Abstract
Objective
Familial autosomal dominant frontotemporal dementia with ubiquitin-positive, tau-negative inclusions in the brain linked to 17q21-22 recently has been reported to carry null mutations in the progranulin gene (PGRN). Hereditary dysphasic disinhibition dementia (HDDD) is a frontotemporal dementia with prominent changes in behavior and language deficits. A previous study found significant linkage to chromosome 17 in a HDDD family (HDDD2), but no mutation in the MAPT gene. Longitudinal follow-up has enabled us to identify new cases and to further characterize the dementia in this family. The goals of this study were to develop research criteria to classify the different clinical expressions of dementia observed in this large kindred, to identify the causal mutation in affected individuals and correlate this with phenotypic characteristics in this pedigree, and to assess the neuropathological characteristics using immunohistochemical techniques.
Methods
In this study we describe a detailed clinical, pathological and mutation analysis of the HDDD2 kindred.
Results
Neuropathologically, HDDD2 represents a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions (FTLD-U). We developed research classification criteria and identified three distinct diagnostic thresholds, which helped localize the disease locus. The chromosomal region with the strongest evidence of linkage lies within the minimum critical region for FTLD-U. Sequencing of each exon of the PGRN gene led to the identification of a novel missense mutation, Ala-9 Asp, within the signal peptide.
Interpretation
HDDD2 is an FTLD-U caused by a missense mutation in the PGRN gene that cosegregates with the disease and with the disease haplotype in at-risk individuals. This mutation is the first reported pathogenic missense mutation in the signal peptide of the PGRN gene causing FTLD-U. In light of the previous reports of null mutations and its position in the gene, two possible pathological mechanisms are proposed: (1) the protein may accumulate within the endoplasmic reticulum due to inefficient secretion; and (2) mutant RNA may have a lower expression because of degradation via nonsense-mediated decay.
Frontotemporal dementia (FTD) is characterized by early behavioral and personality changes, language deterioration, and later in the course of the disease, dementia and parkinsonism.1 FTD shows familial aggregation with a family history of dementia present in 41% of FTD cases.2 Late pyramidal and extrapyramidal signs are present in some cases. Neuropathologically, cases of sporadic and familial frontotemporal lobar degeneration (FTLD) show stereotypical features: atrophy of the frontal and temporal lobes with variable involvement of the basal ganglia that is characterized by neuronal loss, status spongiosus, and reactive astrocytosis.3,4 FTLD may be caused by a wide spectrum of disorders including those characterized by abnormal glial and neuronal inclusions of aggregated microtubule-associated protein tau, diseases with ubiquitin inclusions, and a minority that do not show any detectable abnormal cellular aggregates.1 Some of the FTLD disorders, including Pick's disease, progressive supranuclear palsy, corticobasal degeneration, argyrophilic grain diseases, and FTD with parkinsonism linked to chromosome 17, collectively are referred to as taupathies. The majority of FTLD, however, is not characterized by tauopathies, but instead contain other abnormal protein aggregates. These disorders include FTLD with ubiquitin-positive, tau-negative inclusions (FTLD-U), also called FTLD-motor neuron disease type,1 neuronal intermediate filament inclusion disease,5 and inclusion body myopathy with Paget's disease and frontotemporal dementia.6 Some FTLD cases have no discernible inclusions and are called dementia lacking distinctive histopathology.7
Initial genetic studies of familial FTD with evidence of linkage at 17q21 and tau-immunoreactive inclusions identified mutations in the gene encoding microtubule-associated protein tau (MAPT).8 However, several FTD families with significant linkage (logarithm of odds [LOD] score > 3) to the same region did not show any mutations in the coding and flanking intronic sequences of the tau gene.9–11 Subsequent neuropathological studies using ubiquitin immunohisto-chemistry identified these cases as FTLD-U.11 Recently, null mutations in the progranulin gene (PGRN) have been described in several FTLD-U kindreds showing linkage to chromosome 17.12,13
PGRN was first characterized as a putative family of growth factors constitutively expressed in several tissues.14 PGRN is implicated in cell proliferation, wound repair, and anchorage independent growth.15 Interestingly, when overexpressed, it causes tumorigenesis.15 In brain, PGRN is expressed in Purkinje cells, pyramidal cells of the hippocampus, and some cerebral cortical neurons.16
We have previously described HDDD2 as an autosomal dominant kindred with prominent dysphasia and behavioral disturbances and characterized neuropathologically, before ubiquitin immunohistochemistry, as FTLD.17,18 Previously, no mutation in any of the known dementia-causing genes was identified in this kindred.3,18 Since 1993, we have performed longitudinal assessments in consenting family members to further characterize the clinical and neuropathological characteristics of HDDD2. In addition to the new neuropathological findings reported here, we have also reevaluated linkage data in a larger series of family members and undertaken mutation analysis of the recently identified FTLD-U gene, progranulin.
Subjects and Methods
Subjects
All procedures reported were approved by the Washington University School of Medicine Human Studies Committee. Family members older than 18 years were included after informed consent was obtained. All individuals were interviewed by an experienced research clinician or nurse, using a modified version of the Family History Interview developed by the Consortium to Establish a Registry for Alzheimer's Disease (CERAD).19 For individuals with neurological symptoms, the visit included a clinical and neurological examination and a consensus history derived from the individual or nearest relatives, or both. When possible, available medical records also were analyzed. HDDD2 is a large, multiply affected, multigeneration kindred with FTLD-U segregating in one branch of the pedigree. Several individuals both within the FTLD-U branch and also in the extended kindred exhibit different clinical presentations including dementia of the Alzheimer's type (DAT), rather than classic dysphasia. This variability in clinical phenotype raises the possibility that the disease in this family exhibits variable expressivity, or that the dementias observed in this kindred have more than one cause.
Genetic Analysis
High molecular weight DNA was extracted from blood or brain tissue according to standard procedures. DNA was available for 82 (18 affected) family members with complete clinical information. Because the primary goal of this study was to determine the genetic cause of HDDD, we established clinical and pathological criteria to classify individuals with definite, probable, or possible HDDD (Table 1). These criteria were based on the presence of frontal lobe signs and prominent early memory loss in family members plus confirmation of FTLD-U (or FTLD-motor neuron disease [MND] type) using established neuropathological diagnostic criteria.
Table 1.
Main Features and Research Criteria Used for Definite, Probable, and Possible Hereditary Dysphasic Disinhibition Dementia Classification
| Signs and Symptoms of Hereditary Dysphasic Disinhibition Dementia |
| A. Cognitive impairment of gradual onset and progressive course |
| B. Family history of dementia with frontal features |
| C. Early personality/behavior impairment: either a, b, c, or d are present and prominent within the first 3 years of noted cognitive impairment |
| a. Change in personality |
| b. Impaired executive functions |
| c. Apathy |
| d. Repetitive/compulsive behavior |
| D. Early language impairment: either a or b are present and prominent within the first 3 years of noted cognitive impairment |
| a. Reduced language output: less spontaneous speech, inability to finish sentences (within 3 years), or mutism (within 5 years) |
| b. Verbal perseveration, stuttering, echolalia, pardysphasias (within 3 years) |
| Supportive features: |
| E. Pyramidal/extrapyramidal signs |
| a. Focal hyperreflexia or increased tone, hemiparesis, Babinski's sign, without evidence of stroke |
| b. “Primitive” frontal reflexes |
| c. Bradykinesia/hypokinesia, resting tremor, parkinsonian gait, unilateral extremity neglect |
| Research Criteria for Disease Classification |
| I. Definite HDDD: Pathology (macroscopic findings: frontal lobe atrophy, enlarged lateral ventricles, thinning of the cortical ribbon, pale and gliotic underlying white matter; microscopic findings: neuronal loss, gliosis and superficial spongiosis in the frontal lobe, neuronal loss in the hippocampus, presence of ubiquitin-positive intraneuronal inclusions) plus criteria for probable/possible HDDD or DSM-IV for AD dementia |
| II. Probable HDDD: A, B, C, and D required; memory impairment is not the prominent feature at disease onset |
| III. Possible HDDD: A and B required; C or D required; memory impairment could be prominent at disease onset |
HDDD = hereditary dysphasic disinhibition dementia; DSM-IV = Diagnostic and Statistical Manual, fourth edition; AD = Alzheimer's disease.
For linkage analysis, we used information from a total of 161 individuals spanning 4 generations from the FTLD-U part of the pedigree.
Statistical Analysis
The informativeness of the pedigree was established by simulation studies (FastSlink v2.51 program),20 which yielded a LOD score of 6.35. Three diagnostic thresholds were tested to maximize the linkage information. In model I, affected individuals were defined as those with a diagnosis of definite or probable HDDD. Family members with a diagnosis other than definite or probable HDDD and all asymptomatic individuals younger than 77 years were considered as phenotype unknown. The remaining family members were considered as unaffected (nondemented individuals older than 77 years). In model II, all individuals with a lifetime diagnosis of definite or probable HDDD were considered affected; remaining family members were considered as phenotype unknown. In model III, all individuals with HDDD (definite plus probable plus possible) were considered affected; remaining family members were considered as phenotype unknown.
DNA Sequencing
All the exons and the intron–exon boundaries of PGRN were amplified using gene-specific primers. Direct sequencing of the amplified fragments was performed using Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Wellesley, MA) and standard protocols. For most of the fragments, the primers used for sequencing were the same as those used for polymerase chain reaction amplification (primer sequence available on request). Sequence variants were tested for segregation with the disease and screened in a set of 50 unrelated age-matched population control subjects. To test the effect of the mutation in other forms of dementia in this kindred, we screened DAT individuals from both within the FTLD-U branch and the extended pedigree.
Neuropathological Procedures
Brain tissue was obtained with the consent of the next of kin and with approval of Washington University Human Studies Committee. Autopsies were performed according to established Alzheimer's Disease Research Center Neuropathology Core protocols.21 In brief, brain tissue was preserved in buffered 10% formalin, and when available, tissue was frozen at –80°C for biochemistry and molecular genetic studies. Tissue samples were obtained from the frontal, temporal, parietal, and occipital lobes, basal ganglia, thalamus, amygdala, hippocampus, midbrain, pons, medulla oblongata, and cerebellum. The spinal cord was unavailable for examination.
Histology and Immunohistochemistry
Routine stains included hematoxylin and eosin and a modified Bielschowsky silver impregnation. After antigen retrieval methods, immunohistochemistry was performed using antibodies to ubiquitin (Chemicon International, Temecula, CA), tau (PHF1; gift of Dr P. Davies), α-synuclein (Zymed Laboratories, San Francisco, CA), β-amyloid (4G8; Signet Laboratories, Dedham, MA), α-internexin (Zymed Laboratories, San Francisco, CA), valosin-containing protein (gift of Dr C.-C. Li), and progranulin (R&D Systems, Minneapolis, MN) on representative areas. Cases were diagnosed according to established and other neuropathological criteria where they exist.1,5,6 Alzheimer's disease (AD)–type changes were assessed using the staging of Braak and Braak22 and the diagnostic criteria of Khachaturian,23 the CERAD,24 and the National Institute on Aging-Reagan Institute criteria.25
Results
Clinical Features
Although the HDDD2 kindred originated in Central Europe, all of the subjects included in this study were born and reside in the United States. About 80% of these subjects live in three adjacent counties in the southern United States. The HDDD2 pedigree has 161 members, spanning 4 generations. Among the demented individuals, identified through clinical examination, medical report, or family report,18 18 met criteria for probable or definite HDDD (8 were HDDD definite), 8 met criteria for possible HDDD, and 2 were diagnosed with DAT. Table 2 summarizes the clinical profile of the HDDD individuals.
Table 2.
Clinical Description of Individuals with Hereditary Dysphasic Disinhibition Dementia (HDDD2) Kindred
| Characteristics | HDDD (n = 26) |
|---|---|
| Sex, F/M | 13/13 |
| Mean onset of disease (range), yr | 63.5 (52–77) |
| Mean duration of disease (range), yr | 7.2 (6–11) |
| Mean age at death (range), yr | 70.2 (59–86) |
HDDD = hereditary dysphasic disinhibition dementia.
Genetic Analysis
LINKAGE ANALYSIS OF 17q21
Significant evidence of linkage was observed with markers on 17q using a whole genome scan (data not shown). High-density screening was subsequently performed between D17S798 and D17S784 to identify the putative disease locus. This interval included the region previously linked to HDDD218 and several other FTD families with ubiquitin-positive, tau-negative inclusions.9,10,11
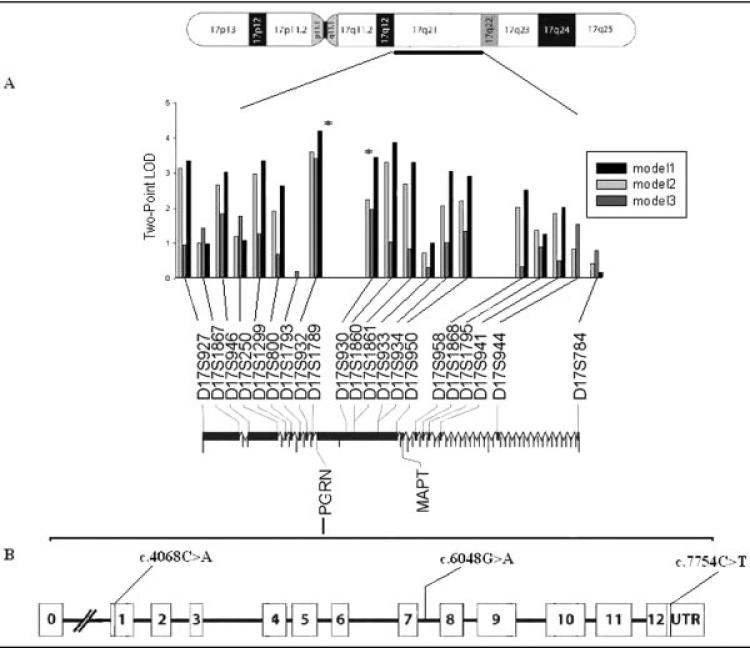
The highest two-point LOD score was observed at D17S932 (LOD = 4.20, θ = 0) for model I. Several flanking markers also yielded conclusive evidence of linkage in the absence of recombination (Fig 1A). Under stringent criteria, model II yielded a similar LOD score for all markers, but slightly lower than model I. This decline in LOD score was even more pronounced at all loci when the analysis was performed using model III, except D17S932 and D17S930 (see Fig 1A). Multipoint analysis yielded evidence of linkage (LOD > 3.00) in the interval containing D17S932 and D17S930, suggesting the possible localization of the causal gene for FTLD-U.
Fig 1.
(A) Two-point linkage screen with chromosome 17 specific markers using three diagnostic thresholds. Markers are placed relative to their physical distance from pter. (B) Schematic map showing the exons of PGRN gene. We identified three novel sequence variants: missense mutation (c.4068C>A) and single nucleotide polymorphisms (c.6048G>A, c.7754C>T). The PGRN gene is located in between D17S932 and D17S930 (0.2Mb) and 1.7Mb upstream microtubule-associated protein tau (MAPT). LOD = logarithm of odds.
DIRECT DNA SEQUENCING
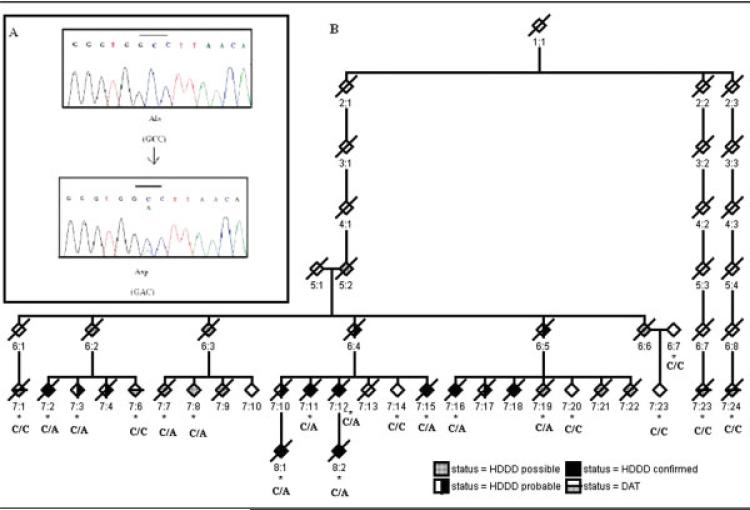
The close proximity of PGRN to the most informative markers (see Fig 1B) and a recent report of null mutations in the PGRN gene led us to examine the gene together with other candidate genes in the region. Mutation detection was performed using the Sequencher package v4.6 (Gene codes pvt Ltd, Ann Arbor, MI). A novel heterozygous missense mutation Ala-9 Asp (A9D) was found in exon 1 (Fig 2A). The mutation was found to cosegregate with the disease phenotype and at-risk individuals with the disease haplotype (see Fig 2B). The mutation was not present in unaffected family members who did not carry the disease haplotype or in matched population control subjects, implying that the mutation is likely to be pathogenic. The mutation was also absent from individuals who had DAT within the FTLD-U branch and in the extended kindred (see Fig 2B).
Fig 2.
(A) Electropherograms showing the heterozygous missense mutation with the wild-type (top) andaCtoA substitution mutation (bottom) in a healthy individual and a patient, respectively. (B) Mutation (Ala-9-Asp) of the PGRN gene in HDDD2 kindred. Mutation status is shown below the symbols of the pedigree. All information that could identify individuals in the pedigree has been removed. Asterisk indicates availability of DNA.
Neuropathological Findings
MACROSCOPY
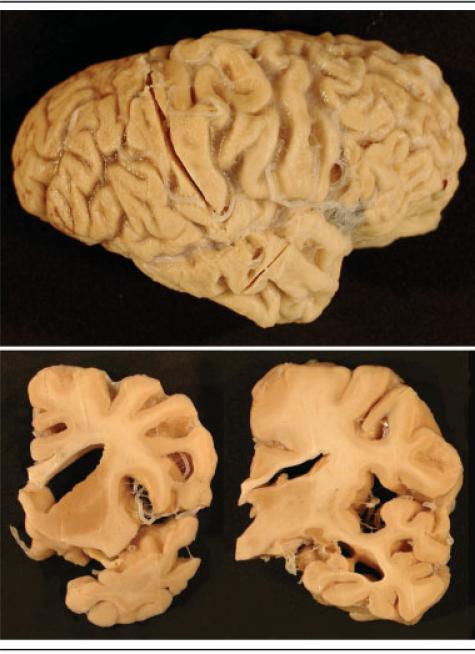
The neuropathology of the first four cases of HDDD2 that came to autopsy has been reported previously.18 Here, we describe an additional four cases and review all eight cases for contemporary immunohistochemical and diagnostic criteria. The mean brain weight was 1,044gm (range, 800 – 1,270gm). Atrophy varied from moderate to severe and was most pronounced in the frontal lobe in all HDDD cases (Fig 3). The temporal lobe was severely affected in the majority of cases, as was the parietal lobe, but to a lesser degree. In about half of the cases, the ventricular dilatation and narrowing of gyri in the frontal and anterior temporal lobes was comparable with that seen in classic cases of Pick's disease. There was variable atrophy of the hippocampus ranging from severe to relatively well preserved (see Fig 3). There was some atrophy of the basal ganglia in about half of the cases. The brainstem and cerebellum were macroscopically unremarkable in most cases, and the pigmented nuclei were generally well preserved.
Fig 3.
The right cerebral hemisphere of chromosome 17–linked frontotemporal dementia with ubiquitin-positive, tau-negative inclusions with PRGN mutation (top). There is pronounced atrophy of the frontal, temporal, and parietal lobes. Coronal slices (bottom) demonstrate marked dilatation of the lateral ventricle and narrowing of gyri (bottom left) and thinned corpus callosum, increased space in the Sylvian fissure and inferior horn of the lateral ventricle, but the hippocampus is relatively well preserved (bottom right).
MICROSCOPY
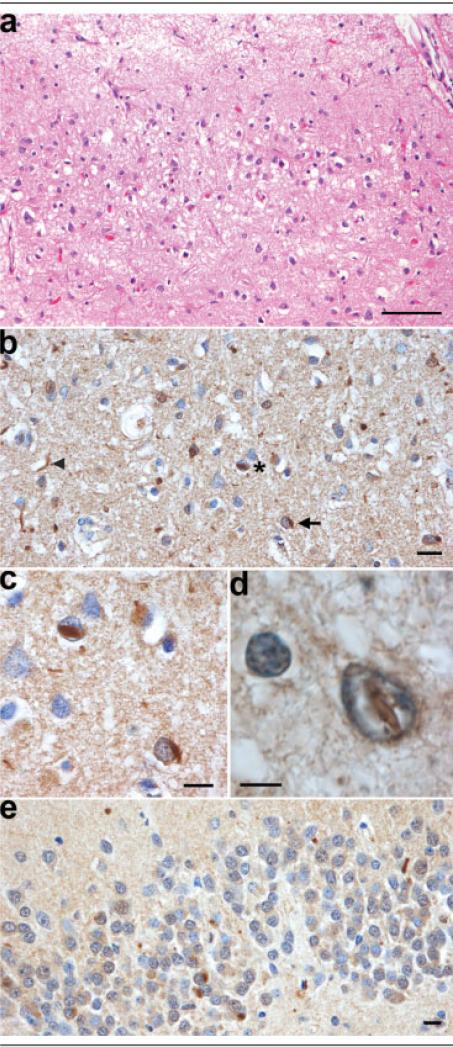
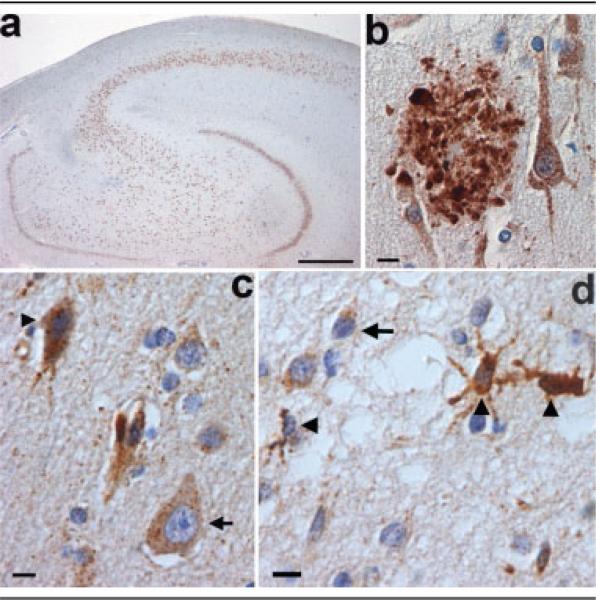
The stereotypical histological features of all FTLDs were present in all eight cases: severe neuronal loss, status spongiosus, and reactive astrocytosis in affected areas. The distribution and severity of pathology varied from case to case, but invariably, the frontal lobe was the most severely affected. There was also pronounced neuronal loss in neocortex of the temporal and, to a lesser degree, the parietal lobe. The degree of neuronal loss was variable, and in the most severe cases, both association and motor cortices were affected. In places, the cortical ribbon was completely devoid of neurons in the most severe cases. More typically, microvacuolation could be seen in laminae II and III (Fig 4). The hippocampus was, generally, less atrophied and showed less neuronal loss than is commonly seen in AD. Neuronal loss from the thalamus was observed in a minority of cases. The nuclei of the brainstem and cerebellum were generally unremarkable. The characteristic lesions of MND, ubiquitin-positive skein-like inclusions and Bunina bodies in motor neurons, and corticospinal tract degeneration were absent. The signature lesions of FTLD-U were present in seven of eight cases: ubiquitin-positive, tau-negative neuronal cytoplasmic inclusions (NCIs); dystrophic neurites (DNs); and neuronal intranuclear inclusions (NIIs) (see Figs 4B–E). In one case, after repeated antigen retrieval methods, no ubiquitin-positive inclusions were detected. These three types of inclusions were seen most readily in the frontal lobe, but their presence was also noted in the temporal and parietal neocortex and basal ganglia; the occipital lobe did not exhibit this pattern of pathology.
Fig 4.
Microscopy of chromosome 17–linked frontotemporal dementia with ubiquitin-positive, tau-negative inclusions and PRGN mutation. (A) Neuronal loss, microvacuolation, and astrocytosis in laminae II and III of the anterior cingulate gyrus. Hematoxylin and eosin staining. (B) Ubiquitin-positive neuronal inclusions: a neuronal cytoplasmic inclusion (arrow), a neuronal intranuclear inclusion (asterisk), and a dystrophic neurite (arrowhead) in lamina III of the middle frontal gyrus. (C) High-power photomicrograph of the neuronal cytoplasmic and intranuclear inclusions in (B). (D) Fusiform ubiquitin-positive neuronal intranuclear inclusion in lamina III of the parietal lobe. (E) Neuronal cytoplasmic inclusions in the dentate fascia. (B–E) Ubiquitin immunohistochemistry. Scale bars = 100μm (A); 10μm (B, C, E); 5μm (D).
Progranulin immunohistochemistry showed a predominantly neuronal cytoplasmic pattern of expression in normal brain (Fig 5A). High levels of expression were also observed in microglial cells (see Fig 5D), and much lower levels in oligodendrocytes and astrocytes. In cases of HDDD2 with AD pathology, the DNs of neuritic plaques were intensely labeled and were identical to those seen in a case of AD (see Fig 5B). Neurofibrillary tangles and diffuse β-amyloid plaques were not labeled by the anti-progranulin antibody (data not shown). Progranulin immunohistochemistry failed to demonstrate progranulin as a component of the ubiquitin-positive, tau-negative NCIs, NIIs, or DNs of familial FTLD-U with PRGN mutation (HDDD2) (see Figs 5C, D).
Fig 5.
Progranulin immunohistochemistry in normal brain, Alzheimer's disease, and familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions (FTLD-U) with PRGN mutation. Progranulin is expressed at high levels in neurons of the pyramidal cell layer of the hippocampus and granule neurons of the dentate fascia in normal brain (A). Progranulin is expressed in the dystrophic neurites of a neuritic plaque of Alzheimer's disease (B). More intense staining of progranulin is seen in the cytoplasm of a degenerating, pyknotic neuron (arrowhead) than in a normal neuron (arrow) in FTLD-U with PRGN mutation (C). The ubiquitin-positive inclusions of the middle frontal gyrus of familial FTLD-U with PRGN mutation (see Figs 4B–D) are not labeled by an antibody to progranulin (D). The cytoplasm of microglial cells (arrowheads) are stained, but a ubiquitin-positive neuronal intranuclear inclusion (arrow), cytoplasmic inclusions (see Figs 4B–D), and dystrophic neurites are unlabeled. Progranulin immunohistochemistry. Scale bars = 1mm (A); 10μm (B, C); 10μm (D).
In none of the six cases where DNA was available was there an APOE4 genotype, yet a feature of the HDDD2 kindred is the presence of additional AD pathology in half of the cases, consistent with neurofibrillary tangle stage greater or equal to IV, according to Braak and Braak.22 In one patient who survived to 82 years, there was sufficient pathology for the diagnosis of AD according to Khachaturian's criteria23 and “probable AD” by CERAD criteria.24 In those cases with AD pathology, the distribution was atypical. The highest densities of lesions (neurofibrillary tangles and β-amyloid plaques) were seen in temporal, parietal, and occipital lobes, the frontal lobe being relatively devoid of this pathology. The relative absence of AD pathology from the frontal lobes indicates that this neurodegenerative disease occurs secondarily to FTLD-U and is probably age related, although synergistic effects of these two neurodegenerative processes cannot be excluded.
Discussion
This study details the clinical, pathological, and mutation analysis of the HDDD2 kindred. The HDDD syndrome resembles the so-called frontal variant of FTD with ubiquitin-positive intraneuronal inclusions.26 The presence of ubiquitin-positive, tau-negative inclusions in the HDDD2 kindred places the disorder within the group of chromosome 17–linked FTD kindreds with neuronal ubiquitin-positive, tau-negative inclusions.9–11
The clinicopathological criteria used to classify the patients into different groups (possible, probable, and definite HDDD) proved to be useful in our genetic analyses. Use of different phenotypic thresholds helped localize the candidate region on 17q21. The recently identified causal gene PGRN is located 0.2Mb in between D17S932 and D17S930. These two markers showed significant evidence of linkage under all three diagnostic thresholds tested, suggesting a common disease locus for them. In contrast, inclusion of the DAT individuals in the analysis did not improve the linkage information, implying a different cause. All affected members of the kindred are heterozygous for a novel missense mutation c.4068C to A in exon 1, resulting in an Ala-9 to Asp substitution, which introduces a charged amino acid into the signal peptide. Furthermore, none of the DAT individuals carried the mutation, confirming that the DAT observed in this kindred has a different cause than HDDD. The location of this mutation is in, or near, the binding site of the signal recognition particle, which targets proteins to the endoplasmic reticulum membrane.27 This putative mechanism introduces a change in the amino acid side chain, which could radically affect protein secretion whereas not affecting the level of the mRNA. However, in light of the functional null alleles it may be possible that the amino acid substitution may degrade the mutant RNA by nonsense-mediated decay, causing variation in protein expression like a null mutation.12,13,28
We also confirm that HDDD2 has the characteristic features of all FTLDs: frontal and temporal lobe atrophy with severe neuronal loss, status spongiosus, and reactive astrocytosis in affected areas. In addition, the signature lesions of FTLD-U are also present: ubiquitin-positive, tau-negative NCIs and DNs. As in most familial cases of FTLD-U and occasional sporadic cases, a third site of ubiquitin pathology was seen in HDDD2: NIIs. Both NCIs and DNs are readily seen in affected neocortex and basal ganglia, and NCIs may also be seen with varying density in the granule cells of the dentate fascia (see Fig 4E). The NCIs of FTLD-U are indistinguishable from those seen in some cases of MND and MND with dementia.29 In the majority of HDDD2 brains, ubiquitin-positive NIIs were seen and, although less abundant than NCIs or DNs, were found in affected cortex (see Figs 4C, D) and subcortical nuclei. NIIs have been reported in a number of familial cases of FTLD-U, but they are not specific to familial cases because they are also seen in sporadic cases.30 Progranulin immunohistochemistry failed to detect the ubiquitin-positive inclusions of familial FTLD-U, an observation that is consistent with the numerous failed attempts to detect a pathological protein in the inclusions of familial and sporadic FTLD-U. The mechanism by which mutant progranulin causes inclusion formation in this disease remains an enigma.
The biological function of PGRN in neurons is not well characterized; however, the basal gene expression of this growth factor suggests that it is multifunctional with important roles in several neuronal signaling pathways15 and neurodegeneration.16,31 The identification of PGRN as a cause of neurodegeneration also highlights the link between tumorigenesis and neurodegeneration. Overexpression of PGRN in vitro leads to uncontrolled cell growth in several cell types, whereas null alleles cause neurodegenerative disease.16,31 Similarly, mutations in the presenilins cause FAD, whereas knock-out of Presenilin (PS) in the skin leads to tumor formation.32,33 The heterozygous missense mutation we have identified could prevent normal insertion of progranulin into the endoplasmic reticulum membrane preventing secretion and leading to an accumulation of the precursor protein in the endoplasmic reticulum. The pathological mechanism could also include degradation of the mutant RNA through nonsense-mediated decay.28
The search for the genetic basis of FTLD-U has been ongoing for more than a decade; this report, together with other positive findings, brings new insight to the disease pathogenesis and will help in the identification of therapeutic agents.
Acknowledgments
This work was supported by the Progressive Supranuclear Palsy Association (PSP Europe; P.P.), the NIH (National Institute on Aging, P50 AG05681 and PO1 AG03991, J.C.M.), McDonnell Center for Molecular and Cellular Neurobiology (N.J.C.), Barnes Jewish Foundation (A.M.G.), Pilot and Feasibility program of the Washington University Center for Genome Science, supported by the Danforth Foundation (A.M.G.), a Fogarty International Postdoctoral fellowship (TW 0511-05, O.M.), and the J. William Fulbright Foreign Scholarship Board (68428174, K.M.L.) a FORD Foundation Predoctoral Fellowship (J.S.K.K.).
We thank the family members of the HDDD2 kindred for their collaboration in the project. We also thank C. Lendon, T. Lynch, L. Harrell, J. Renner, N. Sharrow, and D. McKeel for studying HDDD2 members during the first years of the project and Francis Busfield for studying this family. We also acknowledge the support of the Clinical, Psychometrics, and the Genetics Cores of the Washington University Alzheimer's Disease Research Center (ADRC) and the expert technical assistance of K. Paulsmeyer, K. Carter, D. Carter, C. Groomes, Fei Zang and Kevin Mayo.
References
- 1.McKhann GM, Albert MS, Grossman M, et al. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol. 2001;58(11):1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 2.Rizzu P, Van Swieten JC, Joosse M, et al. High prevalence of mutations in the microtubule-associated protein tau in a population study of frontotemporal dementia in the Netherlands. Am J Hum Genet. 1999;64(2):414–421. doi: 10.1086/302256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Lund and Manchester groups Clinical and neuropathological criteria for frontotemporal dementia. J Neurol Neurosurg Psychiatry. 1999;57:416–418. doi: 10.1136/jnnp.57.4.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun A. Frontal lobe degeneration of non-Alzheimer type. I. Neuropathology. Arch Gerontol Geriatr. 1987;6(3):193–208. doi: 10.1016/0167-4943(87)90021-5. [DOI] [PubMed] [Google Scholar]
- 5.Cairns NJ, Grossman M, Arnold SE, et al. Clinical and neuropathologic variation in neuronal intermediate filament inclusion disease. Neurology. 2004;63(8):1376–1384. doi: 10.1212/01.wnl.0000139809.16817.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watts GD, Thorne M, Kovach MJ, et al. Clinical and genetic heterogeneity in chromosome 9p associated hereditary inclusion body myopathy: exclusion of GNE and three other candidate genes. Neuromuscul Disord. 2003;13(78):559–567. doi: 10.1016/s0960-8966(03)00070-1. [DOI] [PubMed] [Google Scholar]
- 7.Knopman DS, Mastri AR, Frey WH, et al. Dementia lacking distinctive histologic features: a common non-Alzheimer degenerative dementia. Neurology. 1990;40(2):251–256. doi: 10.1212/wnl.40.2.251. [DOI] [PubMed] [Google Scholar]
- 8.Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 9.Mackenzie IR, Baker M, West G, et al. A family with tau-negative frontotemporal dementia and neuronal intranuclear inclusions linked to chromosome 17. Brain. 2006;129:853–867. doi: 10.1093/brain/awh724. [DOI] [PubMed] [Google Scholar]
- 10.Rosso SM, Kamphorst W, de Graaf B, et al. Familial frontotemporal dementia with ubiquitin-positive inclusions is linked to chromosome 17q21–22. Brain. 2001;124(Pt 10):1948–1957. doi: 10.1093/brain/124.10.1948. [DOI] [PubMed] [Google Scholar]
- 11.Rademakers R, Cruts M, Dermaut B, et al. Tau negative frontal lobe dementia at 17q21: significant finemapping of the candidate region to a 4.8 cM interval. Mol Psychiatry. 2002;7(10):1064–1074. doi: 10.1038/sj.mp.4001198. [DOI] [PubMed] [Google Scholar]
- 12.Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia inked to chromosome 17. Nature. 2006 Jul 16; doi: 10.1038/nature05016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Cruts M, Gijselinck I, van der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. nature. 2006 Jul 16; doi: 10.1038/nature05017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Bateman A, Belcourt D, Bennett H, et al. Granulins, a novel class of peptide from leukocytes. Biochem Biophys Res Commun. 1990;173(3):1161–1168. doi: 10.1016/s0006-291x(05)80908-8. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Bateman A. Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res. 1999 Jul 1;59(13):3222–3229. [PubMed] [Google Scholar]
- 16.Daniel R, He Z, Carmichael KP, et al. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48(7):999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- 17.Morris JC, Cole M, Banker BQ, et al. Hereditary dysphasic dementia and the Pick-Alzheimer spectrum. Ann Neurol. 1984;16(4):455–466. doi: 10.1002/ana.410160407. [DOI] [PubMed] [Google Scholar]
- 18.Lendon CL, Lynch T, Norton J, et al. Hereditary dysphasic disinhibition dementia: a frontotemporal dementia linked to 17q21–22. Neurology. 1998;50(6):1546–1555. doi: 10.1212/wnl.50.6.1546. [DOI] [PubMed] [Google Scholar]
- 19.Silverman JM, Raiford K, Edland S, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part VI. Family history assessment: a multicenter study of first-degree relatives of Alzheimer's disease probands and nondemented spouse controls. Neurology. 1994;44(7):1253–1259. doi: 10.1212/wnl.44.7.1253. [DOI] [PubMed] [Google Scholar]
- 20.Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21(3):405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 21.Berg L, McKeel DW, Jr, Miller JP, et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55(3):326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 22.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 23.Khachaturian ZS. Diagnosis of Alzheimer's disease. Arch Neurol. 1985;42(11):1097–1105. doi: 10.1001/archneur.1985.04060100083029. [DOI] [PubMed] [Google Scholar]
- 24.Mirra SS, Heyman A, McKeel D, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 25.Hyman BT, Trojanowski JQ. Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute Working Group on diagnostic criteria for the neuropathological assessment of Alzheimer disease. J Neuropathol Exp Neurol. 1997;56(10):1095–1097. doi: 10.1097/00005072-199710000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Nestor P, Hodges J. Non-Alzheimer dementias. Semin Neurol. 2000;20(4):439–446. doi: 10.1055/s-2000-13176. [DOI] [PubMed] [Google Scholar]
- 27.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 28.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5(2):89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 29.Mackenzie IR, Feldman HH. Ubiquitin immunohistochemistry suggests classic motor neuron disease, motor neuron disease with dementia, and frontotemporal dementia of the motor neuron disease type represent a clinicopathologic spectrum. J Neuropathol Exp Neurol. 2005;64(8):730–739. doi: 10.1097/01.jnen.0000174335.27708.0a. [DOI] [PubMed] [Google Scholar]
- 30.Bigio EH, Johnson NA, Rademaker AW, et al. Neuronal ubiquitinated intranuclear inclusions in familial and non-familial frontotemporal dementia of the motor neuron disease type associated with amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2004;63:801–811. doi: 10.1093/jnen/63.8.801. [DOI] [PubMed] [Google Scholar]
- 31.Malaspina A, Kaushik N, Belleroche J. Differential expression of 14 genes in amyotrophic lateral sclerosis spinal cord detected using griddled cDNA arrays. J Neurochem. 2001;77(1):132–145. doi: 10.1046/j.1471-4159.2001.t01-1-00231.x. [DOI] [PubMed] [Google Scholar]
- 32.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 33.Xia X, Qian S, Soriano S, et al. Loss of presenilin 1 is associated with enhanced beta-catenin signaling and skin tumorigenesis. Proc Natl Acad Sci U S A. 2001;98(19):10863–10868. doi: 10.1073/pnas.191284198. [DOI] [PMC free article] [PubMed] [Google Scholar]