Abstract
Background and Aims
A knowledge of natural populations' breeding systems is important in order to implement in situ and ex situ management and conservation practices. Using microsatellite markers, three Oryza glumaepatula populations from Brazil were studied to determine the breeding system and genetic structure parameters of this species.
Methods
Each population represented by ten families with ten individuals per family was studied using eight microsatellite primers. Families of the Rio Xingu population (XI) were obtained from the greenhouse, whereas families from Rio Solimoes (SO) and Rio Paraguay (PG) were collected from the wild. Amplified products electrophoresed on non-denaturing polyacrylamide gels were visualized with a silver staining procedure. The mating system parameters were analysed based on the mixed mating model (software MLTR) while genetic structure analyses of the three populations and their families were performed using the FSTAT software.
Key Results
The mean numbers of alleles per loci were 2·5, 3·9 and 2·5, respectively for the XI, PG and SO populations. Compared with their families, higher values for the observed heterozygosity and gene diversity were estimated for the parental populations. The subdivision (based on RST) and inbreeding (FIS) in the SO and PG populations had similar effects, while inbreeding was the main effect in the families. Multilocus outcrossing rates varied from 0·011 to 0·223 in the three populations, indicating divergence in the outcrossing rates among O. glumaepatula populations. For the species (considering SO and PG populations together) an intermediate value was observed (t̂m=0·116). Biparental inbreeding varied from 0·008 to 0·123, contributing to the selfing rate in these populations. More than 50 % of the outcrossing occurred between related individuals.
Conclusions
The results indicated divergence in the mating system among O. glumaepatula populations, with consequences for conservation practices. The mating system of this species was classified as mixed with a predominance of self-fertilization.
Key words: F-statistics, genetic structure, mating system, microsatellites, Oryza glumaepatula, outcrossing rate, Poaceae, reproduction, wild rice
INTRODUCTION
The mating system plays a crucial role in the genetic composition of populations, as it determines the frequency of the individual genotypes in subsequent generations, with great influence on the distribution and content of the genetic variation within and among populations (Hamrick et al., 1979; Hamrick and Godt, 1989; Brown, 1990) and affects population evolutionary factors such as gene flow and selection (Hamrick, 1989). Various factors may affect the breeding system: size and density of populations, mode of pollination and pollinator viability (Franceschinelli and Bawa, 2000), flowering synchronicity and phenologic patterns (Hall et al., 1996), degree of genetic structure of populations (Ennos and Clegg, 1982; Franceschinelli and Bawa, 2000) and presence of self-incompatibility systems (Murawasky and Hamrick, 1991).
The Oryza genus presents different reproductive systems (outcrossing, inbreeding, intermediate and vegetative reproduction) (Oliveira, 2002). At present, interest is concentrated on the neotropical species Oryza glumaepatula calling for information of its reproductive system to improve in situ and ex situ conservation practices. Oryza glumaepatula is one of 21 wild species of the genus, of AgpAgp genome, found in various parts of Latin America, from 23°S in Brazil up to 23°N in Cuba (Vaughan et al., 2003). It is the only diploid wild American species and its importance relies on its use in crosses with O. sativa in plant breeding programmes (Brondani et al., 2001) aiming at introgression of important traits from the wild species and amplifying the genetic basis of the cultivated crop. In Brazil, there are O. glumaepatula populations in the Amazon, the Pantanal Matogrossense and smaller hydrographic basins such as those of the States of Goias, Tocantins and Roraima (Oliveira, 1993, 1994; Brondani et al., 2005). This species presents annual, bi-annual or perennial populations, depending of its geographical location (Oliveira, 1993; Akimoto et al., 1998), growing along the riverbeds and margins, presenting behaviour typical of weeds or colonizing plants. As their culms are broken and their bodies released to float on the surface, they are dispersed on the rivers by the force of wind and water, mostly downstream (Akimoto et al., 1998), but sometimes upstream (Black, 1950), founding new populations or clustering to those already existing.
A series of studies have indicated that O. glumaepatula is an inbreeding species (Akimoto et al., 1998; Buso et al., 1998; Ge et al., 1999; Oliveira, 2002). Inbreeding species have the tendency to be more homogeneous within populations and more differentiated among populations, and can form pure lines within families increasing the genetic differentiation among families generating subdivision within the population, with the risk that the fixation of specific alleles may confer little or no flexibility in response to environmental changes. In a more dramatic scenario, these species may become completely extinct due to allele fixation and loss of genetic diversity (Richards, 1997; Hedrick, 2001; Frankham, 2003). On the other hand, under favourable conditions these plants may present an aggressive behaviour (typical of weeds) rapidly establishing themselves in the locality (Holsinger, 2000). In relation to conservation, this type of species needs special care as the highest possible number of populations should be identified to increase the chances of conserving different alleles, thus preserving the diversity existing within the species with greater efficiency.
Microsatellites have been intensively used in studies of plant population genetics due to its reproducibility (Ferreira and Grattapaglia, 1998), being considered neutral markers and more informative for this type of study (Brondani et al., 2005). Other traditional markers have provided inadequate statistic estimates of the diversity among groups of threatened species due to the low existing variation (Hedrick, 2001), especially in populations with low diversity at the DNA level or when its detection is not possible with these other markers (Paetkau et al., 1995).
The main objective of this study was to analyse the mating system and genetic structure parameters in three O. glumaepatula populations originated from three different geographic regions in Brazil using microsatellite markers.
MATERIALS AND METHODS
Materials and sampling procedure
Three Oryza glumaepatula Steud. populations were studied belonging to the wild rice germplasm collection of the Genetics Department of Escola Superior de Agricultura ‘Luiz de Queiroz’, ESALQ/USP, Piracicaba, São Paulo, Brazil. The populations were selected from three locations: Rio Xingu, a tributary to the Amazon from the State of Mato Grosso, collected at Lake Piulaga (12°14′S and 53°35′W) (XI); Rio Solimoes, State of Amazonas, collected at Lake Manacapuru (03°11′S – 60°47′W (SO); and Rio Paraguay, State of Mato Grosso do Sul, collected in the Pantanal (19°01′S – 57°30′W) (PG; Fig. 1). From each population ten maternal families were evaluated, with ten individuals per family, totalling 100 individuals/population and 300 individuals as a whole. Seeds of these families were collected from the wild, except those of the families in the XI population, which were collected from a greenhouse in Piracicaba, SP. The collections from the families in the wild were done in a boat, as the plants were growing in the water, along river banks, and each family was represented by a single panicle. Therefore, to avoid the risk of collecting two panicles from the same mother plant (genet), a distance of approx. 10–20 m was left between samples. This caution was taken for each population sampled. Seeds were germinated in the laboratory at 27 ± 5 °C, and then transplanted to greenhouse pots where the plants were grown to adult stage.
Fig. 1.
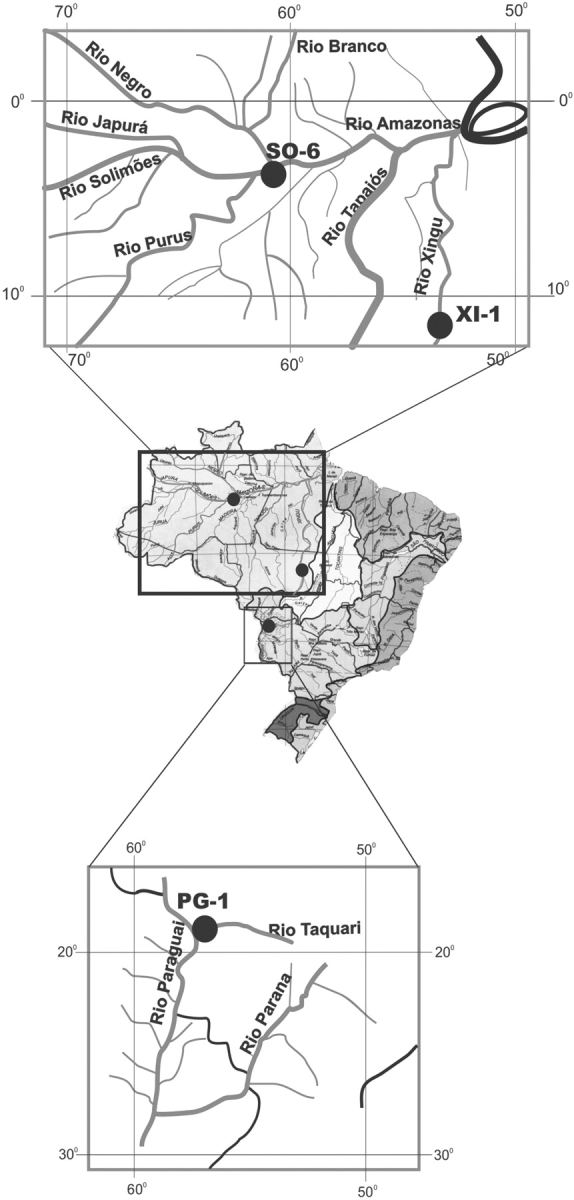
Geographical location of the Oryza glumaepatula maternal populations (SO, Rio Solimoes; XI, Rio Xingu; PG, Rio Paraguay).
Microsatellite analysis
Total genomic DNA was extracted from adult lyophilized leaves using a CTAB procedure according to Hoisington et al. (1994), simplified by Karasawa (2005), consisting mainly in the exclusion of NaOAc (sodium acetate) and all the following steps. DNA was quantified in a 4 % polyacrylamide gel using a silver staining procedure.
Eight primer pairs developed by Brondani et al. (2001) for O. glumaepatula were selected for amplification in this study: OG-22, OG-26, OG-27, OG-29, OG-36, OG-39, OG-42 and OG-63. All primers were first submitted to the basic programme with 56 °C for primer annealing according to the PCR reactions described by Brondani et al. (2001). Primers that did not amplify satisfactorily in all populations in this basic programme were submitted to new amplification cycles with the highest and lowest annealing temperatures (Table 1).
Table 1.
Microsatellite sequences of primer pairs developed for Oryza glumaepatula with the chromosome where the loci was identified (C), allele size range (pb), number of alleles per locus (A), observed heterozygosity (Ho) and annealing temperatures (Ta)
| SSR locus | Primers sequences* | C | pb | A | Ho | Ta (°C) |
|---|---|---|---|---|---|---|
| OG-22 | (F) GCCATCCATTCTTACCAG | 12 | 175–185 (SO) | 3 | 0·000 | 56 |
| (R) CACAGGTGTGGTGCTCA | 190–225 (PG) | 7 | 0·067 | |||
| 180–200 (XI) | 5 | 0·242 | ||||
| OG-26 | (F) CATGGTGCCGATTACGGT | 10 | 95–100 (SO) | 2 | 0·012 | 60 |
| (R) CATCTCCATCGCGGTCAT | 95–110 (PG) | 4 | 0·103 | |||
| 95–110 (XI) | 4 | 0·050 | ||||
| OG-27 | (F) TCGGACGTGGCATATGA | 9 | 140–165 (SO) | 4 | 0·010 | 54 |
| (R) CTGTTCCGAGCGAGAGT | 140–210 (PG) | 6 | 0·117 | |||
| – (XI) | – | – | ||||
| 0G-29 | (F) GACCAGTTCACCATGCAG | 1 | 100–105 (SO) | 2 | 0·000 | 60 |
| (R) GAGTGAGGCAGCAAGACA | 90–130 (PG) | 7 | 0·021 | |||
| 85–105 (XI) | 4 | 0·051 | ||||
| 0G-36 | (F) AACGTTCATCGGTTCTGG | 4 | 170–195 (SO) | 5 | 0·000 | 56 |
| (R) TGCTTGCCAGGTTATTCC | 180–195 (PG) | 4 | 0·098 | |||
| 160–170 (XI) | 3 | 0·280 | ||||
| OG-39 | (F) GCGTACTAGGCCATGATA | 3 | 250–270 (SO) | 2 | 0·000 | 56 |
| (R) TCCACGTAAGAACACTCG | 270–285 (PG) | 4 | 0·150 | |||
| 250–265 (XI) | 2 | 0·000 | ||||
| OG-42 | (F) TGCAGGCTCTGAGCTAC | 5 | 420–430 (SO) | 2 | 0·000 | 56 |
| (R) AGAACAGATCTTGCCGTC | 425–445 (PG) | 4 | 0·010 | |||
| 420 (XI) | 1 | 0·000 | ||||
| OG-63 | (F) CAGGGGACAAGCACATA | 2 | 115–145 (SO) | 4 | 0·000 | 56 |
| (R) TAGACGATGTCGAGAAGG | 100–145 (PG) | 8 | 0·202 | |||
| 130–135 (XI) | 2 | 0·041 |
*Sequences of primer pairs developed by Brondani et al. (2001).
For each PCR reaction, 30 ng of genomic DNA were used in a 12 µL volume containing 0·3 µm of each primer, 0·25 mm of each dNTP, 1·5 mm of MgCl2, 10 mm Tris–HCl and 0·6 unit of Taq DNA polymerase enzyme (Gibco BRL). The reactions were performed on a Primus 96 thermocycler with 4 min initial denaturation at 94 °C, 30 cycles of (1 min at 94 °C, 1 min at 54 °C, 56 °C or 60 °C, 1 min elongation at 72 °C), followed by a final elongation of 5 min at 72 °C. Amplified products were electrophoresed on 6 % non-denaturing polyacrylamide gels run vertically (120 V for 3 h). Amplified fragments were visualized using a silver staining procedure.
Genetic structure
Genotypic frequencies of the parental populations and progenies were submitted to FSTAT software (Goudet, 1995) to estimate the number of alleles per locus (A), observed heterozygosity (Ho), gene diversity (He) and fixation index (f) [f = 1 – (Ho/He)]. F statistics (Weir, 1996) were estimated considering the two populations collected from the wild (PG, SO). The mutation process in microsatellite loci is not in line with the expectations under the infinite alleles model with low rates. Therefore, the analogue of the FST parameter (Slatkin, 1995) developed specifically for microsatellite data (RST) was also estimated. The linkage disequilibrium test was performed among the microsatellite loci with FSTAT software.
Mating system determination
The mating system was analysed based on the mixed mating model of Ritland and Jain (1981) using the MLTR program (Ritland, 2002), based on the following assumptions: (a) each mating event is due to random outcrossing (with probability t) or self-fertilization (with probability s = 1 – t); (b) the probability of outcrossing is independent of the maternal genotype; (c) the pollen pool is homogeneous over all maternal plants; (d) there is no selection between fertilization and the time of assay for progeny genotypes; and (e) alleles at different loci segregate independently (Ritland and Jain, 1981). Based on these assumptions the following parameters were estimated: multilocus outcrossing rate (t̂m), single-locus outcrossing rate (t̂s), outcrossing rate between related individuals (t̂m – t̂s), and the correlation of paternity (rp) or proportion of full sibs among outcrossed progeny. These parameters were estimated using maximum likelihood procedures. The Newton–Raphson method was used to solve the likelihood equation for the maximum likelihood estimates, as recommended by Ritland (1996). The number of pollen donors contributing to each family or neighborhood size, was estimated as 1/rp (Ritland, 1989). The inbreeding coefficient of maternal parents (F̂m) was calculated using the MLTR program (Ritland, 2002). t̂m and t̂s values were compared to assess the degree of biparental inbreeding. In the absence of biparental inbreeding, the values will be the same, whereas in the presence of biparental inbreeding, t̂s will be smaller than t̂m because outcrossing events that are not detected at a single locus have a higher probability of being detected as more loci are examined (Ritland, 1996). The standard error of the reported estimates was calculated based on 1000 bootstraps, where the sampling units were plants within progenies for the individual outcrossing rate per parental population, and the families for the population outcrossing rate. The coefficients of co-ancestry (θ) among families within and between populations (for SO and PG) were obtained using the FSTAT program (Goudet, 1995).
RESULTS
Genetic structure
All loci were polymorphic in the three populations, except for the OG-42 locus, which was monomorphic in population XI (Table 1). The eight loci varied from 2 to 5, 0 to 5 and 4 to 8 alleles, respectively, for the families of populations SO, XI and PG, totalling 24, 21 and 44 alleles. The observed heterozygosity (Ho) for each locus was low (Table 1) and varied from 0·000 to 0·012, 0·000 to 0·280 and 0·010 to 0·202, respectively, among the families of SO, XI and PG populations.
The average number of alleles per locus varied from 2·5 to 3·9 for the three populations studied and from 2·4 to 5·6 for their families (Table 2), the highest values were estimated for the PG population and family. The values of the observed heterozygosity (Ho) and gene diversity (He) obtained from the parental populations were higher than those obtained from the families. Comparing the fixation indexes (f) of the parental populations with those of their respective families showed that they were similar in XI, much lower in PG and higher in SO.
Table 2.
Average number of alleles per locus (A), observed heterozygosity (Ho), gene diversity (He) and fixation index (f) for the parental populations (P) and families (F) of Oryza glumaepatula populations
| Populations |
A |
Ho |
He |
f |
||||
|---|---|---|---|---|---|---|---|---|
| P | F | P | F | P | F | P | F | |
| XI | 2·500 | 2·400 | 0·233 | 0·095 | 0·369 | 0·137 | 0·368 | 0·307 |
| PG | 3·875 | 5·600 | 0·100 | 0·098 | 0·569 | 0·175 | 0·824 | 0·440 |
| SO | 2·500 | 3·000 | 0·136 | 0·003 | 0·369 | 0·039 | 0·631 | 0·923 |
The F-statistics were estimated considering the two populations and families collected from the wild (PG and SO) (Table 3). Population XI was not included in this analysis because the seeds of the families were obtained from a greenhouse, in a controlled environment, in the absence of pollinating agents. Therefore, the population size was restricted to the sample of the seed parent collected from the wild. Higher values were obtained for FIT (0·878 and 0·942) and FIS (0·778 and 0·919), respectively, for populations and families (Table 3). Both FST and RST showed higher values for the populations (FST=0·451; RST=0·716) than for the families (FST=0·284; RST=0·418).
Table 3.
Estimate of Wright's F statistics and R ST for the parental populations (P) and families (F) of Oryza glumaepatula originated from Rio Solimoes (SO) and Rio Paraguay (PG) and the confidence intervals for the lower and upper limits at 95 %
|
FIT |
FIS |
FST |
RST |
|||||
|---|---|---|---|---|---|---|---|---|
| P | F | P | F | P | F | P | F | |
| All loci | 0·878 | 0·942 | 0·778 | 0·919 | 0·451 | 0·284 | 0·716 | 0·418 |
| Lower | 0·794 | 0·914 | 0·655 | 0·883 | 0·299 | 0·192 | 0·680 | 0·397 |
| Upper | 0·953 | 0·964 | 0·902 | 0·952 | 0·607 | 0·386 | 0·751 | 0·439 |
The linkage disequilibrium test showed that locus OG-22 was in disequilibrium with four of the other loci (OG-27, OG-29, OG-36 and OG-39) and OG-27 was also in disequilibrium with loci OG-36 and OG-63 (Table 4). Therefore, the analyses were performed with and without these two loci to verify the effect on the results. In general, the results of both analyses were within the 95 % confidence interval.
Table 4.
Significance of the linkage disequilibrium estimates for eight loci in Oryza glumaepatula populations originated from Rio Solimoes (SO) and Rio Paraguay (PG)
| OG-22 | OG-26 | OG-27 | OG-29 | OG-36 | OG-39 | OG-42 | OG-63 | |
|---|---|---|---|---|---|---|---|---|
| OG-22 | – | |||||||
| OG-26 | n.s. | – | ||||||
| OG-27 | ** | n.s. | – | |||||
| OG-29 | * | n.s. | n.s. | – | ||||
| OG-36 | * | n.s. | * | n.s. | – | |||
| OG-39 | * | n.s. | n.s. | n.s. | n.s. | – | ||
| OG-42 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | – | |
| OG-63 | n.s. | n.s. | * | n.s. | n.s. | n.s. | n.s. | – |
**P<0·01, *P<0·05, n.s., non significant.
Mating system
Multilocus outcrossing rates (t̂m) were low (0·011, 0·160 and 0·223, respectively, for populations SO, XI and PG, and 0·116 considering populations SO and PG together, representing the species) and significantly different from 1·0, as verified by the standard error (Table 5). These values indicated the existence of variation in the outcrossing rates of the different O. glumaepatula populations, ranging from absolute autogamy to a mixed system with predominance of self-fertilization. The single-locus estimates (t̂s) were also low for the species (0·047) and populations (0·003, 0·078 and 0·100, respectively for SO, XI and PG) and significantly different from 1·0, reinforcing the condition of an inbreeding species. The estimates of the t̂m−t̂s values were also low for the species (0·069) and populations (0·008, 0·083 and 0·123, respectively for SO, XI and PG) but positive and significantly different from zero, indicating that biparental inbreeding contributed to the selfing rate of O. glumaepatula populations.
Table 5.
Mating system parameters in three Oryza glumaepatula populations: multilocus outcrossing rate (t̂m), single-locus outcrossing rate (t̂s), outcrossing rate between related individuals (t̂m – t̂s), correlation of paternity (rp), neighbourhood (1/rp), inbreeding coefficient of maternal parents (F̂m) and the coefficient of co-ancestry (θ) for populations and species (considering the SO and PG populations together)
| Parameters | SO | XI | PG | Species (SO and PG) |
|---|---|---|---|---|
| Number of families | 10 | 10 | 10 | 20 |
| Number of individuals | 100 | 100 | 100 | 200 |
| t̂m | 0·011 (0·010)* | 0·160 (0·057) | 0·223 (0·040) | 0·116 (0·030) |
| t̂s | 0·003 (0·003) | 0.078 (0·032) | 0·100 (0·024) | 0·047 (0·014) |
| t̂m – t̂s | 0·008 (0·007) | 0·083 (0·029) | 0·123 (0·025) | 0·069 (0·019) |
| rp | 0·999 (0·261) | 0·564 (0·255) | 0·765 (0·182) | 0·668 (0·178) |
| 1/rp | 1·00 | 1·77 | 1·31 | 1·50 |
| F̂m | 0·846 (0·06) | 0·399 (0·120) | 0·631 (0·100) | 0·768 (0·056) |
| θ | 0·932 (0·028) | 0·711 (0·077) | 0·748 (0·052) | 0·285 (0·054) |
*Numbers in brackets refer to the s.e.
The maternal inbreeding coefficient (F̂m) agrees with the other estimates indicating the presence of a high inbreeding level in the species (0·768). A high inbreeding level was also observed for the SO seed parent (0·846), being intermediate for PG (0·631) and less intense for the XI parental population (0·399) (Table 5). The paternity correlation (rp) gives support to the results, showing higher correlation for the SO families. As observed by the maternal inbreeding coefficient and by the multilocus and single-locus rates, this population is composed of families of absolute autogamy, with the seed parent donating both male and female gametes (ovule and pollen grains), which agrees with the number of pollen donors contributing to each family (1/rp) being 1 for these families. These values also agree with the coefficients of co-ancestry within populations (θ), which was 0·932.
The correlation of paternity (rp) for XI was the lowest one, indicating that this population possesses a higher number of pollen donator parents. The XI coefficient of co-ancestry within this population (θ) was approximately the same for the PG population, suggesting that the families were more closely related than inferred by the observed outcrossing rates. In the PG population, the paternity correlation (rp) was 0·765, indicating the presence of a slightly lower number of pollen donator parents (1/rp) in relation to the XI population, a value of 1·31 (Table 5). Also, the coefficient of co-ancestry (θ) was practically the same as for the XI population. As the XI population showed a lower outcrossing rate (t̂m) than PG, a higher coefficient of co-ancestry (θ) should be presented. In the case of PG, although it presents a higher outcrossing rate (t̂m), the presence of biparental matings also caused the increase in the coefficient of co-ancestry (θ). When considering the coefficient of co-ancestry between SO and PG populations, a much lower value was estimated (0.285), which is expected when comparing different populations.
The mean outcrossing rates (t̂) evaluated per locus in the families of each population (Table 6) indicated the averages of 0·3 %, 7·7 % and 9·3 %, respectively, for SO, XI and PG. These values agree with the single-locus outcrossing values (t̂s) in Table 5. On the other hand, when analysing the mean values observed for each locus in the three populations, a variation in the outcrossing rates, of 1·6 % to 7·7 %, was verified, indicating a good consistency of the data, agreeing with the condition of predominance of inbreeding in this species.
Table 6.
Outcrossing rate (t̂) estimates per locus and per Oryza glumaepatula population
| Locus | SO | XI | PG | Mean |
|---|---|---|---|---|
| OG-22 | 0·001 (0·000)* | 0·177 (0·075) | 0·049 (0·052) | 0·076 |
| OG-26 | 0·001 (0·023) | 0·085 (0·067) | 0·110 (0·156) | 0.065 |
| OG-27 | 0·017 (0·016) | – | 0·136 (0·062) | 0.077 |
| OG-29 | 0.001 (0·024) | 0·150 (0·235) | 0·025 (0·016) | 0·059 |
| OG-36 | 0·001 (0·000) | 0·001 (0·036) | 0·084 (0·039) | 0·029 |
| OG-39 | 0·001 (0·000) | 0·001 (0·000) | 0·186 (0·059) | 0·063 |
| OG-42 | 0·001 (0·000) | 0·001 (0·000) | 0·047 (0·038) | 0·016 |
| OG-63 | 0·001 (0·000) | 0·122 (0·037) | 0·103 (0·056) | 0·075 |
| Mean | 0·003 | 0·077 | 0·093 | 0·058 |
*Numbers in brackets refer to the standard errors.
DISCUSSION
Oryza glumaepatula is considered an autogamous species by some authors (Akimoto et al., 1998; Ge et al., 1999; Oliveira, 2002); others consider that its reproductive system is mixed with a predominance of selfing (Brondani et al., 2005; Karasawa, 2005), but no study at the family level has so far supported this assertion. It is known that there is variation in the outcrossing rates among populations of this species, as estimated by the apparent outcrossing rate (Karasawa, 2005). To investigate this variation in more detail, a study based on families was performed using samples from three distinct Brazilian regions (Rio Solimoes, Amazonas State; Rio Xingu, Mato Grosso State; Rio Paraguay, Mato Grosso do Sul State. It should be stressed that in one of the populations (XI) the families were collected in greenhouse conditions, which precludes direct comparison with the other two (SO and PG, whose families were collected from natural populations). The objective of including XI in this study was to assess change in values, or phenotypic plasticity, of outcrossing rates caused by growing the plants in an environment completely devoid of pollinating agents whatsoever (either wind or animals) in comparison to values obtained under natural conditions. Thus, although the data for the three populations are presented and discussed together in the beginning for the sake of conciseness of display, a special discussion on XI is shown afterwards.
The samples of populations PG and SO were not representative in relation to the number of alleles per locus, which is evident in the higher amount of alleles present in the families collected from the wild. However, the values obtained for the number of alleles per locus of the XI population, whose family was obtained in a controlled environment, were similar between the population and families. A drastic reduction was verified in the levels of observed heterozygosity (Ho) and gene diversity (He) in the families, while the fixation index (f) remained unaltered for the XI population and decreased in the PG families, indicating that this population probably practices a higher level of outcrossing than the one estimated based on a sample of the population (sampling error, not representative). However, the SO families showed a high fixation index, which may be reflecting their preference for self-fertilization or an environmental factor that might have caused the absence of pollinators at the time for anthesis.
The F-statistics evaluated for the two populations collected from the wild (SO and PG) showed high levels of total inbreeding (FIT), both in the populations and their respective families. It was also verified that both subdivision (based on RST) and inbreeding (FIS) had the same effect on total inbreeding for the populations, while in the families the highest effect was promoted by inbreeding (FIS) due to the mating system. Based on these observations, it is concluded that the probable mating system of this species is that in which self-fertilization predominates. As the families of population XI were obtained in an artificial environment (a greenhouse), they were not included in the above analysis, to avoid misinterpretations. However, its mating system was assessed, as each population and respective families were analysed separately by the MLTR software developed by Ritland (2002). It must be emphasized that the information obtained on the XI population does not reflect the real situation from the wild. A certain level of mortality and a probable effect of selection were observed in the XI population when the families were multiplied in the greenhouse. The mortality of 14 individuals, from a total of 17 individuals produced from a given family was observed. The reasons for this fact were further investigated, where it was verified that this mother plant was monomorphic for six of the eight loci assessed. It was also verified that the other mother plants with no mortality presented at least five polymorphic loci (from a total of eight). These observations may be indicating a possible selection effect against inbreeds.
In this study, the allele frequencies in pollen and ovules were considered equal, given the selfing condition of the species. A test assuming different frequencies in pollen and ovules was also performed, but no statistical difference resulted. Thus, the basic condition of Ritland and Jain's mixed model (Ritland and Jain, 1981), which assumes equality between pollen and ovule allele frequencies, was not violated.
The multilocus outcrossing rates (t̂m) observed in O. glumaepatula populations were low in the three populations and for the species when considering the SO and PG families together. These rates, assessed at the locus level (t̂) for the families of each population, agree with the mean single-locus rate (t̂s) of the three populations, which shows low levels of outcrossing. The mean for the three populations was also coherent among the loci, since the variation, from 1·6 % to 7·7 %, was low in comparison with the other indices.
The values of the single- and multilocus outcrossing rates of population SO are similar to those found by Morishima and Barbier (1990) for the annual population of O. rufipogon (t̂s=0·05 – 0·18 and t̂m=0·043), using eight isozyme loci. However, the perennial O. rufipogon populations had single-locus outcrossing rates higher than those described in this study (t̂s=0·35∼0·92 and 0·65∼1·00; t̂m=0·506 and 0·559, respectively, for NE88 and CP20); in this study, however, the multilocus rates of XI and PG were intermediate.
Of the small fraction of multilocus outcrossing (t̂m), 50 % or more occurred between related individuals (t̂m – t̂s), viz. 0·008 out of 0·011, 0·083 out of 0·160 and 0·123 out of 0·223, for the families of populations SO, XI and PG, respectively (Table 5), which must have contributed for the high co-ancestry coefficients observed. As an aid to comparison, it is known that in panmictic populations θ assumes values of 0·125 for half-sibs, 0·25 for full-sibs and 0·50 for selfed progeny. Therefore, the multilocus outcrossing rates (t̂m), the difference between single- and multilocus outcrossing rates (t̂m−t̂s), the co-ancestry coefficient values (θ) and the maternal inbreeding levels indicate that selfing is more frequent in these populations and most progeny originates from the union of pollen and ovule from the same plant.
The indirect estimate of the paternity correlation (rp=0·999, 0·564, 0·765 and 0·668, respectively, for SO, XI, PG, and for the species) was high, indicating that a small number of males donated pollen (1/rp=1·00, 1·77, 1·31 and 1·50, respectively, for SO, XI, PG and for the species). Also, 1·10 %, 9·02 %, 17·06 % and 7·7 % of the offspring were full-sibs (t̂mrp) and 0·001 %, 6·98 %, 5·24 % and 3·85 % were half-sibs [t̂m(1−rp)]. The rest of the offspring were produced by selfing the maternal plant.
In a parallel study, population XI was analysed within the Wright's F statistics theoretical framework and compared with samples from the Solimoes, Paraguay and other basins (Karasawa, 2005). The apparent outcrossing rate of population XI thus estimated (t̂a=0·459) was twice or three times as high as the estimates of the other populations, suggesting a relatively strong tendency in this population to avoid selfing. Under greenhouse conditions, pollen grains have two main fates: they either adhere to the stigma of the same flower that produced them; or fall down under the sole effect of gravity and air resistance, which may make them move slightly sideways. In a crowded greenhouse, the superposition of panicles of neighbouring plants allows the production of half-sibs and full sibs, but the absence of pollinating agents induces a heavy bias towards selfing, even in typically allogamous plants such as maize. This may be the reason why the outcrossing rate estimated for XI was much lower under greenhouse conditions than under natural conditions (even taking into account that the estimating methods were distinct and normally yield different values), suggesting the existence of considerable phenotypic plasticity for the character. Even so, the greenhouse-grown XI population showed outcrossing rates much higher than those obtained for the naturally pollinated SO, and not much lower than PG. The number of pollen donors of XI (1·77) is remarkably high in comparison to the other two populations and the coefficient of co-ancestry (0·711) is lower. These estimates show that, even in adverse conditions, XI avoids inbreeding more efficiently than the other two populations.
The three populations represent three ecotypes and each grows at different altitudes in their original habitats (0 m in the Amazon, 400 m in the Savana and 100 m in the Pantanal, respectively, for SO, XI and PG). The populations in the Solimoes are predominantly annual while those from the other two rivers have a higher degree of perenniality. Oryza glumaepatula populations may cluster on the river banks, like XI, or be widely distributed over the entire river basin, as the PG populations in the Pantanal Matogrossense. The type of vegetation on the banks, the growth habit, the density, the dispersal mechanism, and the level of stem prostration over the water surface may have a major influence on pollinator access and activity, and consequently on the outcrossing rates.
Since the inflorescences have hermaphroditic flowers with long, protracted anthers, characters typical of allogamous plants, one would expect higher levels of outcrossing. Both field observations and experimental data on floral biology, documenting the role of zoophyly in the reproduction of O. glumaepatula are needed. Species pollinated by insects or other animals have a wide distribution of outcrossing rates among individuals, which may range from absolute autogamy to absolute allogamy, but anemophylous plants rarely show intermediate values (Vogler and Kalisz, 2001). It is also known that irregular visits of pollinators favour autogamy (Kalisz et al., 2004).
The reproductive system plays a crucial role in the amplification and recombination of the genetic variability of populations. Consequently, the random mating deviations observed in O. glumaepatula have important consequences for conservation and breeding. Ex situ conservation of populations with deviations from panmixia demands larger samples than those recommended for populations under Hardy–Weinberg equilibrium (i.e. lacking inbreeding) because these deviations cause the sample effective size to decrease. Also, the sampling of as many families as possible within each population should be considered, because of the greater variation between than within families. In the same vein, for in situ conservation it is necessary to identify as many populations as possible to keep most of the existing diversity, because populations tending to autogamy generally have great interpopulational diversity, due to the reduction in or absence of gene flow by pollen.
The present data show that the reproductive system of this species is variable both among and within populations, ranging from complete selfing to a mixed system with predominance of selfing.
A problem caused by recurrent selfing is the loss of diversity through allele fixation, which reduces the flexibility of the species in adapting to environmental conditions and may lead to population extinction in the long term (Richards, 1997; Hedrick, 2001; Frankham, 2003).
ACKNOWLEDGEMENTS
This research was supported by grants provided by the National Council of Scientific and Technological Development (CNPq).
LITERATURE CITED
- Akimoto M, Shimamoto Y, Morishima H. Population genetic structure of wild Oryza glumaepatula distributed in the Amazon flood area influenced by its life-history traits. Molecular Ecology. 1998;7:1371–1381. [Google Scholar]
- Black GA. Os capins aquáticos da Amazônia. Boletim Técnico do Instituto Agronômico do Norte. 1950;19 Notas sobre a flora neotrópica III. [Google Scholar]
- Brondani C, Brondani RPV, Rangel PHN, Ferreira ME. Development and mapping of Oryza glumaepatula-derived microsatellite markers in the interspecific cross Oryza glumaepatula × O. sativa. Hereditas. 2001;134:59–71. doi: 10.1111/j.1601-5223.2001.00059.x. [DOI] [PubMed] [Google Scholar]
- Brondani RPV, Zucchi MI, Brondani C, Rangel PHN, Borba TCO, Rangel PN, et al. Genetic structure of wild rice Oryza glumaepatula populations in three Brazilian biomes using microsatellite markers. Genetica. 2005;125:115–123. doi: 10.1007/s10709-005-4916-4. [DOI] [PubMed] [Google Scholar]
- Brown AHD. Genetic characterisation of plant mating system. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer; 1990. pp. 145–162. [Google Scholar]
- Buso GSC, Rangel PH, Ferreira ME. Analysis of genetic variability of South American wild rice populations (Oryza glumaepatula) with isozymes and RAPD markers. Molecular Ecology. 1998;7:107–117. [Google Scholar]
- Ennos RA, Clegg MT. Effect of populations substructuring on estimates of outcrossing rate in plant population. Heredity. 1982;48:283–292. [Google Scholar]
- Ferreira ME, Grattapaglia D. Introdução ao uso de marcadores moleculares em análise genética. 3rd edn. Brasília: Embrapa/Cenargen; 1998. [Google Scholar]
- Franceschinelli EV, Bawa KS. The effect of ecological factors on the mating system of a South American shrub species (Helicteres brevispira) Heredity. 2000;84:116–123. doi: 10.1046/j.1365-2540.2000.00636.x. [DOI] [PubMed] [Google Scholar]
- Frankham R. Genetics and conservation biology. Comptes Rendue Biologies. 2003;326:22–29. doi: 10.1016/s1631-0691(03)00023-4. [DOI] [PubMed] [Google Scholar]
- Ge S, Oliveira GX, Schaal BA, Gao LZ, Hong DY. RAPD variation within and between natural populations of the wild rice Oryza rufipogon from China and Brazil. Heredity. 1999;82:1–7. doi: 10.1046/j.1365-2540.1999.00516.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [http://www2.unil.ch/izea/softwares/fstat.html. ] [Google Scholar]
- Hall P, Walker S, Bawa KS. Effect on forest fragmentation on genetic diversity and mating system in a tropical tree, Pithecollobium elegans. Conservation Biology. 1996;10:757–768. [Google Scholar]
- Hamrick JL. Isozymes and the analysis of genetic structure in plant populations. In: Soltis DE, Soltis PS, editors. Isozymes in plant biology. London: Chapman and Hall; 1989. pp. 87–105. [Google Scholar]
- Hamrick JL, Godt MJW. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer; 1989. pp. 43–63. [Google Scholar]
- Hamrick JL, Linhart YB, Mitton JB. Relationship between life history characteristic and electrophoretically detectable genetic variation in plants. Annual Review of Ecology and Systematics. 1979;10:173–200. [Google Scholar]
- Hedrick PW. Conservation genetics: where are we now? Trends in Ecology and Evolution. 2001;16:629–636. [Google Scholar]
- Hoisington D, Khairallah M, Gonzalez-de-Leon D Applied Molecular Genetics Laboratory. Laboratory protocols: CYMMYT. Mexico: CYMMYT; 1994. [Google Scholar]
- Holsinger KE. Reproductive systems and evolution in vascular plants. Proceedings of the National Academy of Science of the USA. 2000;97:7037–7042. doi: 10.1073/pnas.97.13.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisz S, Vogler DW, Hanley KM. Context–dependent autonomous self-fertilization yields reproductive assurance and mixed mating. Nature. 2004;430:884–887. doi: 10.1038/nature02776. [DOI] [PubMed] [Google Scholar]
- Karasawa MMG. Análise da estrutura genética de populações e sistema reprodutivo de Oryza glumaepatula por meio de microssatélites. Brazil: University of São Paulo; 2005. PhD Thesis, Escola Superior de Agricultura ‘Luiz de Queiroz’. [Google Scholar]
- Morishima H, Barbier P. Mating system and genetic structure of natural populations in wild rice Oryza rufipogon. Plant Species Biology. 1990;5:31–39. [Google Scholar]
- Murawasky DA, Hamrick JL. The effect of the density of flowering individuals on the plant mating systems of nine tropical tree species. Heredity. 1991;67:167–174. [Google Scholar]
- Oliveira GCX. Padrões de variação fenotípica e ecologia de Oryzae selvagens da Amazônia. Brazil: University of São Paulo; 1993. MS Dissertation – Escola Superior de Agricultura ‘Luiz de Queiroz’. [Google Scholar]
- Oliveira GCX. Geographic distribution of wild Oryza species in Brazil. In: Morishima H, Martins PS, editors. Investigations of plant genetic resources in the Amazon basin with emphasis on the genus Oryza. Japan: Mishima; 1994. pp. 10–15. Report of 1992/93 Amazon Project. [Google Scholar]
- Oliveira GCX. A molecular phylogenetic analysis of Oryza L. based on chloroplast DNA sequences. Louis, EUA: University of Saint; 2002. PhD Thesis. [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology. 1995;4:347–354. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Richards AJ. Plant breeding systems. London: Chapman & Hall; 1997. [Google Scholar]
- Ritland K. Correlated matings in the partial selfer. Mimulus guttatus. Evolution. 1989;43:848–859. doi: 10.1111/j.1558-5646.1989.tb05182.x. [DOI] [PubMed] [Google Scholar]
- Ritland K. MLTR: multilocus mating system program. 1996. Version 1.1. Available from the author. [Google Scholar]
- Ritland K. Extensions of models for the estimation of mating systems using n independent loci. Heredity. 2002;88:221–228. doi: 10.1038/sj.hdy.6800029. [DOI] [PubMed] [Google Scholar]
- Ritland K, Jain SK. A model for estimation of outcrossing rate and gene frequencies using n-independent loci. Heredity. 1981;47:35–52. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA, Morishima H, Kadowaki K. Diversity in the Oryza genus. Current Opinion in Plant Biology. 2003;6:139–146. doi: 10.1016/s1369-5266(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Vogler DW, Kalisz S. Sex among flowers: the distribution of plant mating systems. Evolution. 2001;55(1):202–204. doi: 10.1111/j.0014-3820.2001.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II: methods for discrete population genetic data. Sunderland: Sinauer Associates, Inc; 1996. [Google Scholar]


