffinet 4737), P. pyriforme 2 (Goffinet 9276), Pyramidula tetragona (Ros et al. 15/3/1997) – all Funariaceae; Chamaebryum pottioides (vanRooy 9747200 1), Gigaspermum repens 1 (Schofield 90527) Gigaspermum repens 2 (Tyshing s.n.), Lorentziella imbricata (Schinini 24785, NY), Oedipodiella australis (Thouvenot s.n) – all Gigaspermaceae); Bryobartramia novae-valesiae (Magill and Schelpe 3218a), Bryobrittonia longipes (Ignatov 1997, NY), Encalypta armata (Goffinet 5613, DUKE) – all Encalyptales. Catascopiaceae, which were resolved with poorly supported affinities to Funariales by Goffinet et al. (2001) have now been shown to share a common ancestry with Dicraniidae (Quandt et al., 2007) and hence are not sampled here. All vouchers are deposited in the herbarium of Duke University (DUKE), unless otherwise indicated. Material adequate for DNA extraction could not be obtained for several exotic and monospecific genera of Funariaceae or for Costesia Thér. (Gigaspermaceae).
DNA extraction, PCR amplification and sequencing
DNA was extracted using the NucleoSpin® Plant kit from Macherey Nagel (Düren, Germany) following the manufacturer's protocol. The inversion breaks the sequence between the RNA polymerase β chain gene (rpoB) and the gene encoding the tRNA-Cys (trnCGCA) at the 5′ end, and between the ribosomal protein S11 (rps11) and the cytochrome b6/f complex subunit IV (petD) genes at the 3′ end. To test whether the 71-kb inversion in the gene order that characterized the Physcomitrella genome is present in other taxa, the region spanning both sides of the breakage point at the ends of the inversion was targeted with primers originally designed by Sugiura et al. (2003) (rps11F: TTTTGTTCGTGATGTAACTCCTATG; rpoBR: CTACCATAGCATCCTCAGTAGATT) and several newly designed primers (petN-F2: CCATTAAAGCACCCCAAGC; Giga-petD-R2: GGTTAGGTATTGGAGCAGC; petD-FunF: CCTTTCCGTCGTCCAGTAG; rps11-Fun: CATAATGGRTGTRGRCCTCC; rpoB-Fun: GGAATACTTCCAATRAATATAG; rpoBR-2: GATAATCTATTAAAGGAATACTTCC and trnC-Fun: GCAATCCTCTGCCTTACCAC). The primers were used in various combinations reflecting the gene arrangements at the ends of the potentially inverted region and the gene order at the 3′ end of the plesiomorphic gene order (see Fig. 1A). The amplification was performed in 25 µL with one unit of Hot Master Taq polymerase (Eppendorf AG, Westbury, NY, USA), 1 µL each of a 10 mm solution of each primer, 1 µL of a 10 mm solution of dNTPs, and a 99·9% pure solution of dimethyl sulfoxide (only with rps11F-Giga-petD-R2). The annealing temperature was optimized for individual combinations as follow: 52°C for rps11-Fun with rpoB-Fun, rps11F with Giga-petD-R2 and trnC-Fun with rpoBR2; 55°C for petNF2 with petD-FunF and trnC-Fun with petD-FunF; 56°C for rps11F with rpoBR. The amplification followed the same profile in each case: 95°C for 1 min followed by 30 cycles of denaturation (1 min at 95°C), annealing (1 min), extension (1 min at 72°C), and a final extension at 72°C for 7 min. Amplicons were purified using the NucleoSpin® ExtractII kit from Macherey Nagel following the manufacturer's instructions.
Fig. 1.
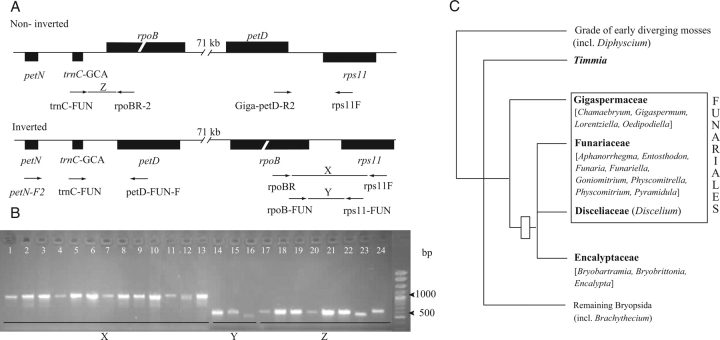
Distribution and phylogenetic significance of the 71-kb inversion in mosses. (A) Location of primers used to target the regions spanning both ends of the inverted region of the large single copy unit of the chloroplast genome in mosses. Non-inverted arrangement is typical of mosses. Inverted gene order was described for Physcomitrella. Note that the polarity of the genome is defined with reference to the Marchantia genome. Genes that are drawn above the line are transcribed left to right, and genes that are drawn below the line are transcribed right to left. See text for primer information. (B) Results from screening taxa of Funariidae for their chloroplast genome architecture based on PCR using primers flanking the putative break points of the inversion. Lanes 1–16: amplicons of region X or Y spanning the 3′ end of inversion in Funariaceae, Disceliaceae and Encalyptaceae: 1, Aphanorrhegma serrata; 2, Entosthodon bonplantii; 3, E. laevis; 4, E. serratus; 5, Funaria hygrometrica; 6, F. flavicans; 7, Funariella curviseta; 8, Physcomitrella patens; 9, Physcomitrium pyriforme; 10, P. lorentzii (Funariaceae); 11, Bryobartramia novae-valesiae; 12, Bryobrittonia longipes; 13, Encalypta armata (Encalyptaceae); 14, Goniomitrium acuminatum; 15, Pyramidula tetragona (Funariaceae); 16, Discelium nudum (Disceliaceae). Lanes 17–24: amplicons of region Z spanning the 5′ end of putative inversion in Gigaspermaceae and other mosses: 17, Chamaebryum pottioides; 18, Gigaspermum repens 1; 19, Gigaspermum repens 2; 20, Lorentziella imbricata; 21, Oedipodiella australis (Gigaspermaceae); 22, Timmia megapolitana; 23, Diphyscium foliosum; 24, Brachythecium salebrosum. (C) Putative phylogenetic relationships within Funariidae (in bold, sensu Goffinet and Buck, 2004), based on Goffinet and Cox (2000); Goffinet et al. (2001), Cox et al. (2004) and Werner et al. (2007). Polytomies identify currently unresolved relationships. The open bar identifies most parsimonious reconstruction for the occurrence of the inversion during the diversification of Funariidae.
All amplicons were sequenced using the PCR primers and these reactions were performed using the ABI PRISM® BigDye™ Terminators ver. 1·1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) optimized for half- or quarter-size reactions. Sequencing products were purified using Sephadex G-50 (Amersham, Piscataway, NJ, USA) gel filters, and then separated by capillary electrophoresis using an ABI Prism™ 3100 Genetic Analyser. Nucleotide sequences were edited using Sequencher 3·1·1 (Gene Codes Corporation, Ann Arbor, MI, USA), entered in PAUP* version 4·0b10 for Macintosh-PPC (Swofford, 2002) and manually aligned to those published for Physcomitrella (Sugiura et al., 2003) or other mosses representing a range of lineages lacking the inversion and the rpoA gene (Goffinet et al., 2005), in order to define genes and intergenic spacers. All sequences were submitted to GenBank (Table 1).
Table 1.
Length of the spacer regions spanning both break points of the 71-kb inversion in Funariidae and their homologous regions in non-inverted genomes
| rps11-rpoB, 5′ end | petD-petN, 3′ end* | trnC-rpoB, 5′ end | rps11-petD, 3′ end | |
|---|---|---|---|---|
| Funariales | ||||
| Disceliaceae | ||||
| Discelium nudum (Dicks.) Brid. | † | 254† (EF173139) | ||
| Funariaceae | ||||
| Aphanorrhegma serrata (Hook. and Wils.) Sull. | 220 (EF173134) | 698 (EF173158) | ||
| Entosthodon bonplandii (Hook.) Mitt. | 239 (EF173127) | 685 (EF173153) | ||
| Entosthodon laevis (Mitt.) Fife | 220 (EF173128) | 697 (EF173151) | ||
| Entosthodon serratus (Brid.) Fife | 221 (EF173130) | 686 (EF173156) | ||
| Funaria hygrometrica Hedw. 1 | 227 (EF173137) | 667 (EF173160) | ||
| Funaria hygrometrica Hedw. 2 | 227 (EF173138) | 669 (EF173161) | ||
| Funaria flavicans Michx. | 227 (EF173136) | 669 (EF173159) | ||
| Funariella curviseta (Schwägr.) Milde | 220 (EF173129) | 692 (EF173152) | ||
| Goniomitrium acuminatum Hook. and Wils. | 222 (EF173124) | 283† (EF173141) | ||
| Goniomitrium seroi Casas | 222 (EF173125) | — | ||
| Physcomitrella patens (Hedw.) Bruch and Schimp. | 220 (EF173135) | 696 (EF173157) | ||
| Physcomitrium lorentzii C.A. Müller | 220 (EF173133) | 264† (EF173142) | ||
| Physcomitrium pyriforme (Hedw.) Hampe 1 | 221 (EF173131) | 697 (EF173154) | ||
| Physcomitrium pyriforme (Hedw.) Hampe 2 | 220 (EF173132) | 690 (EF173155) | ||
| Pyramidula tetragona (Brid.) Brid. | 231 (EF173126) | 270† (EF175217) | ||
| Gigaspermaceae | ||||
| Chamaebryum pottioides Thér. and Dixon | 264 (EF173146) | 969 (EF175218) | ||
| Gigaspermum repens (Hook.) Lindb. | 285 (EF173143) | |||
| Lorentziella imbricata (Mitt.) Broth. | 279 (EF173145) | |||
| Oedipodiella australis (Wager and Dixon) Dixon | 285 (EF173144) | |||
| Encalyptales | ||||
| Encalypta armata Dusén | 236 (EF173121) | 716 (EF173150) | ||
| Bryobrittonia longipes (Mitt.) D.G. Horton | 232 (EF173123) | — | ||
| Bryobartramia novae-valesiae (Broth. ex G. Roth) I.G. Stone and G.A.M. Scott | 238 (EF173122) | 293† (EF173140) | ||
| OUTGROUP TAXA | ||||
| Diphyscium foliosum (Hedw.) D. Mohr | 264 (EF173147) | 526 (AY911401) | ||
| Timmia megapolitana Hedw. | 303 (EF173148) | — | ||
| Brachythecium salebrosum (Hoffm. ex F. Weber and D. Mohr) Schimp. | 290 (EF173149) | 309 (AY911404) |
All sequences were deposited in GenBank.
–, Amplicon obtained but no sequence available.
* Unless otherwise noted, the petN-petD region includes the petN-trnC spacer, trnC (70 bp) and the trnC-petD spacer.
† trnC-petD spacer only.
RESULTS
Amplification using primer pairs compatible with the gene order at both ends of the inverted fragment described for Physcomitrella yield products for species of seven additional genera of Funariaceae (Aphanorrhegma, Entosthodon, Funaria, Funariella, Goniomitrium, Physcomitrium and Pyramidula), Discelium nudum (Disceliaceae) and for exemplars of the three genera of Encalyptaceae (Fig. 1B). The length of the rps11-rpoB intergenic spacer varied between 220 and 239 nucleotides. The amplicon obtained for Discelium could only be sequenced in the reverse direction. This sequencing reaction yielded a sequence that included a portion of the rpoB gene and much but not all of the rps11-rpoB intergenic spacer. The sequence of the intergenic spacer in these taxa could be unambiguously aligned to the published sequence of Physcomitrella. The two exemplars of Physcomitrium pyriforme differ by a single additional T in a poly-T region. No size variation was observed between two accessions of Funaria hygrometrica. The 5′ end of the inversion was targeted using distinct primer pairs that span only the petD-trnC region or the longer petN-petD region, which includes the petN-trnC spacer, the trnC gene (70 bp) and the trnC-petD spacer. An amplicon was obtained from Goniomitrium seroi and Bryobrittonia longipes, but forward and reverse reactions failed to join in the trnC-petD spacer region. Ambiguous base calls seem to be caused by difficulties in sequencing through polynucleotide or short dinucleotide repeat regions. All other newly generated sequences are complete and align unambiguously with those of Physcomitrella. For none of the exemplars of Funariaceae, Disceliaceae and Encalyptaceae could a PCR product, compatible with a non-inverted architecture, be obtained. Conspecific samples of Funaria hygrometrica differ by two additional adenosines in a poly-A region in the trnC-petD intergenic spacer. Physcomitrium pyriforme 1 differs from the other exemplar of this species by the insertion of three nucleotides and the deletion of one in the petN-trnC intergenic spacers and the insertion of five adenosines in a poly-A region in the trnC-petD spacer.
Members of the four genera of Gigaspermaceae tested negative for the inversion and positive for the non-inverted genome architecture (Fig. 1B). Their gene arrangements in regions homologous, in terms of their position, to the end points of the inverted region in Physcomitrella are compatible with the non-inverted type characteristic of other mosses (Fig. 1B). The fragment of the trnC-rpoB intergenic spacer in Gigaspermaceae varies in length between 264 and 285 bp, which is similar to the range found among the three outgroup taxa screened here (Fig. 1B and Table 1). The sequence of the spacer aligns well across these taxa. Amplification spanning the end point at the 3′ end yielded a single band for Chamaebryum, Lorenziella and Oedipodiella but two for Gigaspermum. The amplicon spanning the rps11 to petD region is much longer than that of most other members of Bryopsida that lack the rpoA gene (results not shown). The rps11-petD amplicon could only be sequenced for Chamaebryum for which it is 969 bp long compared with 197 bp in Tetraplodon mnioides or 720 bp in Tetraphis pellucida (Goffinet et al., 2005). A BLAST search for this sequence yielded no match.
DISCUSSION
Gene order in the plastid genome of embryophytes is considered rather conserved (Raubeson and Jansen, 2005). Alterations in the sequence of genes result either from gene losses due to the transfer to the nuclear genome or small permutations. The inversion of 71 kb of the LSC of Physcomitrella (Sugiura et al., 2003) is the largest inversion documented in plants to date. Initially considered diagnostic of Physcomitrella, and then shown to occur in other members of Funariidae (Goffinet et al., 2005), it is here revealed to characterize the genome of all members of Funariaceae, Disceliaceae and Encalyptales screened in this study. By contrast, species of four genera of Gigaspermaceae, a family traditionally considered closely related to Funariaceae and Disceliaceae with which they compose Funariales, lack the inversion. Recent phylogenetic inferences suggested that Funariaceae and Disceliaceae share a most recent common ancestry with Encalyptales rather than Gigaspermaceae (Goffinet and Cox, 2000; Goffinet et al., 2001). Considered dubious because of the lack of support from nucleotide sequence data alone, this hypothesis was ignored in the most recent classification of mosses (Goffinet and Buck, 2004).
Genomic rearrangements are considered rare and thus phylogenetically highly informative events (Rokas and Holland, 2000). Although this view may be biased due to the paucity of taxa sampled for genomic reconstructions (Goffinet et al., 2005), it may hold true especially for alterations involving large portions of the genome, such as the inversion of a fragment spanning more than half the plastid genome. The inverted order of genes in the genome of Funariaceae, Disceliaceae and Encalyptales is thus likely to be inherited from a common ancestor that did not give rise to Gigaspermaceae, which lack the inversion. The distribution of the inversion is thus compatible with the hypothesis of Encalyptales being closely related to Funariaceae and Disceliaceae and of Funariales (including Gigaspermaceae) being paraphyletic (Goffinet and Cox, 2000).
Ordinal affinities of mosses are primarily established based on their peristome architecture (Vitt, 1984; Buck and Goffinet, 2000; Goffinet and Buck, 2004). However, reduction in sporophyte complexity, and hence in peristome differentiation, is rampant in mosses (Vitt, 1981; Zander, 1993; Buck et al., 2000), and consequently the relationships of taxa with reduced morphologies are drawn from other morphological characters, such as those of the gametophyte. Funariales share few apomorphies in the architecture of their vegetative (gametophytic) plants. Vitt (1982) considered only the lax rectangular cells as diagnostic. The monophyly of Funariales sensu Vitt (1982) and Funariineae sensu Vitt (1984) was first questioned by Goffinet and Cox (2000) who suggested, based on phylogenetic inferences from nuclear and plastid DNA sequences, that Ephemeraceae, a lineage of tiny ephemeral mosses lacking a peristome, should be transferred to Pottiales. Their hypothesis subsequently gained support from ontogenetic studies (Pressel and Duckett, 2005). Gigaspermaceae also share a similar leaf architecture with Funariaceae (Vitt, 1982), but differ in a suite of putative adaptations to xeric environments. The vegetative gametophyte is stoloniferous, with the creeping stems producing short erect branches. The sporophyte may be dehiscent or not, but in either case, the capsule is gymnostomous (lacking a peristome). Fife (1980) implicitly considered that the two families also differ in the structure of the stoma, with two guard cells defining the pore in Gigaspermaceae, whereas a single, incompletely divided guard cell defines the stoma in Funariaceae (Fife, 1980). However, Brotherus (1924) described the stomata of Gigaspermaceae as unicellular, whereas Scott and Stone (1976) and Crum and Anderson (1981) reported the number of guard cells to vary between one and two. In Encalyptales, the stoma are always surrounded by two guard cells (Horton, 1982). The single, so-called doughnut-shaped guard cell could be seen as a synapomorphy for Funariales sensu Vitt (1982), and hence support the monophyly of the order. However, unicellular stomata occur also in Buxbaumia and Polytrichum (Paton, 1957), and hence are not free of homoplasy. Furthermore, polymorphism in the architecture of the stoma in Gigaspermaceae may leave reconstructions of ancestral states equivocal.
The distribution of the inversion in the plastid genome of Funariales is congruent with the hypothesis that the order is paraphyletic as proposed by Goffinet and Cox (2000), based on phylogenetic inferences from variation in the nucleotide sequence of three loci: Funariaceae, Disceliaceae (Funariales) and Encalyptaceae (Encalyptales) share a large inversion in their plastid genome that probably occurred in their common ancestor. Gigaspermaceae (Funariales, sensu Goffinet and Buck, 2004), which lack the inversion, are considered to have diverged earlier. For the classification to reflect a phylogenetic scenario wherein Gigaspermaceae comprise the sister group to the remainder of Funariales and Encalyptales, the circumscription of Funariales could be broadened to include Encalyptaceae or, alternatively, Gigaspermaceae could be excluded from Funariales and accommodated in their own order. A third possibility would be to recognize a paraphyletic Funariales; however, the absence of an unambiguous morphological character uniting Gigaspermaceae to Funariales provides no foundation for such concept. Encalyptales differ from Funariales in virtually all aspects of the vegetative morphology and in the architecture of the peristome. Merging the two orders would obscure the wide morphological divergence between these lineages and hence should be avoided. The exclusion of Gigaspermaceae from Funariales is not significantly incongruent with the phylogenetic signal of any morphological character. Hence we recommend addressing the paraphyly of Funariales sensu Goffinet and Buck (2004) by placing Gigaspermaceae in their own order, Gigaspermales Goffinet, Wickett, O. Werner, Ros, A.J. Shaw and C.J. Cox ord. nov. (Plantae terrestres stoloniferae ramis brevibus erectis, folia unicostata cellulis laxis laevibus, peristomium destitum; Type genus: Gigaspermum Lindb., Öfversigt af Förhandlingar: Kongl. Svenska Vetenskaps-Akademien 21: 599. 1865).
A hypothesis of a shared ancestry for Gigaspermales, Funariales and Encalyptales emanates, if only with weak support, from various phylogenetic analyses of nucleotide sequences (Goffinet and Cox, 2000; Goffinet et al., 2001; C. J. Cox et al., Natural History Museum, London UK, unpubl. res.). This combined lineage exhibits a wide range of morphology, and no unambiguous morphological synapomorphy has been identified. The shared ancestry may be supported by ontogenetic data and, in particular, patterns of cell division in the inner peristome forming layer (Goffinet et al., 1999) but critical developmental studies of the sporophyte of Gigaspermales would be required to substantiate this hypothesis.
In conclusion, the inversion of an extensive fragment of the LSC of the plastid genome is considered to have occurred once (Fig. 1C) in the ancestor to Funariaceae, Disceliaceae and Encalyptales. This genomic change strengthens the weak phylogenetic signal extracted from sequences of two plastid loci and one nuclear locus, whereby Funariales are paraphyletic, with the Funariaceae, Disceliaceae and Encalyptaceae sharing a unique common ancestor that did not give rise to Gigaspermaceae. To reflect such evolutionary history, Gigaspermaceae are accommodated in their own order. Funariales continue to emerge from recent phylogenetic reconstructions (Goffinet and Cox, 2000; Goffinet et al., 2001) as a crown group of an early diverging lineage rather than the closest extant relative of the ancestor to the vast majority of true mosses as hypothesized by Vitt (1984).
ACKNOWLEDGEMENTS
The authors thank Drs Jan van Rooy (Pretoria National Herbarium, South Africa), Maria Gibson (Deakin University, Australia) and Louis Thouvenot (Perpignan, France) for sharing material of Chamaebryum, Gigaspermum and Oedipodiella, respectively. We extend our gratitude to Dr William Buck (New York) for providing a Latin diagnosis and to Dr Robert Magill (Missouri Botanical Garden) for confirming the protologue information. This study was made possible by grants from the US National Science Foundation through awards DEB-0089633 to B.G. and DEB-0089131 to A.J.S. and from a Large Grant awarded to B.G. by the Research Foundation at the University of Connecticut.
LITERATURE CITED
- Brotherus VF. Musci (laubmoose). 1. Hälfte. In: Engler A, Prantl K, editors. Die natürlichen Pflanzenfamilien. edn 2. Vol. 10. Leipzig: 1924. [Google Scholar]
- Buck WR, Goffinet B. Morphology and classification of mosses. In: Shaw AJ, Goffinet B, editors. Bryophyte biology. Cambridge: Cambridge University Press; 2000. pp. 71–123. [Google Scholar]
- Buck WR, Goffinet B, Shaw AJ. Testing morphological concepts of orders of pleurocarpous mosses (Bryophyta) using phylogenetic reconstructions based on trnL-trnF and rps4 sequences. Molecular Phylogenetics and Evolution. 2000;16:180–198. doi: 10.1006/mpev.2000.0805. [DOI] [PubMed] [Google Scholar]
- Cox CJ, Goffinet B, Shaw AJ, Boles SB. Phylogenetic relationships among the mosses based on heterogeneous Bayesian analysis of multiple genomic compartments. Systematic Botany. 2004;29:234–250. [Google Scholar]
- Crosby MR, Magill RE, Allen B, He S. A checklist of mosses. St Louis, MO: Missouri Botanical Garden Press; 1999. [Google Scholar]
- Crum H, Anderson LE. Mosses of Eastern North America. 1 and 2. New York, NY: Columbia University Press; 1981. [Google Scholar]
- Doyle JJ, Doyle JL, Ballenger JA, Palmer JD. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Molecular Phylogenetics and Evolution. 1996;5:429–438. doi: 10.1006/mpev.1996.0038. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Systematic Botany. 1995;20:272–294. [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Fife AJ. The affinities of Costesia and Neosharpiella and notes on the Gigaspermaceae (Musci) The Bryologist. 1980;83:466–476. [Google Scholar]
- Fife AJ. A generic revision of the Funariaceae (Bryophyta: Musci). Part 1. Journal of the Hattori Botanical Laboratory. 1985;58:149–196. [Google Scholar]
- Goffinet B, Buck WR. Systematics of the Bryophyta (mosses): from molecules to a revised classification. Monographs in Systematic Botany from the Missouri Botanical Garden. 2004;98:205–209. [Google Scholar]
- Goffinet B, Cox CJ. Phylogenetic relationships among basal-most arthrodontous mosses with special emphasis on the evolutionary significance of the Funariineae. The Bryologist. 2000;103:212–223. [Google Scholar]
- Goffinet B, Shaw AJ, Anderson LE, Mishler BD. Peristome development in mosses in relation to systematics and evolution. V. Orthotrichaceae. The Bryologist. 1999;102:581–599. [Google Scholar]
- Goffinet B, Cox CJ, Shaw AJ, Hedderson TA. The Bryophyta (Mosses): systematic and evolutionary inferences from an rps4 gene (cpDNA) phylogeny. Annals of Botany. 2001;87:191–208. doi: 10.1006/anbo.2000.1318. [DOI] [PubMed] [Google Scholar]
- Goffinet B, Wickett NJ, Shaw AJ, Cox CJ. Phylogenetic significance of the rpoA loss in the chloroplast genome of mosses. Taxon. 2005;54:353–360. [Google Scholar]
- Goremykin V, Hirsch-Ernst KI, Wölfl S, Hellwig FH. The chloroplast genome of the ‘basal’ angiosperm Calycanthus fertilis—structural and phylogenetic analyses. Plant Systematics and Evolution. 2003;242:119–135. [Google Scholar]
- Hackett JD, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, et al. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Current Biology. 2004;14:213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Horton DG. A revision of the Encalyptaceae (Musci) with a particular reference to the North American taxa. Journal of the Hattori Botanical Laboratory. 1982;53:365–418. [Google Scholar]
- Kelch DG, Kriskell A, Mishler BD. Inferring phylogeny using genomic characters: a case study using land plant plastomes. Monographs in Systematic Botany from the Missouri Botanical Garden. 2004;98:3–12. [Google Scholar]
- Kim K-J, Choi K-S, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae) Molecular Biology and Evolution. 2005;22:1783–1792. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- Korpelainen H. The evolutionary processes of mitochondrial and chloroplast genomes differ from those of nuclear genomes. Naturwissenschaften. 2004;91:505–518. doi: 10.1007/s00114-004-0571-3. [DOI] [PubMed] [Google Scholar]
- Kugita M, Kaneko A, Yamamoto Y, Takeya Y, Matsumoto T, Yoshinaga K. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Research. 2003;31:716–721. doi: 10.1093/nar/gkg155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Doyle JJ, Palmer JD. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution. 1990;44:390–402. doi: 10.1111/j.1558-5646.1990.tb05207.x. [DOI] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proceedings of the National Academy of Sciences of the USA; 2002. pp. 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. The Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Hirai A. Identification of the entire set of transferred chloroplast DNA sequences in the mitochondrial genome of rice. Molecular General Genetics. 1993;236:341–346. doi: 10.1007/BF00277131. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, et al. Chloroplast gene organization deduced from the complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. [Google Scholar]
- Olmstead RG, Palmer JD. Chloroplast DNA systematics: a review of methods and data analysis. American Journal of Botany. 1994;81:1205–1224. [Google Scholar]
- Paton JA. The occurrence, structure and functions of the stomata in British bryophytes. Part I. Occurrence and structure. Transaction of the British Bryological Society. 1957;2:228–242. [Google Scholar]
- Plunkett GM, Downie SR. Expansion and contraction of the chloroplast inverted repeat in Apiaceae sub. Apioideae. Systematic Botany. 2000;25:648–667. [Google Scholar]
- Pressel S, Duckett JG. Studies of protonemal morphogenesis in mosses. X. Ephemeraceae revisited; new dimensions underground. Journal of Bryology. 2005;27:311–318. [Google Scholar]
- Quandt D, Bell N, Stech M. Nova Hedwigia Beiheft. 2007. Unravelling the knot: the Pulchrinodaceae fam. nov. (Bryales) (in press) [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992;255:1697–1699. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry RJ, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Cambridge, MA: CAB International; 2005. pp. 45–68. [Google Scholar]
- Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends in Ecology and Evolution. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- Saski C, Lee S-B, Daniell H, Wood TC, Tomkins J, Kim H-G, et al. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Molecular Biology. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Scott GAM, Stone IG. The mosses of southern Australia. London: Academic Press; 1976. [Google Scholar]
- Shaw J, Anderson LE, Mishler BD. Peristome development in mosses in relation to systematics and evolution. III. Funaria hygrometrica, Bryum bicolor, and B. pseudocapillare. Systematic Botany. 1989;14:24–36. [Google Scholar]
- Soltis DE, Soltis PS. Choosing an approach and an appropriate gene for phylogenetic analysis. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants. II. DNA sequencing. Boston: Kluwer Academic Press; 1998. pp. 1–42. [Google Scholar]
- Sugita M, Sugiura C, Arikawa T, Higuchi M. Molecular evidence of an rpoA gene in the basal moss chloroplast genomes: rpoA is a useful molecular marker for phylogenetic analysis of mosses. Hikobia. 2004;14:171–175. [Google Scholar]
- Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M. Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Research. 2003;31:5324–5331. doi: 10.1093/nar/gkg726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 2002. PAUP*, version 4·0. [Google Scholar]
- Thouvenot L. Une seconde station française de Oedipodiella australis (Wag. et Dix.) Dix. var. catalaunica P. de la V. dans les Pyrenées-Orientales. Bulletin de la Société Botanique du Centre-Ouest, nouvelle série. 2000;31:495–500. [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Review in Genetics. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proceedings of the National Academy of Sciences of the USA; 1999. pp. 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitt DH. Adaptive modes of the moss sporophyte. The Bryologist. 1981;83:162–182. [Google Scholar]
- Vitt DH. Sphagnopsida and Bryopsida. In: Parker SP, editor. Synopsis and classification of living organisms. Vol. 305. New York, NY: McGraw-Hill; 1982. pp. 307–336. [Google Scholar]
- Vitt DH. Classification of the Bryopsida. In: Schuster RM, editor. New manual of bryology. Vol. 2. Nichinan, Japan: Hattori Botanical Laboratory; 1984. pp. 696–759. [Google Scholar]
- Werner O, Ros RM, Goffinet B. A reconsideration of the systematic position of the genus Goniomitrium based on chloroplast sequence markers. The Bryologist. 2007;110:108–114. [Google Scholar]
- Zander RH. Genera of the Pottiaceae: mosses of harsh environments. Bulletin of the Buffalo Society of Natural Sciences. 1993;32:1–378. [Google Scholar]
- Zheng D, Nielsen BL, Daniell H. A 7·5-kbp region of the maize (T cytoplasm) mitochondrial genome contains a chloroplast-like trnI(CAT) pseudogene and many short segments homologous to chloroplast and other known genes. Current Genetics. 1997;32:125–131. doi: 10.1007/s002940050256. [DOI] [PubMed] [Google Scholar]