Abstract
Background and Aims
In angiosperms xyloglucan endotransglucosylase (XET)/hydrolase (XTH) is involved in reorganization of the cell wall during growth and development. The location of oligo-xyloglucan transglucosylation activity and the presence of XTH expressed sequence tags (ESTs) in the earliest diverging extant plants, i.e. in bryophytes and algae, down to the Phaeophyta was examined. The results provide information on the presence of an XET growth mechanism in bryophytes and algae and contribute to the understanding of the evolution of cell wall elongation in general.
Methods
Representatives of the different plant lineages were pressed onto an XET test paper and assayed. XET or XET-related activity was visualized as the incorporation of fluorescent signal. The Physcomitrella genome database was screened for the presence of XTHs. In addition, using the 3′ RACE technique searches were made for the presence of possible XTH ESTs in the Charophyta.
Key Results
XET activity was found in the three major divisions of bryophytes at sites corresponding to growing regions. In the Physcomitrella genome two putative XTH-encoding cDNA sequences were identified that contain all domains crucial for XET activity. Furthermore, XET activity was located at the sites of growth in Chara (Charophyta) and Ulva (Chlorophyta) and a putative XTH ancestral enzyme in Chara was identified. No XET activity was identified in the Rhodophyta or Phaeophyta.
Conclusions
XET activity was shown to be present in all major groups of green plants. These data suggest that an XET-related growth mechanism originated before the evolutionary divergence of the Chlorobionta and open new insights in the evolution of the mechanisms of primary cell wall expansion.
Key words: XTH, XET activity, primary cell wall, xyloglucan, cell elongation, growth, Physcomitrella patens, bryophytes, Charophyta, Chlorophyta, Rhodophyta, Phaeophyta
INTRODUCTION
A basic characteristic of vascular plants, mosses and many algae is the substantial post-mitotic increase in volume, known as expansion or elongation, preceding or coinciding with cell differentiation. The mechanisms steering this process are to date largely unknown (Hu et al., 2006). A constant feature, however, is the increase in volume of the vacuole and the increase in surface area of the primary cell wall. The wall is an indispensable and characteristic feature of plant cells. Among many other functions (Albersheim, 1976; Carpita and Gibeaut, 1993) it serves to maintain the shape of plant cells and thereby greatly contributes to the structural integrity and morphology of the entire plant. In angiosperms the primary cell wall (PCW) is composed of cellulose microfibrils that are embedded in a highly hydrated matrix of hemicelluloses, pectins and glycoproteins (Darvill et al., 1980; McNeil et al., 1984; Fry, 1986; McCann and Roberts, 1991; Carpita and Gibeaut, 1993; Pauly et al., 1999). Xyloglucan (XyG) is the major hemicellulose in non-gramineous plants and was shown to be present in all vascular plants (Popper and Fry, 2004). It can form non-covalent hydrogen bonds with cellulose (Hayashi et al., 1987) and is therefore believed to tether adjacent cellulose microfibrils, thereby forming a load-bearing cellulose/XyG network in plant cell walls (Fry, 1989; McCann et al., 1990). During turgor-driven cell wall expansion this network needs to be modified to allow slippage of the cellulose microfibrils, but without losing the integrity of the cellulose/XyG network as this may lead to detrimental cell lysis (Marga et al., 2005). Two enzyme families are considered to play a major part in this highly controlled mechanism of cell wall loosening: expansins and xyloglucan endotransglucosylase/hydrolases (XTHs). Expansins are thought to disturb the hydrogen bonds between XyG and the cellulose microfibrils and hence create the possibility for slippage of the fibrils (McQueen-Mason and Cosgrove, 1994; Cosgrove, 2000; Sampedro and Cosgrove, 2005). XTHs, by contrast, break (=hydrolase or XEH action) or break and rejoin the tethering XyG with another available XyG (=endotransglucosylase or XET action). This action also allows the reorganization of the XyG within the (growing) wall (Thompson and Fry, 2001), as the incorporation of newly secreted XyG (Fry et al., 1992; Nishitani and Tominaga, 1992; Rose et al., 2002). In angiosperms, XTHs were indeed shown to play major roles during plant growth and differentiation (Campbell and Braam, 1999; Albert et al., 2004; Matsui et al., 2005; Vissenberg et al., 2005). As XTHs are so far the only characterized and well-described enzyme group displaying transglucosylation activity in the plant cell wall, the study of their presence and function in lower plants will shed new light on how the PCW and the PCW elongation mechanism originated, evolved and functions today in the angiosperms.
In our previous work the presence of XET action was demonstrated in vivo in roots of representatives of all vascular plants down to one of the most primitive vascular land plant orders, the Selaginellales (Vissenberg et al., 2003). The conservation of the XTH amino acid sequence and function throughout vascular plant evolution was also described in a detailed study of a heterologously expressed XTH of Selaginella kraussiana, Sk-XTH1 (Van Sandt et al., 2006). Like expansins (Sampedro et al., 2006), XTHs are thus part of an ancient cell wall reorganizing machinery that originated even before the evolutionary divergence of vascular plants (Vissenberg et al., 2003; Van Sandt et al., 2006).
As sequencing of the Physcomitrella patens genome is in progress searches were made for the presence of Physcomitrella XTH genes in silico and potential XTH sequences were analysed. Recently the primary cell wall composition of the bryophytes and charophytes was described (Popper and Fry, 2003). In accordance with this work the presence of XET activity in the three major groups of bryophytes and in charophytes was investigated, and some representatives of the chlorophytes, rhodophytes and phaeophytes were also included. Owing to autofluorescence of the tissues, the in vivo assay used previously to study XET activity in roots (Vissenberg et al., 2000, 2001, 2003, 2005), is unfeasible in bryophytes and algae. To overcome this problem tissue prints on XET test papers were used, as described by Fry (1997). The activity thus found in vitro is merely indicative for its presence in vivo, as xyloglucans are absent in the Charophyta and Chlorophyta (Popper and Fry, 2003). However this technique allows the localization of enzymes that are capable of incorporating xyloglucan oligosaccharides into a xyloglucan matrix and thus displaying XET activity. These enzymes can include XTHs or XTH-like enzymes. In accordance, searches were made for the presence of an XTH or XTH-like gene in one of the closest relatives of the vascular plants, Chara vulgaris. Combining these data with recent findings on the phylogenetic relationship of XTHs and family GH16 hydrolases, the possible origin and evolution of the XTH–xyloglucan interacting mechanism itself as well as the evolution of PCW elongation in general are discussed.
MATERIALS AND METHODS
Plant material
Physcomitrella patens (Hedw.) Bruch & Schimp. was grown in Petri dishes on minimal medium (Schaefer, 2001) covered with cellophane discs. Plants were grown in a growth chamber (TCPS, Werchter, Belgium) at 26 °C, 16-h light per day (Sylvania, cool white), with a photosynthetic photon flux of 50–80 µmol m−2 s−1. Seven-day-old gametophytes were used to prepare RNA and tissue homogenates.
Most bryophytes used for tissue printing were collected in the wild in Hechtel-Eksel (Belgium), Phaeoceros carolinianus (Michx.) Prosk. was collected in Boom (Belgium), Anthoceros agrestis Paton nom. cons. prop. near Ghent (Belgium), Chara vulgaris L. in the National Botanical Garden (Meise, Belgium) and marine algae were collected in Wimereux (France). All bryophytes and algae were directly processed in the lab upon arrival to minimize stress and changes in growth conditions.
Analysis of the Physcomitrella database
The Selaginella kraussiana XTH amino acid sequence, Sk-XHT1 (accession no. AY580314), was used in a protein blast against the draft Physcomitrella patens database (http://www.cosmoss.org; Lang et al., 2005). The retrieved sequences were analysed in expasy (http://www.expasy.org) and aligned using ClustalW (http://www.ebi.ac.uk/clustalw/).
3′ RACE PCR
RNA from P. patens was prepared using a slightly modified Plant Concert RNA protocol (Invitrogen). The RNA was kept in 50 % isopropanol overnight, at −20 °C, to achieve optimal precipitation and a maximal extraction yield. Five micrograms of total RNA was reverse-transcribed using Superscript II RNase H-Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. The 3′-end of the CAC43709 cDNA (http://www.cosmoss.org) was subsequently obtained by 3′ RACE using a gene-specific internal primer and an oligo-dT26. The full-length cDNA sequence of CAC43709 was constructed in silico and subsequently amplified from total cDNA using a specific primer set and a High Fidelity (Roche) proofreading enzyme mix.
Preparation of XET test paper
The XET and control test papers were made as described by Fry (1997). A filter paper (Whatman no. 1) was dipped into a solution of 0·5 % 1-1-1 trichloro-2-methylpropan-2-ol and 1 % 200-kDa tamarind xyloglucan, and dried overnight. The dried papers were subsequently dipped into a 5 µm XLLGol-sulforhodamine (XGO-SRs) in 70 % acetone solution to become XET test papers, or into a 5 µm trisaccharide-SR solution (=non-XET substrate) to serve as control papers. They were all left to dry in the dark before use. The trisaccharide-SR preparation contained two main fluorescent compounds (one major and one minor), both of which ran on a silica-gel thin-layer chromatography plate (in BuOH/HOAc/H2O 2 : 1 : 1) between maltose-SR and maltotriose-SR, suggesting that they are trisaccharides with at least one residue smaller than Glc. The material used for preparation of these SR derivatives was a minor fraction of small oligosaccharides obtained by cellulase digestion of tamarind xyloglucan. Their exact structures are unknown, but one may possibly be Xyl-Glc-Glc. They do not act as XET acceptor substrates for a crude preparation of cauliflower enzymes (S. Fry, University of Edinburgh, pers. comm.).
Dot-spot
Physcomitrella patens gametophytic tissue (7 g) was homogenized with several 15-s pulses (IKA Labortechnik, Staufen, Germany) in 300 mm sodium acetate, 20 mm calcium chloride, 1 mm dithiothreitol at pH 5·5 adjusted with sodium hydroxide (Iannetta and Fry, 1999). XET activity was tested by spotting 5 µL of the homogenate onto both a control and an XET test paper, which were then incubated in a sealed acetate envelope and kept overnight at room temperature. XET activity is defined as the transfer of the non-labelled xyloglucans to fluorescent acceptor-substrates (XGO-SRs). Non-reacted fluorescent xyloglucan oligosaccharides and trisaccharides were washed away after the assay in 90 % formic acid/ethanol/water (1 : 1 : 1) for 1 h followed by a 5-min wash in water. XET activity is seen as the remaining fluorescence on the XET test paper upon UV illumination. Images were taken using an Olympus C-5050 ZOOM digital camera with identical settings (1/10 shutter time, F 2.0 diaphragm) to allow comparison of the resulting fluorescence.
Tissue print
Both XET and control test papers used for the tissue prints were initially dipped into 25 mm MES buffer adjusted to pH 5·5. Fresh tissue of bryophytes and algae was subsequently pressed onto the test papers (see above) and sealed into an acetate envelope. This ‘sandwich’ was incubated in the dark at room temperature for 3 d under a constant pressure of 3·2 g cm−2 to ensure that the tissue was kept in contact with the test papers. Pictures of the print were taken before and after it was washed (as described above), using daylight and UV illumination. In some tissue prints, chlorophyll was transferred to the blot, especially at the sites were the tissue was cut. This contaminant, however, was largely washed away in formic acid/ethanol/water. The remaining autofluorescence was extremely weak and of a different colour than that of the SR substrates. It thus did not interfere with the observations. The pigments of brown and red algae were not as easily removed by washing and hence caused some problems in analysing the presence of XET activity. An overlay of the tissue and fluorescence images allowed sites with (high) leachable XET activity to be located exactly.
Searching a putative XTH or XTH-like transcript in Chara
RNA of Chara vulgaris was prepared and reverse transcribed as described for Physcomitrella above. The resulting cDNA served as template in a PCR reaction using a degenerate primer based upon variations present in the catalytic domain of angiosperm XTHs and an oligo-dT26 primer [annealing temperature of 50 °C during 45 s]. A 480-bp cDNA sequence was amplified, sequenced and analysed in silico (http://www.expasy.org/tools/dna.html, http://elm.eu.org/). Its homology to XTHs and 1,3-,1,4-β-d-endoglucanases was analysed using ClustalW (http://www.ebi.ac.uk/clustalw/).
RESULTS
Putative XTH cDNAs in P. patens and XET activity
Blast analyses of the Physcomitrella genome with the amino acid sequence of the most primitive XTH thus far characterized, i.e. the lycopodiophyte XTH, Sk-XTH1 (Van Sandt et al., 2006), identified one putative full-length XTH (CAC43710) and 17 fragmentary putative XTH ESTs. An identical experiment using the different Arabidopsis XTH amino acid sequences gave an identical set of hits. Some of the 18 hits differed in only a few amino acids and probably originated from sequencing errors. Seven ESTs were suggested to be part of Arabidopsis thaliana XTH precursor genes (Rensing et al., 2005; http://www.cosmoss.org). Yet only one EST (CAC43709; cDNA: AX172659) and one full-length sequence (CAC43710; cDNA: AX172661) included a variant of the catalytic domain, which is one of the criteria for a gene to become annotated as ‘putative XTH’. The cDNA sequence of AX172659 was completed using 3′ RACE PCR and the deduced amino acid sequences were analysed in silico.
Both moss amino acid sequences showed high homology with known XTH amino acid sequences of numerous vascular plants. Most domains and some well-positioned amino acids, characteristic of vascular plant XTHs (Campbell and Braam, 1998; Johansson et al., 2003, 2004; Henriksson et al., 2003; Van Sandt et al., 2006), were found in both moss cDNAs. Yet some notable differences were present in the amino acid composition of the catalytic domain. For clarity, in Fig. 1A both Physcomitrella amino acid sequences were aligned with one representative vascular plant XTH, i.e. Sk-XTH1 from the lycopodiophyte Selaginella kraussiana (Van Sandt et al., 2006). A secretion signal at the N-terminus allowed both enzymes to be secreted in the apoplast (Fig. 1A, lower-case letters). The catalytic domain of both putative Physcomitrella XTHs (Fig. 1A, bold) differed in two (CAC43709, CEFDFEFLG) and three (CAC43710, YELDMEFLG) amino acids, respectively, compared with the ‘average’ functional site DEIDFEFLG, conserved among most XTHs of seed plants (Nishitani, 1997) and present in family 16 glycoside hydrolases (Henrissat et al., 2001). However, both glutamic acid residues and the second aspartic acid residue of the motif (xExDxExxx), which play a crucial role in the cleavage of the donor substrate in family GH16 enzymes, were maintained. As in most vascular plant XTHs, the catalytic domain of both moss amino acid sequences was immediately followed by a potential N-glycosylation site (N-X-T/S, Shakin-Eshleman et al., 1996) (Fig. 1A, dashed line). There was a notable variation in the C-terminal part of both putative Physcomitrella XTHs and in vascular plant XTHs. However, important residues involved in the maintenance of three-dimensional stability (Fig. 1A, underlined) and the recognition of the acceptor substrates in higher plant XTHs were also encoded by both moss cDNAs (Fig. 1A, superimposed). The conservation of most of the XTH identity motifs made both Physcomitrella amino acid sequences putative XTH enzymes. Thus, the in silico data suggested the presence of at least two potential XTH-encoding cDNAs in the bryophyte P. patens.
Fig. 1.
(A) Alignment of the amino acid sequence of a lycopodiophyte XTH, Sk-XTH1 and two Physcomitrella patens putative XTH amino acid sequences, CAC43709 and CAC43710. The alignment of the amino acid sequence of Sk-XTH1, CAC43709 and CAC43710 is shown in single letter code. The predicted secretion signal peptide of each sequence is marked in lower-case letters, while the conserved catalytic site, shared with the active site of the β-endoglucanase from Bacillus licheniformis, is presented with bold amino acid residues. The underlined (dashed) amino acids represent the possible N-linked glycosylation sites. Conserved amino acids possibly involved in the stability of the enzyme and in acceptor-substrate recognition are underlined. Tyrosines, putatively involved in the recognition of the acceptor substrate, are superscripted. (B) Dot-spot of homogenized Physcomitrella tissue on XET (left) and control (right) test paper. XET activity is seen as an orange spot on the XET test paper (left). As a control for specificity of the assay, fluorescently labelled non-XET substrate was used, yielding no fluorescence when homogenized Physcomitrella tissue was spotted (right).
A dot-spot of homogenized Physcomitrella tissue on XET test paper revealed a bright fluorescent spot in Fig. 1B, resulting from the specific incorporation of fluorescently labelled XyG oligosaccharides (X), whereas no fluorescence was seen on control test paper (C). This confirmed the presence of at least one functional XTH in Physcomitrella.
XET activity is present at growth sites in bryophytes
The presence of XET activity in other bryophytes was assayed by tissue printing the gametophyte and/or sporophyte of 15 species, representing members of the three major groups of bryophytes, i.e. the Bryophyta, the Anthocerophyta and the Marchantiophyta (Table 1). To localize specifically the sites of the tissue in which enzymes, capable of xyloglucan transglucosylation, are expressed, an image showing the tissue (left) and the tissue print (right) is shown in Figs 2 and 3. Here the xyloglucan transglucosylation, i.e. XET activity, was visible as an orange fluorescent spot under UV illumination.
Table 1.
List of the bryophyte and algal species assayed for XET activity
| Family | Species | |
|---|---|---|
| BRYOPHYTA | ||
| Class Sphagnopsida | Sphagnaceae | Sphagnum fimbriatum Wilson |
| Order Sphagnales | ||
| Sphagnum palustre L. | ||
| Class Polytrichopsida | ||
| Order Polytrichales | Polytrichaceae | Atrichum undulatum (Hedw.) P.Beauv. |
| Class Bryopsida | ||
| Order Funariales | Funariaceae | Funaria hygrometrica Hedw. |
| Order Dicranales | Dicranaceae | Dicranella heteromalla (Hedw.) Schimp. |
| Order Bryales | Mniaceae | Mnium hornum Hedw. |
| Order Hypnales | Hypnaceae | Brachythecium rutabulum (Hedw.) Schimp.* |
| Hypnum cupressiforme Hedw.* | ||
| Vesicularia reticulata (Dozy & Molkenboer) Brotherus* | ||
| Thuidiaceae | Thuidium tamariscinum (Hedw.) Schimp.* | |
| ANTHOCEROTOPHYTA | ||
| Order Anthocerotales | Anthocerotaceae | Anthoceros agrestis Paton nom. cons. prop. |
| Phaeoceros carolinianus (Michx.) Prosk. | ||
| MARCHANTIOPHYTA | ||
| Class Marchantiopsida | Ricciaceae | Riccia sp. |
| Order Ricciales | ||
| Order Fossombroniales | Pelliaceae | Pellia epiphylla (L.) Corda |
| Class Jungermanniopsida | Geocalycaceae | Lophocolea heterophylla (Schrad.) Dum. |
| Order Jungermanniales | ||
| CHAROPHYTA | ||
| Class Charophyceae | ||
| Order Charales | Characeae | Chara vulgaris L. |
| CHLOROPHYTA | ||
| Class Ulvophyceae | ||
| Order Ulvales | Ulvaceae | Ulva linza L. (L.) J. Agardh. – E1 |
| Ulva lactuca L. – E2 | ||
| Order Cladophorales | Cladophoraceae | Cladophora rupestris (L.) Kützing – E3 |
| RHODOPHYTA | ||
| Class Florideophyceae | ||
| Order Gigartinales | Gigartinaceae | Chondrus crispus Stackhouse – G1 |
| Phyllophoraceae | Gymnogongrus crenulatus (Turner) J. Agardh – G2 | |
| Polyideaceae | Polyides rotundus (Hudson) Greville. – G3 | |
| Order Gracilariales | Gracilariaceae | Gracilaria gracilis (Stackhouse) Steentoft, L.M. Irvine & Farnham – G4 |
| Order Plocamiales | Plocamiaceae | Plocamium cartilagineum (L.) Dixon – G5 |
| HETEROKONTOPHYTA | ||
| Class Phaeophyceae | ||
| Order Fucales | Fucaceae | Pelvetia canaliculata (L.) Decaisne & Thuret – I1 |
| Order Dictyotales | Dictyotaceae | Taonia atomaria (Woodward) J. Agardh – I2 |
| Dictyota dichotoma (Hudson) Lamouroux – I3 | ||
| Order Laminariales | Laminariaceae | Lainaria saccharina (L.) Lmouroux – I4 |
* Pleurocarp mosses. Some algae are designated as in Fig. 3, e.g. E1, E2.
References: Bryophytes: Buck and Goffinet (2000). Hornworts: Stotler and Crandall-Stotler (2005). Hepatics: Crandall-Stotler and Stotler (2000). Charophyta and Chlorophyta: Lewis and McCourt (2004). Rhodophyta: Saunders and Hommers (2004). Heterokontophyta: Andersen (2004).
Fig. 2.
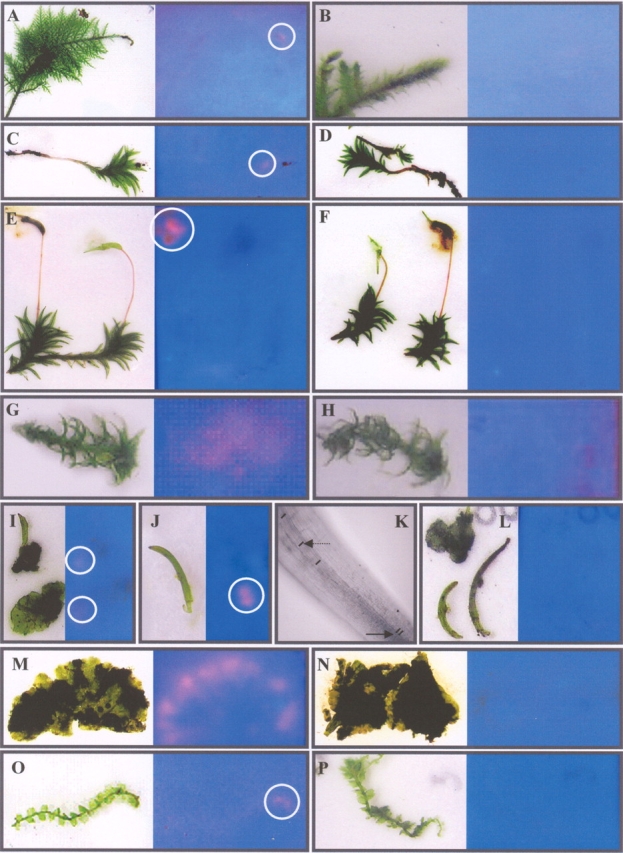
Tissue print on XET (left) and control test paper (right) of representatives of the three major bryophyte lineages. XET activity is present on the XET test papers near the apex of the gametophyte of pleurocarpous Bryophyta: Thuidium sp. (A, encircled) and acrocarpous Bryophyta: Atrichum undulatum (Hedw.) P.Beauv. (C, encircled) and at the site of the mature sporophyte capsule: Atrichum undulatum (Hedw.) P.Beauv. (E). A more diffuse XET-generated signal is visible in Sphagnum fimbriatum (G). A faint spot of XET activity is present at the borders of the anthocerophyte gametohytic thallus of Phaeoceros carolinianus (Michx.) Prosk (I, encircled). A bright spot of XET activity is present directly above the basal meristem, at the site of cell elongation (J) of the sporophyte of Phaeoceros carolinianus (Michx.). This signal is only visible when the sporophyte was removed from the gametophyte and its protecting involucre (I,J). (K) Image of the elongation zone of the sporophyte; arrows point to short and longer cells (see lines representing the cross walls). In the gametophyte of both thalloid (Riccia sp., M) and leafy (Lophocolea heterophylla (Schrad.) Dum., O, encircled) Marchantiophyta an XET spot is visible near the apical cell(s). The images in B, D, F, H, L, N and P represent the fluorescence on trisaccharide-SR control test papers of the XET assays on the left.
Fig. 3.

Tissue print on XET (left) and control test paper (right) of representatives of the Charophyta, Chlorophyta, Rhodophyta and Phaeophyta. In Chara vulgaris L. a fluorescent spot of XET activity is visible at the site where the apex and node are pressed onto the XET test paper (A). At higher magnification a clear XET signal is present at the sites of the branchlets (B). In the Chlorophyta XET activity is visible at the site of the holdfast of Ulva linza (Linnaeus) J. Agardh. in its tubular growth form (this species was previously named Enteromorpha linza) (E1, encircled). No signal was found in the other chlorohytes assayed: Ulva lactuca (E2) and Cladophora rupestris (L.) Kützing (E3). In the assays of both Rhodophyta [G1: Chondrus crispus Stackhouse, G2: Gymnogongrus crenulatus (Turner) J. Agardh, G3: Polyides rotundus (Hudson) Greville, G4: Gracilaria gracilis (Stackhouse) Steentoft, L.M. Irvine & Farnham, G5: Plocamium cartilagineum (L.) Dixon] and Phaeophyta [I1: Pelvetia canaliculata (L.) Decaisne & Thuret, I2: Taonia atomaria (Woodward) J. Agardh, I3: Dictyota dichotoma (Hudson) Lamouroux, I4: Laminaria saccharina (L.) Lamouroux] a substantial amount of autofluorescence is visible, but no XET signal is present on the XET test papers. On the right control prints on trisaccharide-SR test papers of each group are shown (C, D, F, H and J).
The early diversification of the Bryophyta or mosses is linked to spore dispersion and this is reflected in the taxonomy of this group (Goffinet and Buck, 2004). Based on the position of the gametangia and on the branching of the stem, however, two major groups can be distinguished, acrocarps and pleurocarps (Mägdefrau, 1982; Buck and Goffinet, 2000). As growth patterns differ in the two groups, XET activity was assayed in different acrocarp and pleurocarp species (Table 1, pleurocarpous mosses are designated with an asterisk). The expression of XET-displaying enzymes in the pleurocarp mosses corresponded to the site just beneath the apical meristem of the moss (Fig. 2A, see circle), whereas the other internal parts of the gametophyte displayed no visible XET activity on the print. No incorporation of fluorescence was visible in the control assay (Fig. 2B), indicating that the fluorescence in Fig. 2A was indicative of the XET activity of XTHs or XTH-like enzymes. In acrocarp mosses the same pattern of XET activity was visible when a young moss was assayed (Fig. 2C, see circle). However, the XET signal near the acrocarp apex was clearly weaker than that observed in pleurocarp mosses. This was probably caused by the high density of leaves covering the apex of most acrocarps, making it more difficult for the growing part to come into contact with the test paper. Remarkably, no XET activity was visible near the apex when older acrocarp gametophytes, bearing sporophytes, were pressed onto the test paper (Fig. 2E). The remainder of both the young and the old acrocarp gametophytes did not display XET activity (Fig. 2C, E), and nor did the control assays (Fig. 2D, F). Strong XET activity was, however, seen at the sites where the maturing sporophyte capsule of both acrocarp and pleurocarp mosses were pressed onto the XET test paper (Fig. 2E, see circle, example of an acrocarp moss). By contrast, young capsules showed no or only very little XET activity (Fig. 2E, right). This difference in XET activity, however, can be explained by the presence of a calyptra, covering the young capsules. Older capsules completely lost this gametophytic cover (Fig. 2E, left), and were therefore in direct contact with the XET assay paper leaving a bright XET signal. No signal was detected at the site where the seta was pressed onto the paper. In the control assay, the test paper revealed no incorporation of fluorescence (Fig. 2F).
The Sphagnales, or peat mosses, do not belong within the acrocarps or pleurocarps and are completely isolated within the Bryophyta. The gametophyte of a peat moss displayed a more widespread XET signal, from the capitulum down to the youngest branches that are still stretching out to their full length (Fig. 2G, H); this in contrast to the other groups of the Bryophyta.
Similar results were obtained in the Anthocerophyta. Pressing the gametophyte onto the test paper, a faint, but clear incorporation of XGO-SRs was visible at the edges of the thallus (Fig. 2I, see circles), but absent from central parts of the gametophyte (Fig. 2I). When the sporophytes were embedded in the gametophyte and their basis covered by the involucre, no XET signal was visible (Fig. 2I), but when removed from the gametophyte, the sporophyte foot generated a clear spot of XET activity on the test paper (Fig. 2J, see circle). This site of high XET activity corresponded to the zone above the basal meristem of the sporophyte where the epidermal cells elongate from short cells (arrow) to elongated cells (dashed arrow), as can be seen in several cell files at high magnification (Fig. 2K). Some transverse cell walls were marked with a line. No incorporation of fluorescence was visible in the control test paper impregnated with trisaccharide-SR (Fig. 2L).
In the Marchantiophyta both leafy and thalloid liverworts were assayed. In both groups the gametophytes showed clear XET activity. In thalloid liverworts an intense XET signal was visible at the edges of the gametophyte thallus, as seen in a Riccia species (Fig. 2M), but no XET activity was found at central parts of the gametophyte. In leafy liverworts, XET activity was seen near the site of the gametophyte apex (Fig. 2O, see circle). The remainder of the gametophyte showed no XET activity on the test paper (Fig. 2N, P). Sporophytes were not assayed in marchantiophytes.
XET activity is present in green algae
The presence of extractable XET activity was also studied in the Charophyta. In simple charophytic algae, such as the Zygnematales, cell division is often not localized into meristematic regions. Also, the anatomy of these organisms is not particularly suitable for tissue printing. Therefore, Chara vulgaris was analysed. This species has a highly organized thallus with clear spatial segregation of meristematic activity and cell expansion. In Chara clear XET activity was visible at the site where the apex was pressed onto the XET test paper, but also at sites corresponding to other parts of the gametophyte (Fig. 3A, see circles). A clear fluorescent signal was present at the site of the node and at the sites where the branchlets were pressed onto the test paper (Fig. 3B, see circle). The control assays confirmed the specificity of the XET-generated signal (Fig. 3C, D). The cell walls of Chara therefore clearly contain enzymes that act upon the exogenous xyloglucan donor and recognize the XyG acceptor substrates on the XET test paper.
Studying the other major lineage of green plants, the Chlorophyta, the presence of enzymes displaying XET activity was found in the ulvophycean algae Ulva linza, previously known as Enteromorpha linza. At the base, where the foot is attached to substrate, fluorescently labelled xyloglucan was clearly incorporated in the test paper (Fig. 3E, see circle). The absence of incorporation of non-XET substrates (Fig. 3F) confirmed the specificity of the XET activity in Chlorophyta. Other chlorophytes, however, displayed no XET activity (Fig. 3E).
Different representatives of the Rhodophyta (Fig. 3G) and Phaeophyta (Fig. 3I) were also tested for XET activity. A substantial amount of autofluorescence was visible, both in experiments and in controls, hampering the detection of XET activity. However, none of the species assayed appeared to show a signal of XET activity on the prints (Fig. 3H, J).
Searching for an XTH or an XTH-like enzyme in Chara vulgaris
The fluorescence found on the XET tissue print of Chara vulgaris indicated that there are enzymes expressed in the charophycean cell wall that are able to transglucosylate xyloglucans in vitro. Based upon these findings RNA was prepared and searches were made for the presence of XTH or XTH-like transcripts in Chara. 3′ RACE using a degenerate primer, based upon the variations present in the catalytic domain of vascular plant XTHs, resulted in the amplification of a 480-bp fragment, named Chara2. To identify its homology with angiosperm XTHs, the deduced amino acid sequence was analysed in silico. As the evolutionary connection between XTHs and (1,3-1,4)-β-d-glucan endohydrolases was recently demonstrated by Strohmeier et al. (2004), the relationship of Chara2 with both enzyme groups was studied in an alignment (ClustalW) including two monocot XTHs of barley (HvXTH3, HvXTH4), two dicot XTHs of Arabidopsis (AtXTH4, AtXTH22) and two family 16 (1,3-1,4)-β-d-glucan endohydrolases (Fig. 4). Both groups of enzymes shared a homologous catalytic site (shown in italics), which in most XTHs was immediately followed by an N-linked glycosylation site (underlined). Strohmeier et al. (2004) stated that one of the key differences between XTHs and (1,3-1,4)-β-d-glucan endohydrolases is the insertion of three amino acids in XTHs, i.e. the PYX motif (boxed in Fig. 4). This motif is absent in the Chara2 sequence and in the two (1,3-1,4)-β-d-glucan endohydrolases; another key difference is the substitution of a methionine in (1,3-1,4)-β-d-glucan endohydrolases by an aromatic amino acid residue, which is generally a tyrosine in XTHs. Interestingly, this substitution is present in Chara2 (Fig. 4, arrow). In addition, the sequence of Chara2 shaded in grey shows more homology with that of the corresponding acceptor binding loop sequence of XTHs than do the endoglucanase sequences (Fig. 4, acceptor binding loop is shaded in grey, homologous amino acids are marked in white).
Fig. 4.
Alignment of the amino acid sequence of Chara2 with two monocot XTHs of barley (HvXTH3, HvXTH4), two dicot XTHs of Arabidopsis (AtXTH4, AtXTH22) and two family 16 (1,3-1,4)-β-d-glucan endohydrolases. The catalytic site shared by XTHs and (1,3-1,4)-β-d-glucan endohydrolases is shown in italic and the PYX motif, a key difference between XTHs and (1,3-1,4)-β-d-glucan endohydrolases is boxed. The substitution of methionine in (1,3-1,4)-β-d-glucan endohydrolases to an aromatic amino acid residue, mostly a tyrosine, in XTHs is marked with an arrow, while the XTH acceptor binding loop and corresponding sequences in the Chara2 and (1,3-1,4)-β-d-glucan endohydrolases are shaded in grey. Homologous amino acids are marked in white.
DISCUSSION
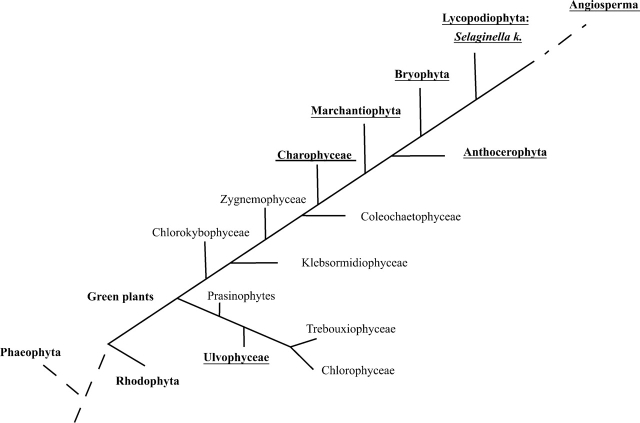
Thus far, XET action has been shown to be present in all vascular plants from the very ‘primitive’ lycopodiophyte Selaginella kraussiana up to ‘more evolved’ angiosperms (Vissenberg et al., 2003). Little is known about the presence of cell-wall-modifying enzymes in even ‘earlier’ land plants and algae. Thus far, extractable XET activity was detected in the liverwort Marchantia and the moss Mnium (Fry et al., 1992), a few partial putative XTH sequences were revealed in the Physcomitrella patens genome (Rensing et al., 2005; http://www.cosmoss.org) and expansin sequences were found in mosses as well (Schipper et al., 2002; Yi et al., 2002). To our knowledge, no data concerning the presence of XET activity in algae have been reported to date. To explore further the presence of XET activity, a broad range of bryophytes were examined and the potential origin of XET activity in green, red and brown algae was studied (Fig. 5).
Fig. 5.
Schematic view of part of the tree of life. The branching order of Phaeophytes, Rhodophytes and the green plants are based upon current DNA sequence data (Palmer et al., 2004). The phylogenetic relationships within the Chlorophyta and Charophyta are according to Lewis and McCourt (2004) and Graham et al. (2000). The branching order of the bryophytes remains uncertain (Shaw and Renzaglia, 2004). Note that the phaeophytes form a separate lineage, completely separate from the rhodophytes and green plants. Lineages studied for the presence of XET activity are shown in black. Those exhibiting XET activity are underlined.
The Bryophyta, Anthocerophyta and Marchantiophyta represent the oldest lineages among extant land plants and together they form the bryophytes (Fig. 5). Given that the Physcomitrella genome is in the process of being sequenced, the XET search started with the Bryophyta, blasting the draft database (http://www.cosmoss.org) with Sk-XTH1 (Van Sandt et al., 2006). This blast result included all hits that were also obtained when blasting with the different Arabidopsis XTHs. At least two of the resulting ESTs included all motifs that are essential for XET activity in vascular plants (Campbell and Braam, 1998; Henriksson et al., 2003; Johansson et al., 2003, 2004; Van Sandt et al., 2006). A functional XET assay on Physcomitrella homogenate was performed to prove the presence of at least one functional XTH protein. XTH and its XET function are thus present in Physcomitrella patens and they are possibly encoded by a multi-gene family. An XTH-related cell-wall-modifying machinery is thus probably present throughout the mosses and perhaps even in evolutionary more primitive phyla.
Specific XET activity was detected in other Bryophyta and in the Anthocerophyta and Marchantiophyta. Furthermore, in both the gametophyte and the sporophyte of the Bryophyta and Anthocerophyta and in the gametophyte of the Marchantiophyta, a clear correlation between the site of growth and the presence of XET activity was demonstrated. In the Bryophyta the pattern of XET activity in acrocarp mosses corresponds nicely with the developmental stage of the apical cell. A fluorescent XET signal is only present when a young gametophyte is pressed onto the test paper. In older acrocarps the apical cell is used to form a gametangium, causing the gametophyte to cease growth. In accordance, no XET activity was found at the site of the gametophyte apex in sporophyte-bearing gametophytes. Another illustration of the correlation between XET activity and growth was found in the sporophytes of the Anthocerophyta. These land plants have a near basal meristem (Renzaglia and Vaugh, 2000; Shaw and Renzaglia, 2004), and XET activity occurs specifically at the elongation site above the basal meristem of the sporophyte. In liverworts, considered to be the most ancient plant lineage (Kenrick and Crane, 1997; Bateman et al., 1998), a clear correlation between XET activity and the sites of growth was observed.
Although significant differences exist in cell wall composition of the major land plant lineages, xyloglucan was found in the primary cell wall of all land plants, including the bryophytes (Popper and Fry, 2003, 2004). The conservation of the XET substrate throughout land plant evolution fits nicely with the conservation of XET function within land plants. It is therefore likely that XET activity is part of an ancient cell-wall-modifying machinery that originated even before the divergence of land plants.
The descent of the embryophytes from a charophyte ancestor (Fig. 5) is among others supported by the presence of cellulose-synthesizing rosettes in both groups (Hotchkiss and Brown, 1987). Charophytic algae were shown to lack xyloglucan (Popper and Fry, 2003). Remarkably, an unambiguous XET signal was present near to or below the meristematic cells of both apex and branchlets of the Chara vulgaris tissue print. Although xyloglucan was found to be absent in charophycean algae (Popper and Fry, 2003), some Chara cell wall enzymes seem to be able to catalyse the incorporation of xyloglucan oligosaccharides into the xyloglucan matrix on the test paper, and thus display XET activity. Recent studies have demonstrated a structural connection between XTHs and xylan endohydrolases. Based upon these findings it was suggested that XTHs could be active not only on cell wall xyloglucan but also on xylans (Strohmeier et al., 2004; Nishitani and Vissenberg, 2006). Interestingly, the enzymatic digests of the Chara AIR (alcohol insoluble residue) resulted in products corresponding with the oligosaccharides of xylan (Popper and Fry, 2004). The presence of genes coding for XTHs or XTH-like enzymes that in vivo transglucosylate xylans or other hemicelluloses in Chara was therefore studied. 3′ RACE, using a degenerate primer based upon the variations present in angiosperm XTHs, resulted in the amplification of a 480-bp fragment, named Chara2. To study its homology with XTHs, Chara2 was aligned with angiosperm XTHs (monocots and dicots). As recent data suggest an evolutionary link between 1,3-1,4-β-d-endoglucanases and XTHs (Strohmeier et al., 2004), 1,3-1,4-β-d-endoglucanases were included in the alignment as well. In addition to a homologous three-dimensional topology of the active site amino acids, as seen for XTHs and endoxylanases, 1,3-1,4-β-d-endoglucanases share the same amino acid composition with XTHs. Both enzyme families are therefore thought to share a common ancestor (Strohmeier et al., 2004). Aligning Chara2 with representatives of both enzyme families revealed that Chara2 shares features of both XTHs and 1,3-1,4-β-d-endoglucanases. The key differences between XTHs and 1,3-1,4-β-d-endoglucanases are the presence of a PYX motif in XTHs and the substitution of a methionine in 1,3-1,4-β-d-endoglucanases by a tyrosine in XTHs (Strohmeier et al., 2004). Chara2 has one of the two possible substitutions (M→Y), positioning it between both groups. The PYX motif, however, is not necessary for the XET function, as AtXTH4, an XTH that was shown to display XET activity, lacks this motif (Campbell and Braam, 1999). An additional difference between XTHs and 1,3-1,4-β-d-endoglucanases is a region in the protein that forms the acceptor binding loop in XTHs. In Chara2 this region is more similar to that of XTHs than in 1,3-1,4-β-d-endoglucanases (Fig. 4, shaded). Again, this positions Chara2 between both groups of enzymes. Together these data suggest that Chara2 is possibly the C-terminal end of an enzyme that is an intermediate between 1,3-1,4-β-d-endoglucanases, present in microbial organisms, and XTHs, seen in vascular plants. This is also reflected in the positioning of the Chara2 amino acid sequence within XTHs and 1,3-1,4-β-d-endoglucanases in a phylogenetic tree (data not shown). The Chara XTH-like enzyme and its interacting donor substrate therefore probably evolved together to an optimal enzyme–substrate interacting model as is seen in higher plants. The evolution to a XyG/XTH interacting mechanism was possibly one of the crucial events allowing the development of land plants.
Remarkably, a tissue print of Ulva linza, an ulvophycean alga belonging to the Chlorophyte lineage of the green plants, also showed a clear XET signal. The fluorescent spot corresponded to a small region above the holdfast of the algae. This holdfast is formed by the basal cell dividing into 3–4 holdfast cells, which elongate and undergo further division (Kim et al., 1991). A clear correlation between cell elongation and transglucosylation activity is therefore even found in the Chlorophyte lineage of green plants. Analysis of the cell wall components from Ulva lactuca and Ulva rigida suggested the presence of cellulose microfibrils associated with XyG-like polysaccharides (Lahaye et al., 1994). The structure of this sulfated glucuronorhamnoxyloglucan (ulvan) is, however, markedly different from that of higher plants (Lahaye and Ray, 1995). In other studies a 1,3-1,4-β-d-glucan endohydrolase digest of the Ulva AIR was shown to contain glucose and xylose (Popper and Fry, 2003), indicating the presence of a mixed linkage glucan with xylose substitutions in the Ulva cell wall. In accordance with the findings in the Charophyta, an ancient chlorophyte XTH-like enzyme possibly interacts with the mixed linkage xyloglucan-like polysaccharide as donor substrate in Ulva linza. Again these findings are supported by the evolutionary connection between 1,3-1,4-β-d-glucan endohydrolases and XTHs.
It is possible that in the green algae transglucosylating enzymes with a broader substrate range were/are present that gave rise to the XTHs with a higher substrate specificity. Recently the reverse story was suggested in monocots, where XTHs are thought to have lost their substrate specificity and now transglucosylate 1,3-1,4-β-d-glucans as well during growth and cell elongation (Strohmeier et al., 2004). Similar to Chara2, these monocot XTHs (HvXTH4) miss some typical XTH features which possibly play a role in narrowing the substrate specificity (Fig. 4, arrows) and have a less conserved amino acid composition of their acceptor binding loop (Fig. 4, shaded). This could be proof of the close relationship and possible interconversions of XTHs and 1,3-1,4-β-d-endoglucanases by small mutations. In addition, a mannan transglycosylase was detected in several plant species (Schroder et al., 2004), but no sequences of responsible enzymes are yet known.
In contrast to Ulva linza, no XET activity was found in Ulva lactuca. This difference in the presence of XET activity is striking. It can be explained by the fact that both organisms have a distinct morphology and growth pattern, being either tubular or planar. The Ulva lactuca specimen studied had a planar form where growth occurs in the entire plant body (http://www.algaebase.org). XET activity would therefore probably cause a very diffuse and hence undetectable XET signal, whereas growth and XET activity is more concentrated and detectable in the holdfast of the tubular growth form, as found in Ulva linza. The absence of a detectable XET signal in Cladophora rupestris, another Ulvophycean, supports this suggestion as the slender filaments could leave only a very faint and thus invisible XET signal on the test paper. This limitation of the technique to visualize XET activity in filamentous or unicellular organisms, as mentioned above, prevented further analysis of the presence of XET-related transglucosylation activity in the other major groups of chlorophytes and charophytes.
The finding of XET-related activity in a chlorophytic species opens new ideas on the understanding of the evolution of green plants and gives new insights into the development of their cell wall. The XTH-modifying machinery therefore probably originated in the ancestors of both charophytes and chlorophytes, before the split of the green plants. The presence of XyG and preference of this hemicellulose as an XTH substrate, as seen in dicots, is probably the result of selecting an optimal enzyme–substrate interacting mechanism to allow efficient cell wall elongation. A future strategy allowing the study of XET-related activity in thread-like and unicellular organisms, such as Mesostigma (see Fig. 5), could further clarify the phylogenetic relationship of the charophytes and chlorophytes and would reveal more details on the evolution of XET activity in green plants.
The presence of XET activity in the major lineages of the green plants raised the question of the presence of XET-related growth mechanism in red and brown algae. No XET activity was detected on tissue prints of different red and brown algae. The colour of the autofluorescence caused by algae pigments differed enough from that of the SR substrates to allow the detection of possible XET activity. Furthermore, the absence of XET activity is in complete agreement with the composition of their primary cell walls. Although both phaeophytes and rhodophytes contain cellulose fibrils, no homologue of XyG, mixed linkage-β-d-glucans and xylans can be found within their cell wall, suggesting another mechanism for cell wall expansion (Graham and Wilcox, 2000).
In summary, the data presented herein have revealed a clear correlation between growth (or cell elongation) and the presence of XET activity in all three major groups of bryophytes, and the presence of at least two potential XTH-encoding cDNAs in the Physcomitrella genome has been demonstrated. For the first time, it has been shown that XET activity is also present at sites of growth in Charophyta and Chlorophyta, suggesting that XET originated even before the evolutionary divergence of the Chlorobionta. In accordance, part of a transcript in Chara that possibly encodes an ancestral XTH enzyme was identified, and the structural and evolutionary link between XTHs, endo-xylanases and 1-3,1-4-β-d-endoglucanases was discussed, explaining the substrate-tolerant behaviour of this ancient transglucosylating enzyme. As no XET activity was detected in the Phaeophyta and Rhodophyta, XET activity is probably a feature unique to green plants.
Supplementary Material
ACKNOWLEDGEMENTS
V.V.S. is funded by a PhD grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT – Vlaanderen). K.V. is a Postdoctoral Fellow of the Fund for Scientific Research – Flanders (FWO – Vlaanderen). This research was partially funded by a University of Antwerp grant (UA-BOF) and a grant from the Fund for Scientific Research – Flanders (FWO – Vlaanderen), grant G.0101.04. We thank S. C. Fry for the labelled oligosaccharides, D. De Beer for his help in finding Phaeoceros carolinianus, Professor S. Hoste, Chairman of the Flemish Study group of Bryology and Lichenology, for his help in the collection of Anthoceros agrestis, Professor W. De Smet for the collection of marine algae and the Fund for Scientific Research – Flanders (FWO – Vlaanderen) for financial support.
LITERATURE CITED
- Albert M, Werner M, Proksch P, Fry SC, Kaldenhoff R. The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite. Cuscuta reflexa. Plant Biology. 2004;6:402–407. doi: 10.1055/s-2004-817959. [DOI] [PubMed] [Google Scholar]
- Albersheim P. The primary cell wall. In: Bonner J, Varner JE, editors. Plant biochemistry. 3rd edn. New York: Academic Press; 1976. pp. 225–274. [Google Scholar]
- Andersen RA. Biology and systematics of heterokont and haptohyte algae. American Journal of Botany. 2004;91:1508–1522. doi: 10.3732/ajb.91.10.1508. [DOI] [PubMed] [Google Scholar]
- Bateman RM, Crane PR, Dimichelle WA, Kenrick PR, Rowe NP, Speck T, Stein WE. Early evolution of land plants: phylogeny, physiology, and ecology of the primary terrestrial radiation. Annual Review of Systematics. 1998;29:263–292. [Google Scholar]
- Buck WR, Goffinet BG. Morphology and classification of mosses. In: Shaw AJ, Goffinet B, editors. Bryophyte biology. Cambridge: Cambridge University Press; 2000. pp. 71–123. [Google Scholar]
- Campbell P, Braam J. Co- and/or post-translational modifications are critical for TCH4 XET activity. Plant Journal. 1998;15:553–561. doi: 10.1046/j.1365-313x.1998.00239.x. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends in Plant Science. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary-cell walls in flowering plants consistency of molecular structures with the physical properties of the wall during growth. Plant Journal. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Crandall-Stotler B, Stotler RE. Morphology and classification of the Marchantiophyta. In: Shaw AJ, Goffinet B, editors. Bryophyte biology. Cambridge: Cambridge University Press; 2000. pp. 21–70. [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Darvill AG, Albersheim P, Delmer DP. The primary cell walls of flowering plants. In: Tobert NE, editor. The biochemistry of plants: a comprehensive treatise. Vol. 1. New York: Academic Press; 1980. pp. 92–162. [Google Scholar]
- Fry SC. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annual Review of Plant Physiology. 1986;37:165–186. [Google Scholar]
- Fry SC. The structure and functions of xyloglucan. Journal of Experimental Botany. 1989;40:1–11. [Google Scholar]
- Fry SC. Novel ‘dot-blot’ assays for glycosyltransferases and glycosyl hydrolases: optimisation for xyloglucan endotransglycosylase (XET) activity. Plant Journal. 1997;11:1141–1150. [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochemical Journal. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet BG, Buck WR. Goffinet BG, Hollowell V, Magill R, editors. Systematics of Bryophyta (mosses): from molecules to revised classification. Molecular systematics of bryophytes. Monographs in Systematic Botany from the Missouri Botanical Garden. 2004;98:205–239. [Google Scholar]
- Graham LE, Wilcox LW. Algae. New York: Prentice Hall; 2000. [Google Scholar]
- Graham LE, Cook M, Busse J. The origin of plants: body plan changes contributing to a major evolutionary radiation. Proceedings of the National Academy of Sciences of the USA. 2000;97:4535–4540. doi: 10.1073/pnas.97.9.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Marsden MPF, Delmer D. Pea xyloglucan and cellulose: VI. Xyloglucan-cellulose interactions in vitro and in vivo. Plant Physiology. 1987;83:384–389. doi: 10.1104/pp.83.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson H, Denman SE, Campuzano DG, Ademark P, Master ER, Teeri TT, Brumer H. N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochemical Journal. 2003;375:61–73. doi: 10.1042/BJ20030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Coutinho P, Davies G. A census of carbohydrate-active enzymes in the genome of. Arabidopsis thaliana. Plant Molecular Biology. 2001;47:55–72. [PubMed] [Google Scholar]
- Hotchkiss AT, Jr, Brown RM., Jr The association of rosette and globule terminal complexes with cellulose microfibril assembly in Nitella translucens (Charophyceae) Journal of Phycology. 1987;23:229–237. [Google Scholar]
- Hu Y, Poh HM, Chua NH. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. The Plant Journal. 2006;47:1–9. doi: 10.1111/j.1365-313X.2006.02750.x. [DOI] [PubMed] [Google Scholar]
- Iannetta PPM, Fry SC. Visualization of the activity of xyloglucan endotransglycosylase (XET) isoenzymes after gel electrophoresis. Phytochemical Analysis. 1999;10:238–240. [Google Scholar]
- Johansson P, Denman S, Brumer H, Kallas AM, Henriksson H, Bergfors T. Crystallization and preliminary X-ray analysis of a xyloglucan endotransglycosylase from Populus tremula×tremuloides. Acta Crystallographica. 2003;59:535–537. doi: 10.1107/s090744490202348x. [DOI] [PubMed] [Google Scholar]
- Johansson P, Brumer H, Bauman MJ, Kallas AM, Henriksson H, Denman S. Crystal structures of a xyloglucan endotransglycosylase reveal details of transglycosylation acceptorbinding. Plant Cell. 2004;16:874–886. doi: 10.1105/tpc.020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick PR, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- Kim KY, Ahn YS, Lee IK. Growth and morphology of Enteromorpha linza (L.) J. Ag. and E. prolifera (Müller) J. Ag. (Ulvales, Chlorohyceae) Korean Journal of Phycology. 1991;6:31–45. [Google Scholar]
- Lahaye M, Ray B. Cell-wall polysaccharides from the marine green alga Ulva “rigida” (Ulvales, Chlorophyta) – NMR analysis of ulvan oligsaccharides. Carbohydrate Research. 1995;283:161–173. doi: 10.1016/0008-6215(95)00407-6. [DOI] [PubMed] [Google Scholar]
- Lahaye M, Jegou D, Buleon A. Chemical characteristics of insoluble glucans from the cell wall of the marine green alga Ulva latuca (L.) Thuret. Carbohydrate Research. 1994;262:115–125. [Google Scholar]
- Lang D, Eisinger J, Reski R, Rensing S. Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism among mosses. Plant Biology. 2005;7:238–250. doi: 10.1055/s-2005-837578. [DOI] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM. Green algae and the origin of land plants. American Journal of Botany. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- Mägdefrau K. Life-forms of bryophytes. In: AJE Smith., editor. Bryophyte ecology. London: Chapman & Hall; 1982. pp. 45–48. [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TL. Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant Journal. 2005;43:181–190. doi: 10.1111/j.1365-313X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, et al. AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant Journal. 2005;42:525–534. doi: 10.1111/j.1365-313X.2005.02395.x. [DOI] [PubMed] [Google Scholar]
- McCann MC, Roberts K. Architecture of the primary cell wall. In: Lloyd CW, editor. The cytoskeletal basis of plant growth and form. London: Academic Press; 1991. pp. 109–129. [Google Scholar]
- McCann MC, Wells B, Roberts K. Direct visualization of cross-links in the primary cell wall. Journal of Cell Science. 1990;96:323–334. [Google Scholar]
- McNeil M, Darvill AG, Fry SC, Albersheim P. Structure and function of primary cell walls of plants. Annual Review of Biochemistry. 1984;53:625–663. doi: 10.1146/annurev.bi.53.070184.003205. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proceedings of the National Academy of Sciences of the USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell walls. International Review of Cytology. 1997;173:157–206. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. Journal of Biological Chemistry. 1992;267:21058–21064. [PubMed] [Google Scholar]
- Nishitani K, Vissenberg K. Roles of the XTH protein family in the expanding cell. In: Verbelen J-P, Vissenberg K, editors. The expanding cell. Berlin: Springer-Verlag (in press); 2006. Plant Cell Monographs-series. [Google Scholar]
- Palmer JD, Soltis DE, Chase MW. The plant tree of life: an overvieuw and some points of view. American Journal of Botany. 2004;91:1437–1445. doi: 10.3732/ajb.91.10.1437. [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinnen S, Kofod LV, York WS, Albersheim P, Darvill AG. A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology. 1999;9:93–100. doi: 10.1093/glycob/9.1.93. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. Primary cell wall composition of bryophytes and charophytes. Annals of Botany. 2003;91:1–12. doi: 10.1093/aob/mcg013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. Primary cell wall composition of pteridophytes and spermatophytes. New Phytologist. 2004;164:165–174. doi: 10.1111/j.1469-8137.2004.01146.x. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Fritzowsky D, Lang D, Reski R. Protein encoding genes in an ancient plant: analysis of codon usage, retained genes and splice sites in a moss. Physcomitrella patens. BMC Genomics. 2005;6:43. doi: 10.1186/1471-2164-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia KS, Vaugh KC. Anatomy development and classification of hornworts. In: Shaw AJ, Goffinet B, editors. Bryophyte biology. Cambridge: Cambridge University Press; 2000. pp. 1–20. [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biology. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. (doi: 10.1186/gb-2005-6-12-242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Carey RE, Cosgrove DJ. Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs. Journal of Plant Research. 2006 doi: 10.1007/s10265-005-0253-z. (doi: 10.1007/s10265-005-0253-z, 2006) [DOI] [PubMed] [Google Scholar]
- Saunders GW, Hommers MH. Assessing red algal supraordinal diversity and taxonomy in the context of contemporary systematic data. American Journal of Botany. 2004;91:1494–1507. doi: 10.3732/ajb.91.10.1494. [DOI] [PubMed] [Google Scholar]
- Schaefer DG. Gene targeting in Physcomitrella patens. Current Opinion in Plant Biology. 2001;4:143–150. doi: 10.1016/s1369-5266(00)00150-3. [DOI] [PubMed] [Google Scholar]
- Schipper O, Schaefer D, Reski R, Fleming A. Expansins in the bryophyte Physcomitrella patens. Plant Molecular Biology. 2002;50:789–802. doi: 10.1023/a:1019907207433. [DOI] [PubMed] [Google Scholar]
- Shakin-Eshleman SH, Spitalnik SL, Kasturi L. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. Journal of Biological Chemistry. 1996;271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- Shaw J, Renzaglia K. Phylogeny and diversification of bryophytes. American Journal of Botany. 2004;91:1557–1581. doi: 10.3732/ajb.91.10.1557. [DOI] [PubMed] [Google Scholar]
- Schroder R, Wegrzyn TF, Bolitho KM, Redgwell RJ. Mannan transglycosylase: a novel enzyme activity in cell walls of higher plants. Planta. 2004;219:590–600. doi: 10.1007/s00425-004-1274-x. [DOI] [PubMed] [Google Scholar]
- Stotler RE, Crandall-Stotler B. A revised classification of the Anthocerotophyta and a checklist of the hornworts of North America, north of Mexico. The Bryologist. 2005;108:16–26. [Google Scholar]
- Strohmeier M, Hrmova M, Fischer M, Harvey AJ, Fincher GB, Pleiss J. Molecular modelling of family GH16 glycoside/hydrolases: potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Science. 2004;13:3200–3213. doi: 10.1110/ps.04828404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Fry SC. Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant Journal. 2001;26:23–34. doi: 10.1046/j.1365-313x.2001.01005.x. [DOI] [PubMed] [Google Scholar]
- Van Sandt VST, Guisez Y, Verbelen J-P, Vissenberg K. Analysis of a xyloglucan endotransglycosylase/hydrolase (XTH) from the lycopodiophyte Selaginella kraussiana suggests that XTH sequence characteristics and function are highly conserved during the evolution of vascular plants. Journal of Experimental Botany. 2006;57:2909–2922. doi: 10.1093/jxb/erl064. [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen J-P, Miller JG, Fry SC. In vivo co-localization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell. 2000;12:1229–1238. doi: 10.1105/tpc.12.7.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Verbelen J-P, Fry SC. Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiology. 2001;127:1125–1135. [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Van Sandt V, Fry SC, Verbelen J-P. Xyloglucan endotransglucosylase action is high in the root elongation zone and in the trichoblasts of all vascular plants from Selaginella to Zea mays. Journal of Experimental Botany. 2003;54:335–344. doi: 10.1093/jxb/erg024. [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato V, Yokoyama R, Verbelen JP, Nishitani K. Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant and Cell Physiology. 2005;46:192–200. doi: 10.1093/pcp/pci013. [DOI] [PubMed] [Google Scholar]
- Yi L, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiology. 2002;128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.