Abstract
The generation of large-scale data sets is a fundamental requirement of systems biology. However, despite recent advances, generation of such high-coverage data remains a significant challenge. We developed a novel pooling-deconvolution strategy that can dramatically decrease the effort required. This “PI-Deconvolution” strategy employs imaginary tagging and allows the screening of 2n probe proteins (baits) in 2*n pools, with n replicates for each bait. Deconvolution of baits with their binding partners (preys) can be achieved by reading the prey’s profile from the 2*n experiments. We validated this strategy for protein-protein interaction mapping using both proteome microarrays and a yeast two-hybrid array, demonstrating that PI-Deconvolution can identify interactions accurately with fewer experiments and better coverage. We also show that PI-Deconvolution can identify protein-small molecule interactions inferred from profiling the yeast deletion collection. PI-Deconvolution should be applicable to a wide range of library-against-library approaches, and can also be used to optimize array designs.
INTRODUCTION
Understanding protein function on a genome-wide scale is one of the central goals of biology1. A fundamental task associated with this goal is the elucidation of cellular functional and interaction networks. Recently, large-scale protein-protein interaction experiments, using yeast two-hybrid2–7 or affinity purification8–10 have generated critical insights into protein function and biological network structure. In addition to the determination of protein interaction networks, systems biology will require the elucidation of other interaction (e.g., protein-small molecule, protein-nucleic acid) and functional (e.g., protein phosphorylation) networks. However, although the need to generate these types of data sets is obvious, our current ability to do so is clearly inadequate.
The challenges of generating protein-protein interaction data sets serve as a good model for what will be required to obtain other interaction and functional network data sets. Because the generation of a complete interaction network one protein pair at a time is labor- (and material-) intensive, simpler alternatives have been sought. The creation of spatially addressable proteome-wide screening platforms (such as the array of yeast two-hybrid strains2 and proteome microarrays11,12), for instance, allows an entire subject library to be screened at once and hits to be decoded without the need for DNA or protein sequencing. However, thousands of array screens are required to cover a yeast-sized interactome, an even more daunting undertaking when replicate screens are needed to improve accuracy and coverage.
To date, pools of 8 baits or more have been used to screen yeast two-hybrid arrays6,13. In these approaches, secondary small-scale screens are subsequently used to deconvolute the hits. A major weakness of this procedure is that final coverage completely depends on the primary pooling screen, which is often performed only once. On the other hand, a primary screen with reduced stringency may generate too many false positives, creating a huge burden on the secondary screens. Furthermore, small-scale secondary screens may not be possible for some platforms such as protein microarrays. Because genome/proteome-wide arrays have the physical capacity to detect far more interactions than those of a typical single protein, an alternative pooling strategy that allows prey-bait deconvolution is possible. This “Pooling with Imaginary tags followed by Deconvolution” (PI-Deconvolution) strategy we describe here reduces the number of screens needed while simultaneously increasing accuracy and coverage. In addition, we suggest a novel method to optimize array design using the same principle.
RESULTS
Description of strategy
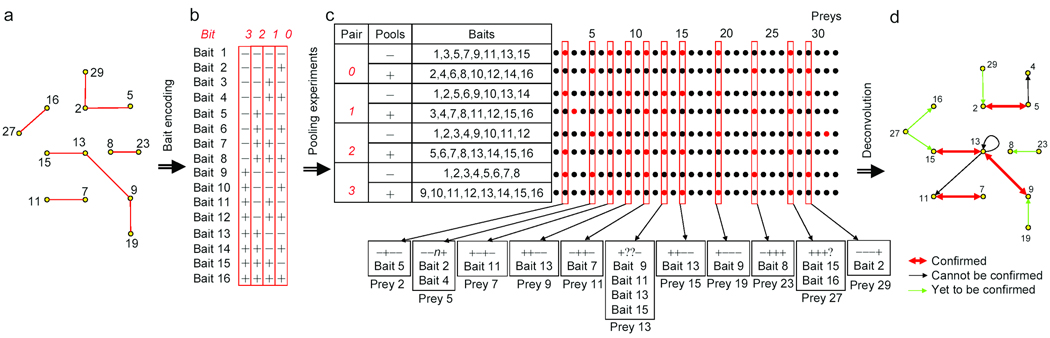
In PI-Deconvolution (Fig. 1), 2n baits are distinguished by their assigned n-bit binary codes, which are text strings consisting of “+” and “−” symbols (Fig. 1b). The baits are assigned to n pairs of experiments (each pair containing one “+” and one “−” experiment) corresponding to the binary bits (Fig. 1c). In each experiment-pair, half of all the 2n baits will be loaded to the “+” experiment pool and the other half to the “−” pool. Any single bait is used once and only once in each pair of experiments. Whether a bait is used for the “+” or “−” experiment in a pair is determined by its symbol in the coding string at the corresponding bit (Fig. 1b–c). For example, bait 6 is represented by string “−+−+”. At bit 2 (third digit from the right in the string), its symbol is “+”. Thus in pair 2, bait 6 is included in the “+” experiment. This “+” experiment also includes all the other baits with a “+” sign in the column for bit 2. Each prey’s interacting bait(s) can be revealed by the prey’s profile in all the n pairs of experiments. For example, prey 2 binds to only bait 5 among baits 1–16. In Fig. 1c, prey 2 is detected in 4 experiments, which are “−” of pair 0, “−” of pair 1, “+” of pair 2, and “−” of pair 3. Accordingly, prey 2 can be represented by the profile “−+−−”, denoting its readout in each of the 4 experiment pairs. Since pair numbering corresponds to bit numbering in the tag of a bait, the prey’s profile can allow a direct track back to its own bait(s). In this case, the profile of prey 2 is identical to the bit tag for bait 5; thus, we know that prey 2 binds to bait 5. With this strategy, 2n baits can be screened in 2*n arrays (Table 1). Pool size will be limited by the technical false negative and false positive rates (discussed below); the flexibility in setting different bit numbers (n) allows the strategy to be applied to different scenarios. Besides significantly decreasing the number of screens needed, a major advantage of PI-Deconvolution is that all the baits are screened n times (Table 1), which allows cross-validation and thus improves both coverage and accuracy of data.
Figure 1.
Scheme for PI-Deconvolution. (a) Graph representation of a hypothetical 32-protein network. Yellow filled circles, proteins (nodes); red lines, interactions (edges). For simplicity, only nodes and edges concerning proteins 1–16 are shown. (b) Proteins 1–16 are used as the first batch of baits to identify their preys. The total 32 proteins can be covered similarly with a second batch of experiments. We encode each bait with a 4-bit +/− string (imaginary coding tag); four bits are enough to uniquely encode 16 (=24) baits. Thus, n bits can encode 2n distinct baits. (c) We prepare 4 pairs of bait pools numbered from 0–3, corresponding to each of the 4 bits. Every pair contains a “+” pool and a “−” pool, each employing 8 baits (half the batch size). Altogether, there will be 8 (2n) experiments (rows) – instead of 16 (2n) – to identify all interacting preys. Each column represents profile of a prey; positive signal (red), negative signal (black). All valid preys (columns outlined in red) and their possible baits are listed. If a prey binds to only one bait in a batch, the prey should be detected only once in each pair of experiments. We use degenerate profile “n” or “?” to indicate neither or both experiments in a pair give a positive call (such as prey 5 or prey 13). Preys with degenerate profiles can still be partially deconvoluted and further narrowing-down can be achieved by reciprocal confirmation. (d) A graph can be drawn according to the result in c.
Table 1.
Number of probings required in PI-Deconvolution.
| Bit # | Batch Size |
Pool Size |
Expt # | Total Expt # | "Depth of coverage" |
|---|---|---|---|---|---|
| (n) | (2n) | (2n-1) | (2n) | (n*N/2n-1) | |
| 4 | 16 | 8 | 8 | 50.0%×N | 4 |
| 5 | 32 | 16 | 10 | 31.3%×N | 5 |
| 6 | 64 | 32 | 12 | 18.8%×N | 6 |
| 7 | 128 | 64 | 14 | 10.9%×N | 7 |
| 8 | 256 | 128 | 16 | 6.25%×N | 8 |
| Conventional single bait strategy | N | 1 | |||
Note: N is the number of proteins in the genome.
Protein interaction mapping with PI-Deconvolution on proteome microarrays
We tested PI-Deconvolution using yeast proteome microarrays11,12, which contain 4,088 purified Saccharomyces cerevisiae proteins (as glutathione S-transferase fusions) immobilized on nitrocellulose-coated glass slides. For this purpose, 15 (~16=24) V5 epitope-tagged bait proteins were employed (Fig. 2a). By first probing the yeast proteome microarrays with each of these 15 proteins individually, we derived a small network of protein interactions (Fig. 2a, bottom). We consider this a “gold standard” network, because all the interactions in the network have been reciprocally confirmed (bi-directional red arrows in Fig. 2a). Pools of 8 baits were prepared (Supplementary Table 1 online) and used to probe the microarrays. All interactions among the 15 proteins were detected and deconvoluted using PI-Deconvolution with only 8 proteome microarrays, and all hits were reproducibly detected 4 times (Fig. 2a). Furthermore, although there were several interacting protein pairs within the mixed pools of baits, they did not appear to affect detection or deconvolution. (Raw chip images will be available for download at “http://labs.pharmacology.ucla.edu/huanglab/”.)
Figure 2.
PI-Deconvolution applied to protein interaction mapping. (a) Yeast proteome microarray screening. 15 bait proteins are encoded as shown and 8 bait pools are prepared accordingly (see also Supplementary Table 1 online). Each image column represents the result of a pooling screen, and each image row represents the same spot of the array. A positive signal indicates the presence of one or more binding proteins in the pool. Signals from “+” pools are false-colored red and “−” pools green. For example, the prey spots representing CMD1 (first row) were positive when probed with the “+” pools of pairs 1 and 2 (in red), and the “−” pools of pairs 0 and 3 (in green). The profile of CMD1 is thus read as “−++−”, which equals the encoding tag for the bait CMK1. The results obtained by the PI-Deconvolution analysis (using 8 arrays) are identical to those obtained from single-bait probing (using 15 arrays). Only reciprocally confirmed interactions (red bidirectional arrows) and self interactions (black arrows) are shown (bottom). Detailed explanation of hit recognition is described in Methods. (b) Yeast two-hybrid array screening. Encoding and pooling schemes of 16 bait strains are shown in Supplementary Table 2 online. The whole library array consists of 16 plates with 384 strains each. Shown are images of one representative library plate screened with 16 baits using PI-Deconvolution; each image is the result of a pooling screen with 8 baits.
Protein interaction mapping with PI-Deconvolution on yeast two-hybrid arrays
We demonstrated the utility of PI-Deconvolution to screen a genome-wide two-hybrid array consisting of ~6,000 yeast strains, each designed to contain one of the ~6,000 S. cerevisiae open reading frames (ORFs) fused to the Gal4 activation domain (AD)2. To this end, we used 16 two-hybrid bait strains that each express a full length ORF fused to the Gal4 DNA binding domain (Fig. 2b). Thirteen of these bait strains have previously been screened against the genome-wide array. Because of experimental variability, these single bait screens required each bait to be screened in duplicate, resulting in a total of 32 screens for the 16 baits2,14. For the PI-Deconvolution format, the 16 bait strains were mixed into 8 pools and screened against the two-hybrid array. In this procedure, two 8-bait pools in the same pair cover all of the 16 baits. Therefore, 4 pairs of PI-Deconvolution screens represent 4 independent screens of all the 16 baits (Supplementary Table 2 online). This protocol is a significant advantage over the individual bait procedure because it reduces the number of screens from 32 to 8, yet each bait is screened in quadruplicate. (Raw data will be available for download at “http://www.gs.washington.edu/”.)
In the 13 single bait screens, 484 preys were observed and defined as two-hybrid positive colonies2,14. Among these positive colonies, 125 arose twice out of the duplicate screens and were termed “reproducible” positives; the other 359 arose once as either false positives, or true positives that due to experimental variability did not yield reproducible results. Further testing is usually required to confirm or reject such non-reproducible hits (for a complete list of all single bait screen hits, see Supplementary Table 3 online). For the PI-Deconvolution screens, each pair of pools identified 153–189 hits, and in total 343 positive colonies were identified (Table 2). The number of “reproducible hits” between any two independent experiment-pairs ranged from 103 to 112. On the other hand, as many as 155 positive colonies were reproducible in at least two experiment-pairs (40–50% higher than considering any two experiment-pairs only, Table 2). This result suggests that, as expected, a higher level of repetition indeed improves coverage. We consider all 155 as reproducible positives from PI-Deconvolution. A complete list of all PI-Deconvolution hits and deconvolution results can be found in Supplementary Table 3 online.
Table 2.
Reproducibility over PI-Deconvolution experiment pairs.
| Overlapped Hits | Pair 0 | Pair 1 | Pair 2 | Pair 3 | All pairs |
|---|---|---|---|---|---|
| Pair 0 | 103 | 107 | 109 | ||
| Pair 1 | 103 | 110 | 110 | ||
| Pair 2 | 107 | 110 | 112 | ||
| Pair 3 | 109 | 110 | 112 | ||
| Total # of hits | 153 | 189 | 179 | 180 | 343 |
|
# of hits overlapping with at least one other pair |
123 | 127 | 130 | 133 | 155 |
|
# of hits having no overlap with other pairs |
30 | 62 | 49 | 47 | 188 |
Further evidence for improved coverage by PI-Deconvolution is provided by comparing the PI-Deconvolution data with the single bait data. The 155 reproducible hits from the PI-Deconvolution data set include 26 (out of 359, 7%) “non-reproducible positives” found in the single bait screens, in addition to recapitulating 71 (of 125, 57%) reproducible single bait positives (Table 3). This result suggests that in the single bait data set, ~30% (26 of 97) of the true positives were not found as reproducible hits. The remaining 58 PI-Deconvolution reproducible positives are either novel interaction partners for these 13 baits or belong to the 3 baits not screened before. In contrast, the 188 PI-Deconvolution non-reproducible hits contain only 2 (of 125, 1.6%) reproducible hits from the single bait data (Table 3), suggesting that the PI-Deconvolution 155 reproducible hits might represent almost complete (saturated) coverage, subject to the detection sensitivity of the current system. The increased coverage is due to the high repetitions inherent in PI-Deconvolution screening.
Table 3.
Comparison between single-bait and PI-Deconvolution Y2H screens.
| PI-Deconvolution screening | Hits not in PI- Deconvolution data |
Total | |||
|---|---|---|---|---|---|
| Reproducible hits (***) | Non reproducible hits | ||||
|
Single bait screening |
Reproducible hits (**) | 71 | 2 | 52 | 125 |
| Non reproducible hits | 26 | 13 | 320 | 359 | |
| Hits not in single bait data | 70 | 173 | |||
| Total | 155* | 188 | |||
This number is smaller than the sum of those above because some hits belong to multiple baits.
Positive in duplicate.
Positive in at least 2 experiment-pairs.
Of the 155 PI-Deconvolution reproducible positives, 57 could be assigned to a single bait (see Supplementary Table 4 online), 34 to two baits, and 51 to four possible baits (see discussion below about further deconvolution of positives assigned to more than one bait). Of the 57 unambiguously deconvoluted preys, 56 belong to the 13 previous screened baits. Of these 56, 38 were previously classified as reproducible positives in single bait screens; 11 were previously classified as non-reproducible positives (i.e., appearing only once in duplicate screens), but can now be considered reproducible because they appeared all four times in PI-Deconvolution screens; and 7 are novel interactions (Supplementary Table 4 online) that had eluded detection in single bait screens. One example of a novel interaction is an interaction between Gac1 and Glc7, which are regulatory and catalytic subunits, respectively, of a type 1 phosphatase (PP1) involved in the regulation of glycogen synthesis15.
In PI-Deconvolution, unambiguous deconvolution requires a prey to have a profile that consists of only “+” or “−” values, which means that it is discovered once and only once in every PI-Deconvolution pair. Ambiguity occurs when a prey’s profile contains “?” (the prey turns up positive in both “+” and “−” experiments of a PI-Deconvolution pair) or “n” (the prey turns up negative in both “+” and “−” experiments of a PI-Deconvolution pair). Such degenerate profiles still cover all possible baits, although they do not allow complete deconvolution. Degenerate profiles can occur because of experimental false-positives (FP) and false-negatives (FN) (Supplementary Table 5 online), or when more than one bait in a batch bind to the same prey (see Fig. 1). It is obvious that more preys will have a “?” profile with the use of larger pools (see Supplementary Notes online). Ambiguous profiles may be further clarified by reciprocal (pair-wise) confirmation. When an interaction can be observed only in one direction, profile ambiguity can be clarified by bait “reshuffling”. For instance, when using n=5, 64 baits will be randomly divided into two 32-bait batches. In a “reshuffling” screen, the baits will be divided differently into two batches. A prey-bait pair will be accepted only when it is positive in both screens.
One reason why PI-Deconvolution data cover only a portion (57%) of the reproducible positives from single bait screens could be that sensitivity is compromised when multiple baits (8 in this case) are pooled. If single bait screens were more sensitive, then the single bait data set should cover more true positives than the pooled bait data set. However, although 52 out of 125 (42%) reproducible positives found in single bait screens were missed in the PI-Deconvolution screens, 70 out of 155 (45%) PI-Deconvolution reproducible hits were not found by the single bait screens (Table 3). Thus, it does not appear that the single bait data set has a better coverage than the PI-Deconvolution data set. These results suggest that intrinsic yeast two-hybrid variability, rather than loss of sensitivity due to pooling, likely underlies the discrepancy between the two data sets.
Application of PI-Deconvolution to fitness screening in identifying drug-resistant mutants
The primary advantage of PI-Deconvolution is that it increases screening efficiency by making better use of the physical capacity of whole-proteome platforms for parallel detection. We tested the PI-Deconvolution approach on an assay independent of protein interaction mapping, namely the identification of yeast mutants resistant to specific drugs. The S. cerevisiae deletion collection is a set of ~4,500 strains, each deleted for one of the non-essential ORFs16,17. We assayed 128 (n=7) strains from this collection for fitness response to rapamycin, which targets the Tor proteins18, and wortmannin, which is a phosphatidylinositol 3-kinase inhibitor19. Using 14 (=2x7) pools each containing 64 (=26) strains (see Supplementary Table 6 online), the PI-Deconvolution approach deconvoluted the two strains previously known to be resistant to rapamycin (fpr1Δ) or to wortmannin (ppg1Δ) (Fig. 3), a screening efficiency that is an order of magnitude higher than when single strains are used. Although higher efficiency could be obtained by setting a higher n value (i.e., screening a larger pool) in PI-Deconvolution, acceptable pool size is also determined by the sensitivity and background of the detection method (as is true for any pooling strategy). In addition, because pooled screening generally relies on the gain of a signal, drug hypersensitivity cannot be scored in a pooling screen using fitness as a readout.
Figure 3.
PI-Deconvolution applied to drug resistance screening of 128 (=27) yeast deletion strains in 14 pools (64 strains per pool). Designated drug resistant strains (fpr1Δ for rapamycin18 and ppg1Δ for wortmannin32) are correctly deconvoluted.
In silico simulation on DIP
In order to predict the performance of PI-Deconvolution in an actual large-scale network, we performed a series of in silico experiments to simulate the performance of PI-Deconvolution on the Database of Interacting Proteins (DIP)20. DIP contains 4,716 proteins and 14,848 interactions, which is close to the estimation that a typical protein binds to 3–10 other proteins21,22. The simulation shows that even with 16 or 32 baits per pool, both reciprocal and reshuffling methods can efficiently clarify the ambiguous profiles under a variety of experimental FP and FN conditions (Fig. 4, curves for accuracy). (For additional simulations, see Supplementary Fig. 1 online.) Importantly, coverage after reciprocal or reshuffling confirmation is still significantly higher than for single bait screens, especially for interactions between low-degree nodes (Fig. 4, curves for coverage). Our simulation also shows that PI-Deconvolution can be applied to random networks as well (see Supplementary Fig. 2 online).
Figure 4.
PI-Deconvolution simulated on yeast interactome (DIP). We assume that all interactions in DIP are correct. Only the interactions that are confirmed reciprocally or after bait-reshuffling are accepted. Both accuracy (fraction of true interactions among the detected ones; blue curves) and coverage (fraction of true interactions detected; red curves) are calculated for interactions between proteins with different node degrees (X-axis). For example, blue and red curves with X-axis value 0–24 showed accuracy and coverage of PI-Deconvolution for interactions between nodes with degrees no more than 24. The curves with “x” indicate the accuracy and coverage by duplicated single bait screening, where only reproducible hits are accepted. We assume 40% of the hits from each array experiment are false positives (FP), and each array experiment will lose 40% of true positives (FN). (a) PI-Deconvolution simulated using BitNum=5 (n=5; 16 baits per pool) and CutOcc=2 (only hits that show up at least twice are accepted). (b) PI-Deconvolution simulated using BitNum=6 (n=6; 32 baits per pool) and CutOcc=2.
PI-Deconvolution for compressing arrays
PI-Deconvolution is a robust alternative experimental design that will save time and resources in proteome-wide protein-protein interaction mapping. Although we describe this strategy as a method to pool multiple baits, the idea of PI-Deconvolution can be also used to re-design prey arrays and maximize the efficiency of single bait screens. For example, the yeast two-hybrid array consists of 16 plates, each containing 384 AD-fusion strains. Because we have shown that 8-bait pools can be used to screen the prey array, it should also be possible to screen a single bait against pools of 8 prey strains. Thus, the 16 plates can be compressed into 8 plates using the PI-Deconvolution scheme (8 AD strains per well). This compressed library can be maintained and screened against single baits, equivalent to screening the original 16-plate array in quadruplicate (total reduction to 12.5%).
DISCUSSION
Pooling and deconvolution designs have been of great interest to various fields23,24 (see also Supplementary Notes online). Although 8-bait or larger pools6,13 have been used for screening yeast two-hybrid arrays, two major concerns exist. First, a significant number of true positives will be lost as false negatives, because only the hits that pass the initial pool screening (usually performed only once) will go to the secondary deconvolution screening. Second, because many false positives can pass the primary screen, the burden of secondary deconvolution screens can be huge. PI-Deconvolution overcomes both limitations. We have demonstrated that, due to the built-in repetitions, PI-Deconvolution improves coverage and accuracy simultaneously. Deconvolution is inherent in the PI-Deconvolution method and involves direct “profile reading” without the necessity of secondary screens. Most hits are at least partially deconvoluted (92% of the hits are narrowed down to at most 4 baits); further deconvolution can be achieved by pair-wise confirmation. Unlike methods requiring secondary screens, PI-Deconvolution is generally applicable to both two-hybrid array and proteome microarray platforms.
Protein networks are best modeled as scale-free networks25, in which the majority of nodes have only a few neighbors while a small number of nodes (“hubs”) have many. As expected, the coverage of interactions between proteins of high connectivity is lower than that between proteins of low connectivity (coverage for highly-connected nodes can be improved when the bit number is decreased, see Supplementary Fig. 1 online). PI-Deconvolution is especially useful for mapping interactions of low-degree nodes, which account for the majority of nodes in protein networks (Supplementary Fig. 2 online) and require the largest number of experiments using traditional single-bait methods.
If average degree increases proportionally with number of nodes, PI-Deconvolution will be able to cover a human interactome network with the same efficiency as for yeast (using same pool size). However, since average degree appears to be conserved among different organisms21,22,26, even fewer experiments may in fact be needed, because a larger pool of baits can be accommodated on a larger proteome array. We suggest that PI-Deconvolution-guided community efforts will greatly accelerate interactome mapping from yeast to human. Likewise, PI-Deconvolution should be amenable to increasing the throughput of high-content and/or high-dimensional screening/mapping projects27,28.
Besides increased screening efficiency, a primary benefit of PI-Deconvolution is better data accuracy due to a high level of repetition. Analogously, communication systems are made accurate through extensive redundancy to reduce noise29 (see also http://www.lucent.com/minds/infotheory/), and this principle has been used in designing DNA microarrays30). The similarity between PI-Deconvolution and binary communication systems should allow the translation of error-correction methods in information and computer science to large-scale biological network analyses. For example, an extra pair of pool screening (e.g., with bait reshuffling) can be performed to provide additional redundancy for error detection, analogous to a parity check in communication systems.
Finally, many topological motifs have been described for complex networks (including electronic circuits, the World Wide Web, cells, the brain, ecological systems, and social networks)31. However, not all motifs occur in biological networks (the same is true for non-biological networks). For example, the 4-node feedback loop (square lattice) design, which is commonly found in electronic circuits, is not a motif in protein interaction networks31. It would be interesting to study to what extent the PI-Deconvolution strategy can be robustly applied to each type of network and each type of subgraph.
In summary, this paper presents a novel pooling and deconvolution strategy (PI-Deconvolution) that is generally applicable to maximize screening efficiency in a wide variety of situations. The essence of PI-Deconvolution is imaginary tagging (binary coding), combinatorial pooling, and built-in deconvolution and cross-validation. The key advantages of PI-Deconvolution include better accuracy, coverage, and efficiency. PI-Deconvolution is very flexible because it can be easily scaled up or down by setting different n values. In addition to protein interaction networks, PI-Deconvolution can be useful for other library-against-library scenarios, as long as most probes in the query library have only a few targets in the subject library. The versatility lies in imaginary tagging, which is universally applicable regardless of the nature of the query (molecules, cells, organisms, etc).
METHODS
Proteome microarray, Y2H array and drug resistance screening
The yeast proteome microarrays12 (Protoarray™ Yeast Proteome Microarray, Invitrogen) were probed with purified V5 epitope-tagged “bait” proteins according to manufacturer’s instruction. For pooling experiments, the final concentration of each probe (bait protein) was 5 ng/µl. To detect the bound bait proteins, the arrays were probed with an Alexa Fluor 647™-labeled anti-V5 antibody, and the array image was acquired using an Axon GenePix 4000B scanner. Yeast two-hybrid array screens were performed as described previously14. For drug resistance screening, the 128 yeast deletion strains containing one each resistant to rapamycin and wortmannin were chosen based on previous genome-wide, single strain fitness data32,33. Pools of strains were tested in clear 96-well plates for fitness response to rapamycin (100 nM) and wortmannin (1µM), using DMSO (drug carrier) as control.
Analysis of proteome microarray data
The amount of yeast protein for each ORF on the array is variable and the approximate equivalent solution protein concentrations are available for each spot. Because there is the potential for proteins present in higher quantity on the array to skew data analysis by having higher signals, an approach was developed that identifies hits from paralleled protein microarray experiments that considers the amount of protein in each spot. Suppose dc is the fluorescence intensity read from spot Xc with concentration c, a subset of intensity data D = {di, i represents all spots with concentrations between 90%*c and 110%*c} will be compared to dc. Let d0 = ∑i di / nc (nc is the number of spots in D), p-value of testing if intensity of spot Xc is higher than d̄ can be calculated as 1 – Φ( ( dc – d0 ) / SD ) , in which SD is the standard deviation of dataset D and Φ(x) is the cumulative probability function of standard normal distribution. We will usually consider spot Xc as a positive hit if its p-value < 0.01. Spot data representing the results of probing with the 15 bait proteins in both single and pooling screens are shown in Supplementary Table 7 online.
Simulation on the DIP network
We assume that all the interactions in the DIP graph are true interactions. In a simulated pooling experiment, a set of preys of a given set of baits will be returned. Experimental false-positive rate (FP) and false-negative rate (FN) are considered in the virtual screening procedures. Let Nneighbor be the number of neighbors of all the baits in a pool according to DIP, Ntp be the number of detected neighborhood, and Nfp be the number of false positives in the virtual screening procedure. A simulated screen will return a set of nodes containing Ntp random nodes from true positives and Nfp nodes from non-neighboring nodes. Knowing that FN = (Nneighbor – Ntp) / Nneighbor and FP = Nfp / (Nfp + Ntp), we can get Ntp = Nneighbor * (1 – FN) and Nfp = FP / (1 – FP) * Ntp. Accuracy (fraction of true interactions among detected interactions) and coverage (fraction of true interactions detected) are two criteria that we use to measure the performance of our strategy. Four parameters are simulated: bit number (BitNum), occurrence cutoff (CutOcc, the number of times a signal is detected to be considered a true positive), experimental false-positive rate (FP, fraction of false positive signals in all positive signals) and false-negative rate (FN, fraction of undetected true positives).
Supplementary Material
Acknowledgment
We thank Harvey Herschman, Chris Miller, Erin O’Shea, Fred Fox, Clem Stanyon, the anonymous reviewers, and members of the J.H. Lab for critical readings and suggestions on the manuscript, an anonymous reviewer for introducing to us the idea of communication systems, Ying Du for assistance with drug screening, and Kevin Scanlan for his faithful support of our work. This research was partially supported by a Singleton Developmental Grant (J.H.), University of California Systemwide Biotechnology Research & Education Program GREAT Training Grant 2005-268 (F.J. and J.H.), and a grant from the National Center for Research Resources of the NIH, P41 RR11823 (S.F.). S.F. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 2.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, et al. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 5.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 8.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 9.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 10.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 12.Michaud GA, et al. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003;21:1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 13.Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RL., Jr A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res. 2003;13:2691–2699. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazbun TR, et al. Assigning function to yeast proteins by integration of technologies. Mol Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Hart H, Cheng C, Roach PJ, Tatchell K. Characterization of Gac1p, a regulatory subunit of protein phosphatase type I involved in glycogen accumulation in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:622–635. doi: 10.1007/s004380100455. [DOI] [PubMed] [Google Scholar]
- 16.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 17.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 18.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 19.Carpenter CL, Cantley LC. Phosphoinositide kinases. Biochemistry. 19990;29:11147–11156. doi: 10.1021/bi00503a001. [DOI] [PubMed] [Google Scholar]
- 20.Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigoriev A. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 2003;31:4157–4161. doi: 10.1093/nar/gkg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bork P, et al. Protein interaction networks from yeast to human. Curr Opin Struct Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Janda KD. Tagged versus untagged libraries: methods for the generation and screening of combinatorial chemical libraries. Proc Natl Acad Sci U S A. 1994;91:10779–10785. doi: 10.1073/pnas.91.23.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agyare FD, et al. Mapping expressed sequence tag sites on yeast artificial chromosome clones of Arabidopsis thaliana DNA. Genome Res. 1997;7:1–9. doi: 10.1101/gr.7.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 26.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 27.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 28.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 29.Barry JR, Lee EA, Messerschmitt DG. Digital communication. Edn. 3rd. Boston: Kluwer Academic Publishers; 2004. [Google Scholar]
- 30.Khan AH, Ossadtchi A, Leahy RM, Smith DJ. Error-correcting microarray design. Genomics. 2003;81:157–165. doi: 10.1016/s0888-7543(02)00032-0. [DOI] [PubMed] [Google Scholar]
- 31.Milo R, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 32.Zewail A, et al. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc Natl Acad Sci U S A. 2003;100:3345–3350. doi: 10.1073/pnas.0530118100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie MW, et al. Insights into TOR function and rapamycin response: Chemical genomic profiling by using a high-density cell array method. Proc Natl Acad Sci U S A. 2005;102:7215–7220. doi: 10.1073/pnas.0500297102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.