Abstract
Background and Aims
Myo-inositol-1l-phosphate synthase (MIPS) catalyses the conversion of d-glucose 6-phosphate to 1-l-myo-inositol-1-phosphate, the first and rate-limiting step in the biosynthesis of all inositol-containing compounds. Inositol phospholipids play a vital role in membrane trafficking and signalling pathways, auxin storage and transport, phytic acid biosynthesis, cell wall biosynthesis and production of stress-related molecules. In the present study, an MIPS cDNA from developing Passiflora edulis f. flavicarpa seeds was characterized and an investigation made into its spatial and differential expression, as well as changes in its transcription during exposure of growing plants to cold and heat stresses.
Methods
The MIPS-encoding gene was isolated by polymerase chain reaction (PCR) methods, and transcript levels were examined using semi-quantitative reverse transcription–PCR (RT–PCR) during seed development and in response to heat and cold stress. In addition, the copy number of the cloned PeMIPS1 gene in the genome of Passiflora edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea was determined by Southern blot analyses.
Key Results
A full-length cDNA clone of the PeMIPS1 from P. edulis was isolated and characterized. Southern blot analyses indicated that the genomic DNA might have diverse sequences of MIPS-encoding genes and one copy of the cloned PeMIPS1 gene in the genomes of P. edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea. RT–PCR expression analyses revealed the presence of PeMIPS1 transcripts in ovules, pollen grains and leaves, and during the seed developmental stages, where it peaked at 9 d after pollination. The PeMIPS1 gene is differentially regulated under cold and heat stress, presenting a light-responsive transcription.
Conclusions
Experimental data suggest that PeMIPS1 transcription plays an important role in the establishment of developmental programmes and during the response of plants to environmental changes. The PeMIPS1 is differentially transcribed during cold and heat stress, presenting a light response pattern, suggesting that it is important for environmental stress response.
Key words: Passiflora, passion fruit, MIPS, myo-inositol-1L-phosphate synthase, gene expression, abiotic stress
INTRODUCTION
Myo-inositol-1l-phosphate synthase (MIPS) (EC 5·5·1·4) and myo-inositol monophosphatase (IMP) (EC 3·1·3·25) are involved in the de novo inositol biosynthesis pathway (Loweus and Loweus, 1983; Loweus, 1990; Raboy, 2002). MIPS catalyses the conversion of d-glucose 6-phosphate to 1-l-myo-inositol-1-phosphate (MIP), the first and rate-limiting step in the biosynthesis of all inositol-containing compounds. Inositol is a negative feedback inhibitor of this conversion (Majumder et al., 1997). IMP catalyses the dephosphorylation of MIP to produce inositol. An alternative route to produce intracellular inositol pools is by recycling or scavenging inositol phosphates through the phosphatidylinositol signalling pathway and by myo-inositol polyphosphate breakdown (for a review, see Downes et al., 2005).
Inositol phospholipids play a vital role in membrane trafficking and signalling pathways, auxin storage and transport, phytic acid biosynthesis, cell wall biosynthesis and production of stress-related molecules (Loewus, 1990; Loewus and Murthy, 2000; Stevenson et al., 2000; Downes et al., 2005). The phosphorylated derivatives are essential as a phosphorus store and as a second messenger in signal transduction. In addition, myo-inositol polyphosphates participate in chromatin remodelling, gene expression and mRNA export (Odom et al., 2000; Shen et al., 2003). Recently, it was demonstrated that the lack of expression of GmMIPS in immature soybean seeds leads to an absence of accumulation of phytate globoids and seed development (Nunes et al., 2006).
The inositol phosphate biosynthesis pathway in developing seeds is poorly understood (Hitz et al., 2002; Shi et al., 2005). Hegeman et al. (2001) found detectable levels of MIPS protein mainly during seed development, indicating high level expression only in early cotyledonary stages. This fact suggests that the conversion of glucose 6-phosphate to MIP occurs earlier in seed development. It is also suggested that inositol and the o-methyl inositol esters function in salt tolerance by protecting cellular structures from reactive oxygen species such as hydrogen peroxide, and by controlling turgor pressure (for a review, see Loewus and Murthy, 2000). The introduction of PcINO1 (MIPS-coding gene from Porteresia coarctata) in tobacco, Oryza sativa and Brassica juncea generated transgenic lines presenting salt tolerance with concomitant increased inositol production (Majee et al., 2004; Das-Chatterjee et al., 2006). MIPS is the first enzyme in a metabolic pathway to d-pinitol, which is a cyclic sugar alcohol involved in the tolerance of drought stress that accumulates to higher concentrations in salt-tolerant legumes (Bohnert and Sheveleva, 1998; Bray et al., 2000).
MIPS coding sequences have been isolated and characterized from a number of plant species, such as Spirodela polyrrhiza (Smart and Fleming, 1993), Arabidopsis thaliana (Johnson, 1994), Citrus paradisii (Abu-Abied and Holland, 1994), Mesembryanthemum crystallinum (Ishitani et al., 1996), Nicotiana tabacum (Hara et al., 2000), Glycine max (Hegeman et al., 2001), Hordeum vulgare (Larson and Raboy, 1999), O. sativa (Yoshida, 1999), Zea mays (Larson and Raboy, 1999), P. coarctata (Majee et al. 2004) and Sesamum indicum (Chun et al., 2003). MIPS-encoding sequences represent multigene families in some plant species. Seven sequences were found in maize (Larson and Raboy, 1999), two in Arabidopsis (Johnson and Sussex, 1995), at least four in soybean (Hegeman et al., 2001) and at least three copies in S. indicum (Chun et al., 2003). The MIPS gene from M. crystallinum may also belong to a multigene family, as suggested by low-stringency Southern analysis (Ishitani et al., 1996). The multiple MIPS genes in plant species may be applied to attune its differential expression to specific physiological functions. The evolution of the MIPS protein/gene among the prokaryotes seems more diverse and complex than amongst the eukaryotes. However, conservation of a core catalytic structure among the MIPS proteins implies an essential function for the enzyme in cellular metabolism throughout the biological kingdom (Majumder et al., 2003).
In this study, a MIPS cDNA from developing Passiflora edulis seeds was characterized and its spatial and differential expression, as well as changes in its transcription during exposure of growing plants to cold and heat stresses were examined. Few studies have characterized MIPS gene expression in tropical species. The yellow passion fruit (P. edulis f. flavicarpa) is rich in ascorbic and malic acids, niacin, riboflavin, carotenoids and alkaloids (mainly harman), and the juice is slightly sedative. The fruit is commercially important in Australia, the USA (Hawaii and Southern Florida), South Africa and Brazil (Knight and Sauls, 2005). Despite its importance, there is no information about genes associated with seed development and that are responsive to environmental stress.
MATERIALS AND METHODS
Plant material
Leaves from P. edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea from the Amazon and Brazilian Cerrado (high-altitude savannah) were collected from the Germplasm Collection at Embrapa Cerrados (Planaltina, DF, Brazil). Fruits from P. edulis f. flavicarpa were collected from plants cultivated in the field. For Southern analyses, leaves were protected from light with aluminium foil for 2 d and harvested. Seeds used for obtaining plants for abiotic stress assays were purchased from Feltrin Sementes (Farroupilha, RS, Brazil). Plant tissues were harvested, frozen immediately in liquid nitrogen and stored at –80 °C until DNA or RNA extraction.
Cloning the PeMIPS1 gene
Total RNA was extracted using the RNAeasy kit (Qiagen, Valencia, CA, USA) from 200 mg of fresh tissue from immature embryos. The remaining genomic DNA was eliminated by DNase digestion of the RNA samples. An 8 µg aliquot of total RNA was used to produce total cDNA using the Superscript III kit (Invitrogen, Carlsbad, CA, USA). Degenerated primer MIPSPeF (5′-TTCATCAATGGCAGCCCHCARAACAC-3′) and MIPSPeR2 (5′-TCACTTGTAYTCCARRATCATGTTATT-3′) (H = A or C or T; R = G or A; Y = C or T) based on conserved regions of available plant MIPS gene were used to amplify an internal sequence from the PeMIPS1 gene. Reverse transcription–polymerase chain reacions (RT–PCRs) were carried out in a thermocycler (PTC-100, MJ Researcher, USA) in 50 µL of solution containing 40 ng of cDNA, 60 mM Tris–SO4 (pH 8·9), 18 mM (NH4)2SO4, 2 mM MgSO4, 250 nM of each dNTP, 200 nM of each primer and 5 U of Platinum Taq DNA Polymerase High Fidelity (Invitrogen). The mixture was treated at 95 °C (5 min) and subjected to 35 cycles of amplification (95 °C for 1 min, 55 °C for 1 min, 68 °C for 1 min) with a final elongation cycle of 5 min at 68 °C. The fragments were cloned into the pCR2·1 TOPO vector for PCR products (Invitrogen) and sequenced by using universal M13 and T7 primers on an automatic sequencer (ABI Prism 3700).
To obtain a full-length PeMIPS1 cDNA sequence, 5′- and 3′-rapid amplifications of cDNA ends (RACE)-PCR were carried out. Gene-specific primers were designed from the internal sequenced fragment and the cDNA end was amplified by using the 5′- and 3′-RACE System (Invitrogen, Carlsbad, CA, USA), using the reverse-specific nested primer MIPSPeRACER (5′-GAGGTGTACTCGTCCATAGC-3′) in combination with the GeneRacer™ 5′-Primer (Invitrogen), and the forward-specific primer MIPSPeRACEF (5′-GTGAACATCCTGACCACGTTG-3′) in combination with GeneRacer™ Oligo dT (Invitrogen). Nested PCRs were carried out as described above. PCR products were cloned into the pCR2·1 TOPO vector (Invitrogen) and sequenced.
Phylogenetics
The relationship between the PeMIPS1 and the MIPS1 genes from another 31 plant species was determined by aligning it with sequences available at GenBank (www.ncbi.nih.nlm.gov) and the TIGR Gene Indices (www.tigr.org). Only complete coding sequences were used for analysis. The alignment was performed using CLUSTAL W (Thompson et al., 1994). Phylogenetic analyses were carried out using the MEGA (Molecular Evolutionary Genetic Analysis) version 3·1 software program (Kumar et al., 2004). Mean frequencies of each nucleotide were similar (T = 23 %, C = 23 %, A = 27 %, G = 26 %), but a high transitions: transversions ratio was observed. Thus, to consider this unequal probability of the substitution types, the Kimura 2-parameter (Kimura, 1980) was used to compute distances between each pair of sequences. Phylogenetic trees were then constructed using the neighbour-joining algorithm (Saitou and Nei, 1987). Bootstrap values were computed using 1000 replicates to evaluate support for the groupings.
RT–PCR expression analysis
Developing seeds (3, 9, 15, 21 and 27 d after pollination), leaves, ovules, pollen grains, stems, leaf glands and petals were removed from plants cultivated under field conditions and used for total RNA extraction as described above. For abiotic stress assays, plants 8 weeks after germination maintained at room temperature and light intensity 10 µmol m−2 s−1 were transferred to a growth chamber (Conviron, Winnipeg, Canada) at 5, 12, 27 and 37 °C, 85 % relative humidity, under darkness or a light intensity of 200 µmol m−2 s−1 for a period of 8 and 16 h. Leaves were collected for total RNA extraction as described. Total RNA was used to produce cDNA using the reverse transcriptase Superscript III (Invitrogen), according to the protocol suggested by the manufacturer. PCRs were carried out as described above, except that Taq DNA polymerase (Invitrogen) was used and that 25 ng of cDNA was used as a template, in reactions with 20 cycles of amplification. The number of amplification cycles was previously optimized in order to stop the reaction at the exponential stage, ensuring that amplification was semi-quantitative. Primers MIPSPeF and MIPSPeR2 were used to amplify a 693 bp fragment from the PeMIPS sequences. As an internal control, primers EF1F (5′-TGTTGCTGTTAAGGATTTGAAGCG-3′) and EF1R (5′-AACAGTTTGACGCATGTCCCTAAC-3′) were utilized to amplify 358 bp within the passion fruit housekeeping gene, PeEFα, elongation factor EF-1α (GenBank accession nos DQ447160 and DQ447161; cloned and sequenced in this work). PCRs with total RNA presented no amplified fragments, while PCR with genomic DNA presented a higher fragment for both PeMIPS1 and EF-1α genes, suggesting the presence of intron(s) within the sequence. Fragments of the MIPS-encoding sequence amplified from expressing plant tissues were cloned into the pGEMT-Easy and sequenced. Experiments were repeated three times.
Genomic Southern blot analysis
Genomic DNA was isolated from leaves using the DNeasy Plant Mini Kit (Qiagen). Southern blotting was carried out as described (Sambrook and Russell, 2001). Genomic DNA (15 µg) was digested with EcoRI, HindIII and XhoI, separated on a 1 % agarose gel and transferred to a nylon membrane (Hybond N+, Amersham Pharmacia Biotech, Buckinghamshire, UK). Hybridization was carried out at 55 °C using the 693 bp internal fragment from the PeMIPS1 gene, labelled with [α-32P]dCTP (3000 Ci mol−1) using a random primer DNA labelling kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Membranes were washed twice in 2 × SSC (1 × SSC = 150 mM NaCl and 15 mM sodium citrate), 0·1 % (w/v) SDS at room temperature for 15 min and once in 1 × SSC, 0·1 % (w/v) SDS at 65 °C for 15 min. Stringent washing was carried out using 0·1 ×SSC, 0·1 % (w/v) SDS at 65 °C for 15 min prior to exposing the membrane for autoradiography.
RESULTS
Isolation of a MIPS-encoding gene from P. edulis
A full-length MIPS cDNA of 1951 bp was isolated from developing passion fruit seeds and entered into the NCBI GenBank database with the accession number DQ489558. The cDNA contains one open reading frame of 1533 bp encoding 510 amino acids. In the 3′-non-translated region, the putative polyadenylation signal sequences (AATAA and ATTAA) were found upstream of the poly(A) tail. The nucleotide sequence of the PeMIPS1 gene-coding region was compared with sequences of MIPS genes from 31 plant species available in the GenBank and TIGR databases and showed a high similarity (from 74 to 86 %). The deduced amino acid sequence was used to compare the amino acid composition of the PeMIPS1 polypeptide with those of other plant MIPS. The analysis revealed a high similarity of 85–93 %. The TargetP 1·1 (Emanuelsson et al., 2000) and ChloroP (Emanuelsson et al., 1999) program algorithms predicted no signal, chloroplast transit or mitochondrial targeting peptides in the N-terminal region of the PeMIPS1.
The PeMIPS1 polypeptide contains four highly conserved motifs found in other plant species: GWGGNNG (domain 1), VLWTANTER (domain 2), NGSPQNTFVPGL (domain 3) and SYNHLGNNDG (domain 4). These motifs are also found in animal and microorganism species, such as Anopheles gambiae (GenBank accession no. XM320685), Drosophila melanogaster (GenBank accession no. AF071103), Homo sapiens (GenBank accession no. AF207640), Xenopus laevis (GenBank accession no. BC077437), Aspergillus fumigatus (GenBank accession no. XM744144), Pichia pastoris (GenBank accession no. AF078915), Entamoeba histolytica (GenBank accession no. Y11270) and Leishmania amazonensis (GenBank accession no. U91965).
Phylogenetic analysis divided MIPS genes into clusters in agreement with taxonomic differentiation (Fig. 1). The rootless phylogenetic tree obtained in the phylogenetic analysis of MIPS genes showed four main branches. The first includes the Pinnus sp, the only gymnosperm species present. The second main branch encompasses the sequences of monocots. The third branch is formed by MIPS from S. polyrrhiza, an aquatic plant. The fourth main branch comprises the sequences of dicotyledonous species. Surprisingly, Triticum aestivum rooted with dicotyledonous species. The branch of the dicotyledonous species was split into six conspicuous observable sub-branches. The main sub-branch is formed by species from the order Fabales (G. max, Tripolium pratense, Lotus japonicus, Medicago truncatula and Phaseolus vulgaris). Passiflora edulis MIPS rooted with Populus sp.; both species are from the order Malpighiales. The other sub-branches are formed by the orders Solanales (Solanum tuberosum, N. tabacum and Lycopersicon esculentum), Lamiales (S. indicum and Avicennia marina), Brassicales (A. thaliana and Brassica napus) and Caryophylales (M. crystallinum and Suaeda maritima) (Fig. 1).
Fig. 1.
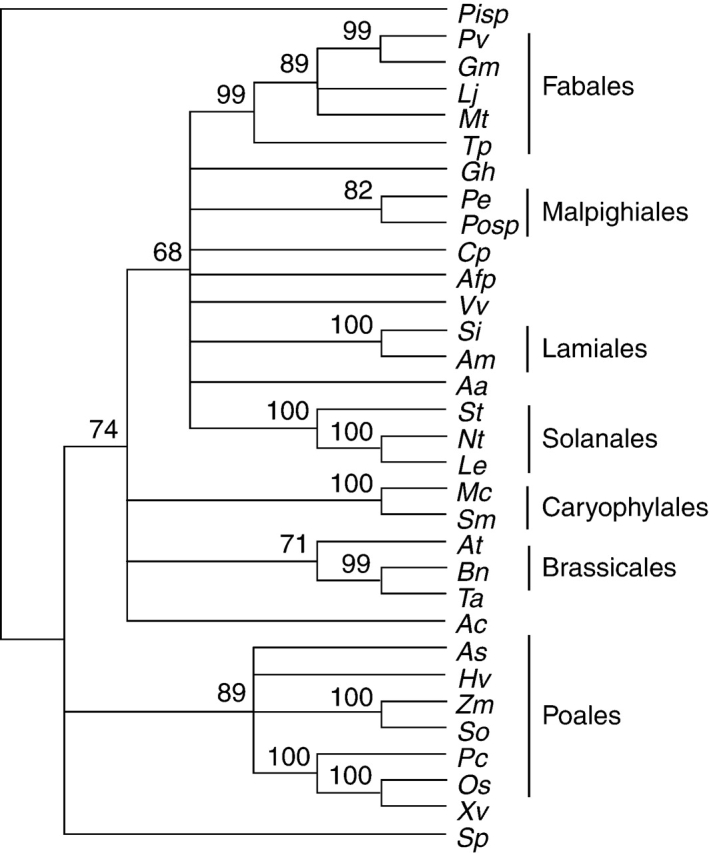
Bootstrap consensus phylogenetic tree of myo-inositol-1-phosphate synthase (MIPS) genes from plant species. The tree includes sequences of MIPS available in GenBank and TIGR databases. Passiflora edulis is highlight in bold, as Pe. Abbreviation of the species and accession number of each sequence are: Aa, Actinidia arguta (AY005128); Ac, Allium cepa (TC248); Afp, Aquilegia formosa × pubescens (TC14836); At, Arabidopsis thaliana (U04876); As, Avena sativa (AB059557); Am, Avicennia marina (AY028259); Bn, Brassica napus (U66307); Cp, Citrus paradise (Z32632); Gm, Glycine max (AY382834); Gh, Gorssypium hirsutum (TC27410); Hv, Hordeum vulgare (AF056325); Lj, Lotus japonicus (TC8275); Le, Lycopersicon esculentum (TC154132); Mt, Medicago trunculata (TC93972); Mc, Mesembryanthemum crystallinum (U32511); Nt, Nicotiana tabacum (AB059557); Os, Oryza sativa (AB012107); Pv, Phaseolus vulgaris (AM048843); Pisp, Pinus sp. (TC64990); Posp, Populus sp. (TC19238); Pc, Porteresia coarctata (AF412340); So, Saccharum officinarum (TC65413); Si, Sesamum indicum (AF284065); St, Solanum tuberosum (TC112573); Sp, Spirodela polyrrhiza (Z11693); Sm, Suaeda maritima (AF433879); Tp, Tripolium pratense (AB236831); Ta, Triticum aestivum (AF120148); Vv, Vitis vinifera (TC45187); Xv, Xerophyta viscosa (AY323824); Zm, Zea mays (AF56326). Bootstrap values >50 % are displayed on the nodes.
Southern analysis of MIPS-encoding genes in five species of Passiflora
Southern blot analyses were performed to determine the sequences related to the PeMIPS1 clone in the Passiflora species genome. Genomic DNA was digested with EcoRI, HindIII and XhoI separately and probed with the 693 bp internal fragment from the PeMIPS1 gene. At low stringency, a large number of strong bands were visible (Fig. 2A), which disappeared when the blot was washed at high stringency (Fig. 2B). Digestions with HindIII presented a distinct pattern among species, showing a polymorphism within the enzyme site. A single hybridizing band was observed with XhoI digestions (Fig. 2B). Results obtained under low-stringency conditions indicated that there might be diverse sequences of MIPS-encoding genes in genomic DNA. The strong signal in one band, approx. 3 kb, obtained under high-stringency conditions after digestion with XhoI indicated that the genome of P. edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea may have one copy of the cloned PeMIPS1 gene.
Fig. 2.
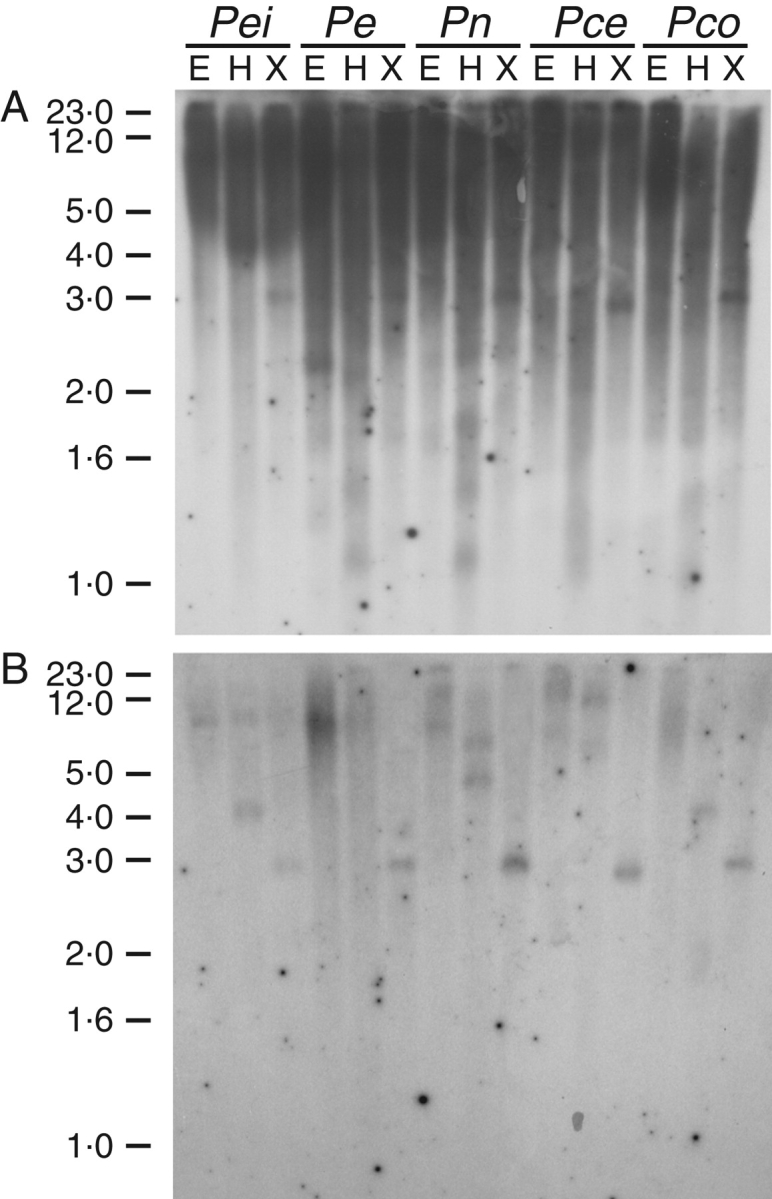
Southern blot analysis of the MIPS gene in the genome of Passiflora species. The genomes of P. edulis (Pe), P. eichleriana (Pei), P. caerulea (Pca), P. nitida (Pn) and P. coccinea (Pco) were digested with EcoRI (E), HindIII (H) and XhoI (X). The membrane was probed with the 693 bp internal fragment from the PeMIPS1 gene at low (A) and high stringency (B) of washing conditions.
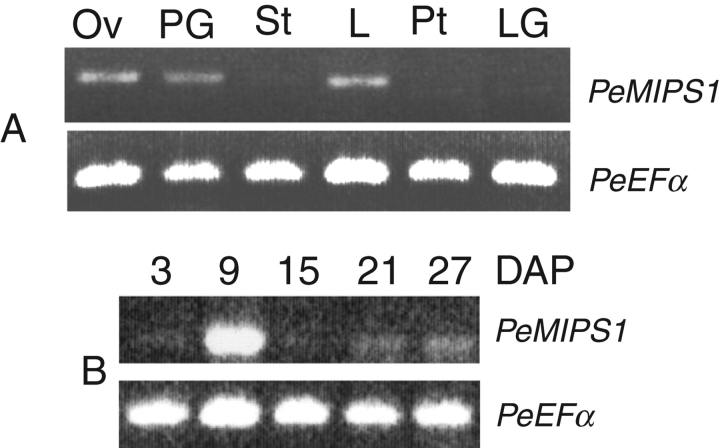
Levels of PeMIPS1 transcripts in organs and developing seeds of P. edulis
RT–PCR expression analyses were carried out to detect endogenous PeMIPS1 transcripts in different P. edulis organs and developing seeds. The results revealed the presence of PeMIPS1 transcripts in ovules, pollen grains and leaves, and no signal could be observed in stems, petals or leaf gland (Fig. 3A). PeMIPS1 transcripts were observed in all seed developmental stages analysed. However, the transcription peaked in seeds at 9 d after pollination (Fig. 3B). As development progressed, MIPS transcript levels decreased. A similar level of the PeEFα (elongation factor EF-1α) housekeeping gene was observed for all RT–PCR amplifications. The 693 bp fragment that corresponds to the PeMIPS1-encoding sequence amplified from expressing tissues was sequenced and revealed 100 % identity with the cloned PeMIPS1 gene.
Fig. 3.
Differential transcription of the PeMIPS1 gene from passion fruit (P. edulis) in different organs (A) and developing seeds (B) 3, 9, 15, 21 and 27 days after pollination (DAP). Ov, ovules; PG, pollen grains; St, stem; L, leaves; Pt, petals; LG, leaf gland. The upper bands are consistent with the expected fragment amplified from the PeMIPS1 gene and the lower band corresponds to transcripts from the PeEFα gene (elongation factor EF-1α; internal control).
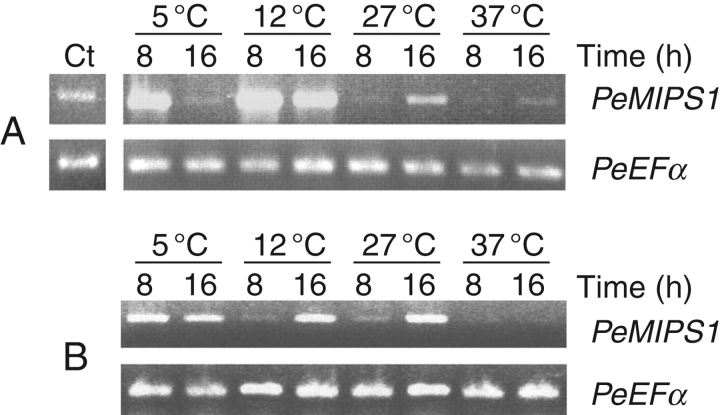
Levels of PeMIPS1 transcripts in response to cold and heat stress under light and dark conditions
The effect of temperature and light on the transcript levels of the endogenous PeMIPS1 gene was studied in growing plants (8 weeks after germination). The results revealed that under dark conditions, PeMIPS1 transcripts were upregulated during a short period under cold stress (5 °C), whereas at higher temperatures (27 and 37 °C), transcription levels increased slightly after 16 h (Fig. 4A). The highest expression levels were observed at 12 °C at 8 and 16 h, with a slight decrease at 16 h. Plants exposed to 5 °C showed PeMIPS induction after 8 h of cold stress but a decrease at 16 h. Enhanced transcription of the PeMIPS1 gene was also observed after 16 h at 27 °C but not at 37 °C, which showed the lowest amounts of transcripts compared with control (Fig. 4A). Under continuous light conditions (Fig. 4B), enhanced PeMIPS1 transcription was observed after 16 h at 12 and 27 °C, when compared with the transcriptional level observed after 8 h (Fig. 4B). PeMIPS1 transcripts were quasi-undetectable at 37 °C.
Fig. 4.
Effects of temperature and light on transcription of the PeMIPS1 gene in leaves of passion fruit plants 8 weeks after germination. Plants were exposed for 8 and 16 h at different temperatures in the dark (A) and under a continuous light intensity of 200 µmol m−2 s−1 (B). A plant before treatment (maintained at room temperature and a light intensity of 10 µmol m−2 s−1) is indicated by ‘Ct’. The upper bands are consistent with the expected fragment amplified from the PeMIPS1 gene and the lower band corresponds to transcripts from the PeEFα gene (elongation factor EF-1α; internal control).
DISCUSSION
The phylogenetic tree constructed on the basis of multiple alignments of MIPS genes shows clear segregation of these genes into monocotyledonous and dicotyledonous. It is possible that the divergence of MIPS genes happened after the divergence of monocotyledons and dicotyledons. Alignment of the DNA sequence of the gene encoding plant MIPS revealed remarkable evolutionary conservation. The presence of conserved sequences and conservative changes observed in a wide range of organisms indicates the central role that this enzyme plays in biological systems (Majumder et al., 2003). MIPS catalyses the synthesis of MIP, which is a precursor to compounds connected to essential cellular functions, such as phosphorus storage, signal transduction, actin remodelling, membrane trafficking, stress protection, hormonal homoeostasis and cell wall biosynthesis (Loewus and Murthy, 2000; Stevenson et al., 2000; Downes et al., 2005).
Genomic Southern blot suggests that several MIPS, MIPS-related genes or pseudogenes might be present in the five studied Passiflora species. However, only one copy of the cloned PeMIPS1 gene was found in the genome of P. edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea. Although several copies of the MIPS-encoding genes have been observed in the plant genome, the exact copy number for each gene from MIPS family members has not been determined for most species. In rice, genomic Southern analysis suggested that a single gene encoding the RINO1 gene was present in the genome (Yoshida et al., 1999). Seven sequences were found in maize (Larson and Raboy, 1999), two in Arabidopsis (Johnson and Sussex, 1995), two in common ice (Ishitani et al., 1996) and at least four in soybean (Hegeman et al., 2001). The multiple MIPS genes in crop plants may be used to attune differential MIPS expression to specific physiological functions.
Transcripts of the PeMIPS1 gene were detected in ovules, pollen grains, developing seeds and leaves of plants cultivated in field conditions. However, no signal was observed in stems, petals or leaf gland. This demonstrated that the expression of the PeMIPS1 gene is organ specific and such differential regulation would coordinate inositol metabolism with cellular growth (Ishitani et al., 1996; Majumder et al., 1997). In soybean, steady-state RNA levels of the GmMIPS1 gene were higher in developing seeds than in other tissues, including flowers, leaves, germinating cotyledons and somatic embryos (Hegeman et al., 2001). In silico northern analyses generated from the soybean expressed sequence tag (EST) database predict an expression pattern of four highly similar MIPS-coding sequences. ESTs from GmMIPS3 were only observed in leaf and bud libraries, while ESTs from GmMIPS1 were only observed in immature cotyledons (Hegeman et al., 2001). In addition, data from 15 EST libraries showed that GmMIPS1 is the preferred gene of the seed, although it is expressed in other tissue types, while GmMIPS2 is expressed in many tissues but not in developing seeds (Hitz et al., 2002). This suggested that the expression of distinct MIPS genes is spatially controlled by regulatory sequences. In S. indicum, SeMIPS1 transcription was observed in leaves, stem, root and developing seeds (Chun et al., 2003). In rice, transcripts of an MIPS-encoding gene (RINO1) accumulated at high levels in developing seeds, but not in leaves, roots or flowers (Yoshida et al., 1999). Our data suggested that the MIPS-encoding sequences might present a distinct pattern when compared with other plants. However, more studies should be conducted to associate MIPS gene transcription with specific environmental conditions.
A marked peak in MIPS RNA level was observed 9 d after pollination, showing that the gene is regulated at the transcriptional level during seed development. Similar results were observed in soybean. GmMIPS transcript was observed in cotyledons at the earliest developmental stages, with a peak in 2–4 mm seeds (Hegeman et al., 2001). In contrast, no recognizable differences in levels of SeMIPS1 gene transcription were found between developing stages of S. indicum seeds (Chun et al., 2003).
The results suggest that conversion of glucose-6-phosphate to MIP occurs earlier in P. edulis seed development. This expression pattern of PeMIPS1 may be correlated with the accumulation of phytate, which is a storage reserve of phosphorus in seeds (Loewus and Murthy, 2000). However, the absence of seed development was recently observed in transgenic soybean plants silencing the GmMIPS1 gene (Nunes et al., 2006). This suggests that MIPS expression during early stages of seed development is closely related to the essentiality of inositol biosynthesis for several biochemical pathways in plants (Downes et al., 2005), rather than phytate accumulation. In this work, all sequenced PCR-amplified fragments presented 100 % identity with the cloned PeMIPS1 gene. Passiflora edulis may have other MIPS-coding genes, and sequences are highly similar; because of this and the fact that sequence-specific primers were not used for RT–PCR analysis, it cannot be definitively concluded that the PeMIPS1 gene is the only MIPS-encoding gene expressed in the organs studied.
RT–PCR analyses were carried out to analyse the effect of cold and heat stress on the transcript levels of PeMIPS1. The results revealed that PeMIPS1 transcripts were upregulated, after a short period (8 h) under cold stress (5 °C), which decreased after 16 h (Fig. 4A). It has been shown that sugars and abscisic acid (ABA) have similar effects on MIPS gene transcriptional levels (Yoshida et al., 2002). These molecules induced transcript accumulation of the MIPS-encoding gene (RINO1) in rice. mRNA accumulation revealed that the highest accumulation level occurred 2–4 h after initiation of the combined treatment, and the signal declined thereafter (Yoshida et al., 2002). In M. crystallinum, salinity stress was observed to induce the upregulation of MIPS mRNA expression 5-fold and free inositol accumulation approx. 10-fold (Ishitani et al., 1996). Interestingly, the present results were similar to those observed in Citrus sinensis submitted to herbivory from xylem-feeding leafhopper and to mechanical damage. Both treatments induced a PeMIPS1 transcript that peaked after 48 h of treatment and was only slightly increased after 96 h (Mozoruk et al., 2006). These results agree with the documented fact that rapid changes in inositol phosphates have been observed in different plant systems with many different stimuli, including osmotic and cold shock, light and biotic elicitors (Perera et al., 2006). Many of the genes regulated by biotic and abiotic factors can also be induced by the phytohormone ABA or by osmotic stress treatment (Ishitani et al., 1997; Seki et al., 2002; Li et al., 2006). Indeed, high and low non-freezing temperatures and osmotic stresses appear to have several features in common.
Under continuous light conditions, PeMIPS1 transcripts were upregulated at most temperatures tested, except at 37 °C (Fig. 4B). In potato, it was observed that light drastically increased StIPS-1 (an MIPS-encoding gene) transcript level in leaves (Keller et al., 1998). It appears that a light- and salt-mediated interplay of protease and kinase system regulates the processing and activation of the chloroplast inositol synthases genes in higher plants (Hait et al., 2002). There are few studies on the possible importance of inositol biosynthesis in light-dependent pathways. A plastid-located pathway from inositols to d-glucuronate and further to l-ascorbate has been postulated (Loewus, 1988). Apparently, l-ascorbate supports chloroplast integrity under stress conditions which require protection against radical oxygen species. Nelson et al. (1998) suggested that myo-inositol acts to facilitate sodium sequestration, protect photosynthesis and sustain membrane biosynthesis. It seems that in the common ice, photosynthesis can simultaneously control root growth via inositol supply and osmotic stress protection via ononitol and pinitol availability in the roots. The sequence analysis of the predicted PeMIPS1 indicated that the N-terminal region contains no signal, chloroplast transit or mitochondrial targeting peptides. It suggests that PeMIPS1 encodes a cytosolic rather than an organelle-addressed MIPS protein. However, the absence of a convincing transit peptide in PeMIPS1 does not preclude its targeting to a plastidic site for inositol synthesis. A MIPS from M. crystallinum that does not contain a transit peptide for chloroplast import presented enhanced activity only in chloroplasts from light-grown, salt-tolerant rice plants (RayChaudhuri and Majumder, 1996). Moreover, experimental data have suggested that a MIPS enzyme from P. vulgaris, which does not have a recognizable transit peptide, is also present in plasma membranes, plastids, mitochondria, endoplasmic reticulum, nuclei and cell walls (Lackey et al., 2003). Additionally, the program PSORT (Prediction of Protein Sorting Signals and Localization Sites in Amino Acid Sequences) predicted a conserved transmembrane motif (CEDSLLAAPIILDLVLLAELSTR), located approx. 68 amino acids from the C-terminus in both P. vulgaris and P. edulis MIPS.
CONCLUSIONS
The experimental data presented here suggest important roles for MIPS not only in basic metabolism, but also in the establishment of developmental programmes and during the response of plants to environmental changes. The results demonstrated that a MIPS-encoding gene cloned from P. edulis, called PeMIPS1, is preferentially expressed in leaves, pollen grains, ovules and seeds 9 d after pollination. The PeMIPS1 is differentially transcribed during cold and heat stress, presenting a light response pattern, suggesting that it is important for the environmental stress response. There is correlation of these results with ecological adaptation of passion fruit, a typical species adapted to tropical and sub-tropical environments that endures winter chills for a short period without injury, but does not tolerate severe or prolonged freezing. Our knowledge of stress proteins/genes is still far from complete, and there are lacunae with respect to behaviour in tropical plant species. Consequently, other stress-responsive genes should be studied, cloned and characterized. Additionally, studies are required for localization of PeMIPS1 in subcellular compartments of distinct plant tissues, as well as MIPS silencing in transgenic plants, to develop a greater understanding of its role during seed development.
ACKNOWLEDGEMENTS
We are grateful to Dr Marcelo Fidelis Braga (Embrapa Cerrados, Brazil) and Fundação Zoobotânica do Distrito Federal (Brasília, Brazil) for providing some of the samples used in this study, and to Professor Dr José Tadeu Abreu de Oliveira (Universidade Federal do Ceará, Brazil) for critical reading of the manuscript.
LITERATURE CITED
- Abu-Abied M, Holland D. The gene c-ino1 from Citrus paradisi is highly homologous to tur1 and ino1 from yeast and Spirodela encoding for myo-inositol phosphate synthase. Plant Physiology. 1994;106:1689. doi: 10.1104/pp.106.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert HJ, Sheveleva E. Plant stress adaptations – making metabolism move. Current Opinion in Plant Biology. 1998;1:267–274. doi: 10.1016/s1369-5266(98)80115-5. [DOI] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E. Response to abiotic stress. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and molecular biology of plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 1158–1203. [Google Scholar]
- Chun JA, Jin UH, Lee JW, Yi YB, Hyung NI, Kang MH. Isolation and characterization of a myo-inositol 1-phosphate synthase cDNA from developing sesame (Sesamum indicum L.) seeds: functional and differential expression, and salt-induced transcription during germination. Planta. 2003;216:874–880. doi: 10.1007/s00425-002-0940-0. [DOI] [PubMed] [Google Scholar]
- Das-Chatterjee A, Goswami L, Maitra S, Dastidar KG, Ray S, Majumder AL. Introgression of a novel salt-tolerant l-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Letters. 2006;580:3980–3988. doi: 10.1016/j.febslet.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends in Cell Biology. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Hait NC, RayChaudhury A, Das A, Bhattacharyya S, Majumder AL. Processing and activation of chloroplast l-myo-inositol 1-phosphate synthase from Oryza sativa requires signals from both light and salt. Plant Science. 2002;162:559–568. [Google Scholar]
- Hara K, Yagi M, Koizumi N, Kusano T, Sano H. Screening of wound-responsive genes identifies an immediate-early expressed gene encoding a highly charged protein in mechanically wounded tobacco plants. Plant and Cell Physiology. 2000;41:684–691. doi: 10.1093/pcp/41.6.684. [DOI] [PubMed] [Google Scholar]
- Hegeman CE, Good LL, Grabau EA. Expression of d-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiology. 2001;125:1941–1948. doi: 10.1104/pp.125.4.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz WD, Carlson TJ, Kerr PS, Sebastian SA. Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiology. 2002;128:650–660. doi: 10.1104/pp.010585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Majumder AL, Bornhouser A, Michalowski CB, Jensen RG, Bohnert HJ. Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant Journal. 1996;9:537–548. doi: 10.1046/j.1365-313x.1996.09040537.x. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhul J-K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD. The Arabidopsis thaliana myo-inositol 1-phosphate synthase (EC 5·5·1·4) Plant Physiology. 1994;105:1023–1024. doi: 10.1104/pp.105.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Sussex IM. 1-l-myo-inositol 1-phosphate synthase from. Arabidopsis thaliana. Plant Physiology. 1995;107:613–619. doi: 10.1104/pp.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Brearley CA, Trethewey RN, Müller-Röber B. Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1d-myo-inositol 3-phosphate synthase. Plant Journal. 1998;16:403–410. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Knight RJ, Sauls JW. The passion fruit. University of Florida IFAS Extension HS60; 2005. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lackey KH, Pope PM, Johnson MD. Expression of 1l-myo-inositol-1-phosphate synthase in organelles. Plant Physiology. 2003;132:2240–2247. doi: 10.1104/pp.103.020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SR, Raboy V. Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: correspondence with a low phytic acid mutation. Theoretical and Applied Genetics. 1999;99:27–36. [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Research. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA. Ascorbic acid and its metabolic products. In: Preiss J, editor. The biochemistry of plants. New York: Academic Press; 1988. pp. 85–107. [Google Scholar]
- Loewus FA. Inositol biosynthesis. In: Morré DJ, editor. Inositol metabolism in plants. New York: Wiley-Liss; 1990. pp. 13–19. [Google Scholar]
- Loewus FA, Loewus MW. Myo-inositol: its biosynthesis and metabolism. Annual Review of Plant Physiology. 1983;34:137–161. [Google Scholar]
- Loewus FA, Murthy PPN. Myo-inositol metabolism in plants. Plant Science. 2000;150:1–19. [Google Scholar]
- Majee M, Maitra S, Dastidar KG, Pattnaik S, Chatterjee A, Hait NC. A novel salt-tolerant l-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice. Journal of Biological Chemistry. 2004;279:28539–28552. doi: 10.1074/jbc.M310138200. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Johnson MD, Henry SA. l-myo-inositol-1-phosphate synthase. Biochimica et Biophysica Acta. 1997;1348:245–256. doi: 10.1016/s0005-2760(97)00122-7. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Chatterjee A, Dastidar KG, Majee M. Diversification and evolution of l-myo-inositol 1-phosphate synthase. FEBS Letters. 2003;553:3–10. doi: 10.1016/s0014-5793(03)00974-8. [DOI] [PubMed] [Google Scholar]
- Mozoruk J, Hunnicutt LE, Cave RD, Hunter WB, Bausher MG. Profiling transcriptional changes in Citrus sinensis (L.) Osbeck challenged by herbivory from the xylem-feeding leafhopper Homalodisca coagulata (Say) by cDNA macroarray analysis. Plant Science. 2006;170:1068–1080. [Google Scholar]
- Nelson DE, Rammesmayer G, Bohnert HJ. Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell. 1998;10:753–764. doi: 10.1105/tpc.10.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes ACS, Vianna GR, Cuneo F, Amaya-Farfán J, Capdeville G, Rech EL, Aragao FJL. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta. 2006;224:125–132. doi: 10.1007/s00425-005-0201-0. [DOI] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Brady S, Muday GK, Boss WF. A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiology. 2006;140:746–760. doi: 10.1104/pp.105.075119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V. Progress in breeding low phytate crops. Journal of Nutrition. 2002;132:503–505. doi: 10.1093/jn/132.3.503S. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri A, Majumder AL. Salinity-induced enhancement of l-myo-inositol 1-phosphate synthase in rice (Oryza sativa L.) Plant, Cell and Environment. 1996;19:1437–1442. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DE, Harp T. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant Journal. 2005;42:708–719. doi: 10.1111/j.1365-313X.2005.02412.x. [DOI] [PubMed] [Google Scholar]
- Smart CC, Fleming AJ. A plant gene with homology to d-myo-inositol-3-phosphate synthase is rapidly and spatially upregulated during an abscisic-acid-induced morphogenic response in. Spirodela polyrrhiza. Plant Journal. 1993;4:279–293. doi: 10.1046/j.1365-313x.1993.04020279.x. [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF. Inositol signaling and plant growth. Trends in Plant Science. 2000;5:252–258. doi: 10.1016/s1360-1385(00)01652-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida KT, Wada T, Koyama H, Mizobuchi-Fukuoka R, Naito S. Temporal and spatial patterns of accumulation of the transcript of myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiology. 1999;119:65–72. doi: 10.1104/pp.119.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida KT, Fujiwara T, Naito S. The synergistic effects of sugar and abscisic acid on myo-inositol-1-phosphate synthase expression. Physiologia Plantarum. 2002;114:581–587. doi: 10.1034/j.1399-3054.2002.1140411.x. [DOI] [PubMed] [Google Scholar]