Abstract
The phytohormone abscisic acid (ABA) regulates the expression of many genes in plants and plays critical roles in stress resistance, and growth and development1-7. Several proteins have been reported to function as ABA receptors8-13 and many more are known to be involved in ABA signaling3,4,14. However, the identities of ABA receptors remain controversial and the mechanism of signaling from perception to downstream gene expression is unclear15,16. Here we show that by combining the recently identified ABA receptor PYR1, with the protein phosphatase 2C ABI1, the serine/threonine protein kinase SnRK2.6/OST1, and the transcription factor ABF2/AREB1, we can reconstitute ABA-triggered phosphorylation of the transcription factor in vitro. Introduction of these four components into plant protoplasts results in ABA-responsive gene expression. The protoplast and test tube reconstitution assays were used to test the function of various members of the receptor, protein phosphatase, and kinase families. Our results suggest that the default state of the SnRK2 kinases is an autophosphorylated, active state and that the SnRK2 kinases are kept inactive by the PP2Cs through physical interaction and dephosphorylation. We found that in the presence of ABA, the PYR/PYL receptor proteins can disrupt the interaction between the SnRK2s and PP2Cs, thus preventing the PP2Cs-mediated dephosphorylation of the SnRK2s and resulting in the activation of the SnRK2 kinases. Our results reveal new insights into ABA signaling mechanisms and define a minimal set of core components of a complete major ABA signaling pathway.
Several ABA receptors have been reported8-13, although many of them remain unconfirmed15-16. Recently, a family of novel START domain proteins, known as PYR/PYLs (pyrabactin resistance1/PYR1-likes, also known as RCARs), were identified as ABA receptors. Several of the PYR/PYLs were shown to interact with and inhibit clade-A PP2Cs (type 2C protein phosphatases)11-13. The PP2Cs (ABI1, ABI2, HAB1, PP2CA/AHG3) negatively regulate ABA responses13. On the contrary, a subfamily of ABA-activated SnRK2s are positive regulators of ABA signaling17-21. Through unknown mechanisms, the inhibition of the negatively acting PP2Cs leads to the successful activation of a subfamily of SnRK2 kinases (SnRK2.2, SnRK2.3 and SnRK2.6 in Arabidopsis), which phosphorylate the basic leucine zipper (bZIP) transcription factors called ABFs/AREBs22-23. The ABFs bind to ABA-responsive promoter elements (ABRE) to induce the expression of ABA-responsive genes1.
The present study was aimed at defining the core components of the ABA response pathway that are both necessary and sufficient for ABA perception, signaling and finally ABA-responsive gene expression. ABA-dependent phosphorylation of ABF2 at amino acid residues S26, S86, S94 and T135 was suggested to be important for stress responsive gene expression in Arabidopsis23. We used transient activation analysis with protoplasts from the snrk2.2/2.3/2.6 triple mutant to determine the role of ABF2 phosphorylation and its dependence on SnRK2s for ABA-responsive gene expression. We have shown previously that the snrk2.2/2.3/2.6 triple mutant is deficient in ABA responses21. As expected, transfection of snrk2.2/2.3/2.6 protoplasts with ABF2 did not induce RD29B-LUC expression even in the presence of ABA, but co-transfection of ABF2 with SnRK2.6 resulted in induction of RD29B-LUC in an ABA-dependent manner (Fig. 1a). Furthermore, ABF2 with alanine substitutions at all of the four phosphorylation sites was inactive, whereas aspartic acid substitutions at these sites led to a constitutively active ABF2 resulting in induction of RD29B-LUC expression even without ABA treatment (Fig. 1a). Co-transfection of Ala-substituted ABF2 with SnRK2.6 led to only a very low level of RD29B-LUC induction (Fig. 1a). Substitution of lysine 50, a conserved residue critical for ATP-binding and kinase activity, with asparagine (K50N) inactivates SnRK2.6 in phosphorylation assays in vitro (our unpublished data). Co-transfection of ABF2 with SnRK2.6K50N did not induce RD29B-LUC expression (Fig. 1a), demonstrating that the kinase activity is necessary for ABF2 activation. Transfection of ABF2 alone in wild type protoplasts induced a low level of RD29B-LUC expression under ABA treatment, which is consistent with the presence of a low basal level of endogenous ABA signaling components in the protoplasts (Supplementary Fig. 1a). These results show that SnRK2.6 mediates ABF2 activation in an ABA-dependent manner, and that ABF2 phosphorylation is sufficient for ABA-induction of RD29B-LUC expression.
Figure 1. Reconstitution of ABA signaling pathway for stress responsive gene expression in Arabidopsis protoplasts.
a, SnRK2-mediated phosphorylation of ABF2 is sufficient for ABA-responsive gene expression. b, Reconstitution of ABA signaling pathway by co-expression of PYR1, ABI1, SnRK2.6 and ABF2. c, econstitution of ABA signaling pathway with different members of the PYR/PYL family. d, Reconstitution using different combinations of the core components. Protoplasts (2 × 104) from the snrk2.2/3/6 triple mutant were used except in (d), where protoplasts from the Col-0 wild-type plants were used. The RD29B::LUC and ZmUBQ::GUS were used as the ABA responsive reporter and internal control, respectively. After transfection, protoplasts were incubated for 5 h under light and in the presence of 0 (open bars) or 5 μM (solid bars) ABA. Error bars, mean ± s.e.m. (n=3).
We next tested the effect of ABI1 and PYR1 on ABA-induction of RD29B-LUC expression. Transfection of ABI1 together with ABF2 and SnRK2.6 resulted in inhibition of RD29B-LUC expression (Fig. 1b, Supplementary Fig. 1a). This shows that ABI1 negatively regulates SnRK2.6- and ABF2-dependent activation of RD29B-LUC expression. Addition of PYR1 together with ABI1, SnRK2.6 and ABF2 enabled ABA-dependent induction of RD29B-LUC expression (Fig. 1b, Supplementary Fig. 1a). However, addition of PYR1P88S that is defective in interaction with and inhibition of PP2Cs12 did not enable ABA-dependent induction of RD29B-LUC expression. The dominant abi1-1 mutation (G180D) disrupts the interaction between ABI1 and PYR112. Like the wild type ABI1, ABIG180D also inhibited the effect of SnRK2.6 and ABF2 on RD29B-LUC expression in response to ABA, but this antagonistic effect could not be overcome by expression of PYR1 (Fig. 1b, Supplementary Fig. 1a). This suggests that the ABIG180D mutant protein retains the inhibitory activity but can no longer be regulated. Thus reconstitution with PYR1, ABI1, SnRK2.6 and ABF2 is sufficient to enable ABA-mediated gene expression in protoplasts, providing in vivo evidence to our previously proposed model of ABA signaling12.
The PYR/PYL family consists of 14 members. Although genetic studies suggested redundancy in their function12, it is not known whether all members can act as ABA receptors and transduce the ABA signal to induce gene expression. To address this question, we reconstituted the ABA signaling pathway with different members of the PYR/PYL family. Our results show that all of the tested PYR1/PYLs could antagonize the ability of ABI1 to inhibit ABA-dependent induction of RD29B-LUC expression in snrk2.2/2.3/2.6 protoplasts, although not all PYR/PYL members were equally effective (Fig. 1c). The results suggest that all of the PYR/PYLs are likely to function as ABA receptors. We also tested reconstitution of the ABA signaling pathway with different combinations of SnRK2 kinases, PP2Cs and receptors, and found that the SnRK2 kinases are inhibited by both the ABI1 and HAB1 PP2Cs, and PYR1 or PYL2 can antagonize this inhibition. The inhibitory effect of ABI1 was stronger than that of HAB1 in the reconstituted ABA signaling system in protoplasts (Fig. 1b-d, Supplementary Fig. 1). The three clade A PP2Cs (ABI1, ABI2 and HAB1) were each capable of interacting with the three SnRK2 kinases (SnRK2.2, SnRK2.3 and SnRK2.6) in yeast two-hybrid (Y2H) assays, although with different intensities. For example, the ABI1 interaction was stronger than those of ABI2 and HAB1 (Supplementary Fig. 2a), which correlate with the level of inhibitory effect of ABI1 and HAB1 in the protoplast assay (Fig. 1d, Supplementary Fig. 1b). A C-terminally truncated SnRK2.6 lacking amino acids 280-362 did not interact with ABI1 (Supplementary Fig. 2a), which is consistent with previous studies demonstrating that deletion of a short C-terminal domain abrogates the interaction between ABI1 and SnRK2.6 in yeast19. Bimolecular fluorescence complementation (BiFC) assays in tobacco show that ABI1 interacts with the SnRK2s in the nucleus as well as the cytosol, and that the C-terminal region of SnRK2.6 is required for the interaction with ABI1 (Supplementary Fig. 2b). Expression of the fusion proteins was verified by immunoblot analysis (Supplementary Fig. 2c). The interaction between ABI1 and SnRK2.6 in vivo was further confirmed by a co-immunoprecipitation assay using the tobacco protein extracts (Supplementary Fig. 2c).
PYR/PYLs inactivate clade A PP2Cs in an ABA-dependent manner11-13. In protoplast transactivation assays, we showed that PYR/PYLs can reverse the inhibitory effect of PP2Cs (Fig. 1, Supplementary Fig. 1). We hypothesized that the PYR/PYLs may prevent the inhibitory effect of the PP2Cs by disrupting the interaction between the PP2Cs and the SnRK2s. We tested whether co-expression of PYLs might disrupt the interaction between PP2Cs and SnRK2s by yeast triple-hybrid assays. First, we reproduced the interaction of the ABI1, ABI2 and HAB1 PP2Cs (fused to the Gal4 activation domain (GAD)) with SnRK2.6 (fused to the Gal4 DNA binding domain (GBD)) by using the pBridge triple-hybrid vector (Supplementary Fig. 3). Next, we cloned into the SnRK2.6-pBridge construct the PYL5 and PYL8, which have been shown to act as potent inhibitors of the PP2Cs13. Nuclear localization of PYL5 and PYL8 in yeast is driven by fusion with a nuclear localization sequence present in the pBridge vector. Co-expression of PYL8 together with GBD-SnRK2.6 abrogated or reduced (depending on the dilution of the yeast culture) the interaction with GAD-ABI1 (Supplementary Fig. 3). Similar results were obtained when GBD-SnRK2.6 and GAD-ABI2 or GAD-HAB1 was tested with either PYL8 or PYL5, respectively (Supplementary Fig. 3). These results show that co-expression of a PYL impairs the interaction of ABI1, ABI2 and HAB1 PP2Cs with SnRK2.6.
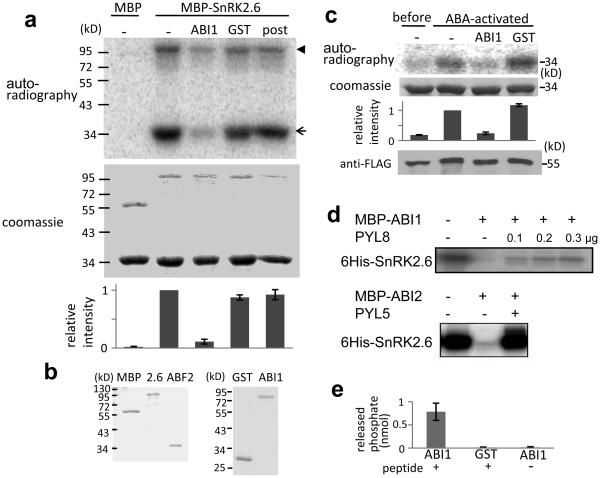
We have reconstituted the apparent entire ABA signaling pathway for stress responsive gene expression by co-expression of the PYR/PYLs, PP2Cs, SnRK2s and ABF2 in Arabidopsis protoplasts (Fig. 1b-d, Supplementary Fig. 1). To verify whether these are the minimal signaling components that are both necessary and sufficient for ABA signaling in the absence of other cellular components, we attempted to reconstitute the pathway in vitro. We constructed recombinant MBP-SnRK2.6, and found that it is capable of phosphorylating an ABF2 fragment as well as autophosphorylation (Fig. 2a and b). Incubation of GST-ABI1 but not GST with SnRK2.6 before the kinase assay substantially decreases ABF2 phosphorylation by the recombinant SnRK2.6 (Fig. 2a). SnRK2.6 pulled down from extracts of ABA-treated plants is also active in phosphorylating ABF2 but SnRK2.6 from untreated plants is not. This phosphorylation is also inhibited by GST-ABI1 (Fig. 2c). ABI1 added after ABF2 phosphorylation by SnRK2.6 is not as effective in reducing the level of phosphorylation (Fig. 2a), suggesting that ABI1 inhibits ABF2 phosphorylation by dephosphorylating SnRK2.6 (Fig. 2a). Indeed, we found that both ABI1 and ABI2 efficiently dephosphorylated SnRK2.6 (Fig. 2d). The autophosphorylated Ser175 is essential for the kinase activity of SnRK2.6 in vitro24. We tested and found that ABI1 can dephosphorylate a synthetic phosphopeptide corresponding to amino acids His170-Pro180 of SnRK2.6, which is phosphorylated at Ser175 (HSQPKpSTVGTP; Fig. 2e). These results suggest that ABI1 may deactivate SnRK2.6 by dephosphorylating Ser175.
Figure 2. ABI1 and ABI2 inhibit SnRK2.6 by dephosphorylation.
a, SnRK2.6 is deactivated by ABI1. MBP or MBP-SnRK2.6 treated without (−) or with GST-ABI1 or GST was incubated with GST-ABF2 fragment (amino acids Gly73 to Gln119) in the presence of [γ32P]-ATP. In the furthest right lane (post), GST-ABI1 was added after phosphorylation of GST-ABF2 fragment by MBP-SnRK2.6. Bands of GST-ABF2 fragment and MBP-SnRK2.6 are indicated by an arrow and an arrowhead, respectively. Radioactivities of GST-ABF2 fragment bands were measured with a phospho-imager and were normalized, taking the radioactivity of the band by MBP-SnRK2.6 without ABI1 treatment as 1 (mean ± s.e.m., n = 5). b, Coomassie staining of purified MBP, SnRK2.6, ABF2, GST and GST-ABI1. c, FLAG-SnRK2.6 extracted from transgenic plants before and after ABA treatment was used instead of MBP-SnRK2.6 in (a). Coomassie staining, autoradiography and relative radioactivities (mean ± s.e.m. n = 5) of GST-ABF2 fragment are shown. Western blotting with anti-FLAG antibody shows FLAG-SnRK2.6 protein amount. d, Autoradiography of autophosphorylated SnRK2.6 showing dephosphorylation of SnRK2.6 by MBP-ABI1 and MBP-ABI2 and the effect of PYL8 and PYL5, respectively, in the presence of 1 μM ABA. e, Phosphate release from synthetic peptide HSQPKpSTVGTP, corresponding to amino acids 170-180 of SnRK2.6.
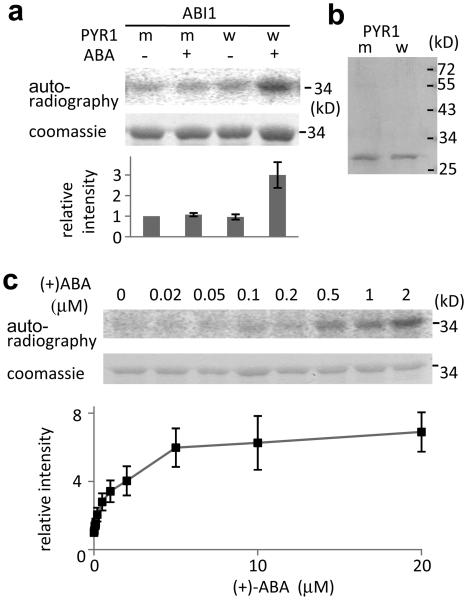
When His-PYR1 is incubated together with GST-ABI1 and MBP-SnRK2.6, SnRK2.6-mediated phosphorylation of ABF2 is significantly recovered in the presence of 2 μM (+)-ABA (Fig. 3a and b). Without ABA, His-PYR1 cannot reverse the inhibitory effect of ABI1 on SnRK2.6-mediated phosphorylation of ABF2 (Fig. 3a). PYR1P88S, which cannot bind to and inhibit ABI112, is not capable of reversing the inhibitory effect of ABI1 even in the presence of ABA (Fig. 3a). We found that in the presence of ABA, PYL8 or PYL5 can prevent the dephosphorylation of SnRK2.6 by ABI1 or ABI2 (Fig. 2d). These data are consistent with results from the protoplast transactivation assays, and show that it is possible to reconstitute ABA activation of ABF2 phosphorylation in vitro. Importantly, ABF2 phosphorylation status in this reconstituted in vitro system responds to ABA in a concentration-dependent manner (Fig. 3c). The apparent EC50 of this response is 0.8 μM, which is similar to the IC50 value for ABA inhibition of seed germination11 and falls within the physiological range of ABA concentrations in plants. Similar ABA responses were observed when ABA-activated SnRK2.6 isolated from plants instead of recombinant SnRK2.6 was used in the reconstitution assay (Supplementary Fig. 4a). Furthermore, reconstitution was also achieved when the PP2C protein HAB1 was used instead of ABI1 (Supplementary Fig. 4b). Our protoplast and in vitro reconstitution results support a model in which PYR1 (and PYLs) binds ABA, and then interacts with and is able to inactivate the PP2Cs. The ABA-bound receptors also disrupt the interaction between the PP2Cs and the SnRK2 kinases. These actions of the receptors prevent the dephosphorylation and thereby relieve inhibition of the SnRK2s by the PP2Cs. The relieved SnRK2s can then phosphorylate ABFs to activate ABA-responsive genes.
Figure 3. The combined effect of ABA, PYR1 and ABI1 on SnRK2.6 phosphorylation of GST-ABF2 fragment in vitro.
a, Reconstitution of ABA regulation of ABF2 phosphorylation. MBP-SnRK2.6 treated with GST-ABI1 and His-tagged wild type PYR1 (w) or mutated PYR1P88S (m) in the absence (−) or presence (+) of 2 μM (+)-ABA was incubated with GST-ABF2 fragment (amino acids Gly73 to Gln119) in the presence of [γ32P]-ATP. Coomassie staining, autoradiography and relative radioactivities of GST-ABF2 fragment are shown. Radioactivities of GST-ABF2 fragment were normalized, taking the radioactivity of the band with PYR1P88S in the absence of ABA as 1 (mean ± s.e.m., n = 5). b, Coomassie staining of PYR1 (w) and PYR1P88S (m). c, ABA dose response. MBP-SnRK2.6, GST-ABI1 and His-PYR1 were incubated with different concentrations of (+)-ABA before the kinase assay using GST-ABF2 fragment as substrate. Coomassie staining, autoradiography and relative radioactivities (taking the radioactivity of the band in the absence of ABA as 1; mean ± s.e.m., n = 9 for < 5 μM, n = 4 for 5 μM or more) of GST-ABF2 fragment are shown.
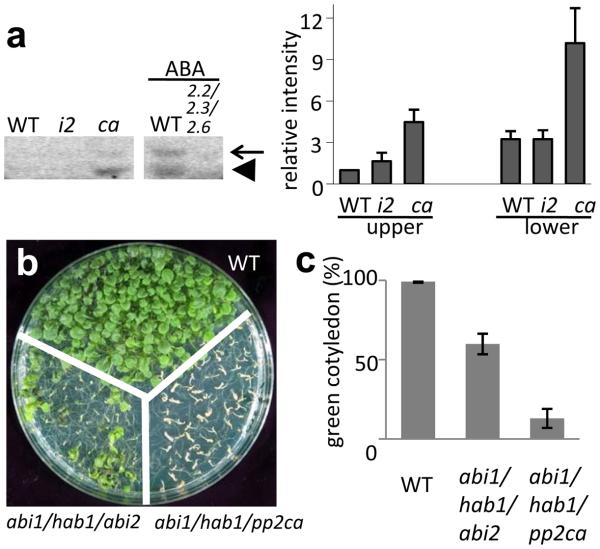
Consistent with our model, we showed previously that the SnRK2s are substantially less activated by ABA in the pyr1pyl1pyl2pyl4 mutant compared with the wild type12. The model also predicts that the SnRK2s may be constitutively activated in mutant plants that are deficient in the PP2Cs. Indeed, the PP2C triple mutant abi1-2hab1-1pp2ca-1 shows a constitutive activation of 42 and 45 kD kinases, which correspond to SnRK2.2/2.3 and SnRK2.6, respectively (Fig. 4a). This mutant displays a constitutive ABA response phenotype in germination and early seedling development (Fig. 4b and c), as reported previously25. In contrast, the PP2C triple mutant abi1hab1abi2 does not have a constitutive ABA response as strong as in abi1-2hab1-1pp2ca-1 (Fig. 4b and c), and does not show a strong constitutive activation of the SnRK2s (Fig. 4a).
Figure 4. Effect of PP2C mutations on ABA response phenotypes and kinase activities of SnRK2s.
a, In-gel kinase assay showing the activities of SnRK2s in the abi1/hab1/abi2 (i2) and abi1/hab1/pp2ca (ca) triple mutants. The snrk2.2/2.3/2.6 was used as a control. GST-fused ABF2 fragment (amino acids Gly73 to Gln119) was used as the phosphorylation substrate. The expected positions of SnRK2.6 and SnRK2.2/2.3 are indicated by arrow and arrowhead, respectively. Radioactivities of the upper and lower bands were normalized, taking the radioactivity of the upper band in WT as 1 (mean ± s.e.m., n = 3). b, The PP2C triple mutants show ABA hypersensitivity during germination and early seedling development. Photograph of plants of the indicated genotypes growing on MS medium with 3% sucrose for 14 d. c, The percentage of seedlings with green cotyledons 6 d after the end of stratification is shown (mean ± s.e.m., n = 3).
To our knowledge, this is the first report of in vitro reconstitution of a phytohormone signal transduction pathway using recombinant proteins. The in vitro reconstitution results are supported by the reconstitution assays in the protoplasts and by genetic analysis. The protoplast reconstitution assays enabled us to test the functions of nearly all members of the PYR/PYL family. Our results suggest that all in the family can function as ABA receptors in inducing gene expression. Although each of the proteins used in the reconstitution assays has been studied previously, it was not known how these components may connect to form a signaling pathway. Our study has revealed significant new insights into the mechanisms of action of these components. Our results suggest that the default state of the SnRK2 protein kinases is an autophosphorylated, active state, and that the SnRK2 kinases are kept inactive by the PP2Cs through physical interaction and dephosphorylation. We found that upon binding to ABA, the PYR/PYL receptor protein can disrupt or reduce the interaction between the SnRK2s and PP2Cs, and prevent the PP2Cs-mediated dephosphorylation of the SnRK2s, thus resulting in the activation of the SnRK2 kinases.
Successful reconstitution with the recombinant proteins implies that we have identified all essential core components of an ABA response pathway from hormone perception to phosphorylation of ABFs. Although ABA signaling in plants has been considered to be very complicated with numerous other proteins involved, our study reveals a surprising simplicity of the pathway and demonstrates that the PYR/PYLs, clade-A PP2Cs, SnRK2s, and ABFs are the only core components to complete the ABA regulation of gene expression. Since there are multiple family members for each of these core components, many combinations of them are possible. The functions of the family members may overlap, but their unique spatial and temporal expression patterns may confer some distinct functions in specific tissues. Extensive genetic analysis will be necessary to determine the in planta importance of specific combinations of the core components.
We suggest that the other proteins previously identified as involved in ABA responses, may function to modulate the expression and/or activities of one or more of the core components defined here. Calcium and reactive oxygen signaling, RNA metabolism and protein degradation are known to have important roles in regulating ABA sensitivity2-4,14,26,27. It will be of great interest to determine how these processes may connect to one or more of the core components to impact ABA responses. It will also be interesting to determine whether other ABA response pathways such as ABA regulation of ion channels in guard cells2,3,6 may also use components of the PYR/PYLs-PP2C-SnRK2 regulatory module and whether additional receptors and core signaling components are involved.
Methods Summary
Transient activity assays were performed in Arabidopsis mesophyll protoplasts from Columbia wild-type or SnRK2.2/2.3/2.621 as described previously (http://genetics.mgh.harvard.edu/sheenweb)28. Transfected protoplasts were incubated for 5 h in light in the presence of 0 (open bars) or 5 μM (+) ABA, and then used for measuring LUC and GUS activities as described previously28. Yeast two-hybrid and triple-hybrid assays, co-immunoprecipitation and BiFC assays were similar to those described previously13. Purification of GST-HAB1, His-PYR1 and His PYR1P88S was carried out as described previously12. GST, GST-ABI1, GST-ABF2 fragment, MBP and MBP-SnRK2.6 constructs were transformed into E. coli Rosetta cells (Novagen) and the recombinant proteins isolated by affinity purification. Purification of MBP-ABI1, MBP-ABI2, His-PYL8, His-PYL5 and His-SnRK2.6 was as described previously13. In-gel kinase assays were performed as described previously20 with the modification that 300 μg protein was loaded for samples without ABA treatment. For germination assays, seeds were plated on MS medium containing 3% sucrose. In each experiment, at least 50 seeds per genotype were stratified at 4 °C for 3 d, and the presence of green cotyledons was scored after 6-day incubation at 23 °C.
Methods
Plant culture
For protoplast isolation, Arabidopsis thaliana ecotype Columbia and snrk2.2/2.3/2.6 triple mutant plants were grown on Jiffy7 soil (Jiffy Products Ltd., Canada) in an environment-controlled chamber (illumination of 75 μmol m−2 s−1, photoperiod of 13 h light/11 dark, 22 °C).
Plasmid constructs for protoplast transient assay
The RD29B promoter region11 was PCR amplified form Columbia genomic DNA and cloned by replacing CBF3 promoter in CBF3-LUC protoplast expression vector. CBF3-LUC, UQ10-GUS, ABI1 and ABI1G180D protoplast expression plasmid vectors were kindly provided by Jen Sheen (Massachusetts General Hospital, Boston, MA 02114). ABI1 was replaced with His-PYR1/His-PYLs/His-PYRP88S, HAB1-myc, ABF2-HA/ABF2 mutant versions, SnRK2-Flag/SnRK2.6K50N sequences. All the plasmids were sequenced to confirm the sequence and avoid cloning errors. β-glucuronidase (GUS) reporter plasmid was used as internal control to normalize transfection efficiency in protoplast assays. Primers used for preparing protoplast expression vectors are as follows:
PRD29B-F: CGGGATCCGTTGAATCTTGCGGAAGCA
PRD29B-R: CATGCCATGGTTCAAGTGAATCAATCAAAC
HAB1-F: CATGCCATGGAGGAGATGACTCCCGCAG
HAB1-R1: CAGATCCTCCTCAGAAATCAGCTTTTGCTCAGGCCTGGTTCTGGTCTTGAACTTTC
HAB1-R2: GCCTGCAGTCATAAGTCTTCTTCGCTTATTAATTTCTGTTCCAGATCCTCCTCAGAAA
ABF2-F: CGGGATCCATGGATGGTAGTATGAATTTGG
ABF2-R: GTCAAGGCCTCCAAGGTCCCGACTCTGTCCT
SnRK2.2-F: CATGCCATGGATCCGGCGACTAATTCAC
SnRK2.2-Flag-R: GTCCTTGTAGTCAGAAGGCCTGAGAGCATAAACTATCTC
SnRK2.2-Flag-Pst-R GCCTGCAGTCACTTGTCATCGTCGTCCTTGTAGTCAGAAG
In SnRK2.2-Flag plasmid, SnRK2.2 was replaced with SnRK2.3 or SnRK2.6.
SnRK2.3-F: CATGCCATGGATCGAGCTCCGGTGACCA
SnRK2.3-R: GTCAAGGCCTGAGAGCGTAAACTATCTCTCCGCT
SnRK2.6-F: CATGCCATGGATCGACCAGCAGTGAGT
SnRK2.6-R: GTCAAGGCCTCATTGCGTACACAATCTCTC
His-PYR1 was constructed with following primers:
His- PYR1-F1: CACCATCACCCATGGATGCCTTCGGAGTTAACACCAG
His-PYR1-F2: CGGGATCCATGAAACATCACCATCACCATCACCCATGGATG
PYR1-R: GTCAAGGCCTTCACGTCACCTGAGAACCACTTC
In His-PYR1 plasmid, PYR1 was replaced with PYLs with following primers:
PYL1-F: CATGCCATGGATGGCGAATTCAGAGTCCTCCTC
PYL1-R: GTCAAGGCCTTTACCTAACCTGAGAAGAGTTG
PYL2-F: CATGCCATGGATGAGCTCATCCCCGGCCGTG
PYL2-R: GTCAAGGCCTTTATTCATCATCATGCATAGGTG
PYL3-F: CATGCCATGGATGAATCTTGCTCCAATCCATG
PYL3-R: GTCAAGGCCTTCAGGTCGGAGAAGCCGTGGA
PYL4-F: CATGCCATGGATGCTTGCCGTTCACCGTCCTTC
PYL4-R: GTCAAGGCCTTCACAGAGACATCTTCTTCTTG
PYL5-F: CATGCCATGGATGAGGTCACCGGTGCAACTC
PYL5-R: GTCAAGGCCTTTATTGCCGGTTGGTACTTCGAG
PYL6-F: CATGCCATGGATGCCAACGTCGATACAGTTTC
PYL6-R: GTCAAGGCCTTTACGAGAATTTAGAAGTGTTCTC
PYL7-F: CATGCCATGGATGGAGATGATCGGAGGAGAC
PYL7-R: GTCAAGGCCTTCAAAGGTTGGTTTCTGTATGA
PYL8-F: CATGCCATGGATGGAAGCTAACGGGATTGAGAAC
PYL8-R: GTCAAGGCCTTTAGACTCTCGATTCTGTCGTG
PYL9-F: CATGCCATGGATGATGGACGGCGTTGAAGG
PYL9-R: GTCAAGGCCTTCACTGAGTAATGTCCTGAGAAG
PYL10-F: CATGCCATGGATGAACGGTGACGAAACAAAGAAG
PYL10-R: GTCAAGGCCTTCATATCTTCTTCTCCATAGATTCTG
PYL11-F: CATGCCATGGATGGAAACTTCTCAAAAATATC
PYL11-R: GTCAAGGCCTTTACAACTTTAGATGAGCCACC
PYL12-F: CATGCCATGGATGAAAACATCTCAAGAACAGCA
PYL12-R: GTCAAGGCCTTTAAGTGAGCTCCATCATCTTC
In vitro mutagenesis
ABF2-HA plasmid was used for in vitro mutagenesis by using The GeneTailor™ Site-Directed Mutagenesis System (Catalog no. 12397-014, Invitrogen) with the following mutagenic primers:
ABF2S26-R: ACCTTGTCTAGTCAACCCTCCACCTCCACCAC
ABF2S26A-F: AGGGTTGACTAGACAAGGTGCTATATACTCGTTG
ABF2S26D-F: AGGGTTGACTAGACAAGGTGATATATACTCGTTG
ABF2S86-R: GCCTTGCCTCTGCAGCTGCAAACCCTCTTG
ABF2S86A-F: CAGCTGCAGAGGCAAGGCGCTTTGACTCTGCCTC
ABF2S86D-F: CAGCTGCAGAGGCAAGGCGATTTGACTCTGCCTC
ABF2S94-R: AAGCGTTCGAGGCAGAGTCAACGAGCCTTG
ABF2S94A-R1: AAGCGTTCGAGGCAGAGTCAAAGCGCCTTG
VC-ABF2S94D-R1: AAGCGTTCGAGGCAGAGTCAAATCGCCTTG
ABF2S94A-F: ACTCTGCCTCGAACGCTTGCTCAGAAGACGGTTG
ABF2S94D-F: ACTCTGCCTCGAACGCTTGATCAGAAGACGGTTG
ABF2T135-R: TTGCTGCCTCTGACTCTGACTCTGACTCTG
ABF2T135A-F: TCAGAGTCAGAGGCAGCAAGCTTTAGGTGAAGTA
ABF2T135D-F: TCAGAGTCAGAGGCAGCAAGATTTAGGTGAAGTA
Protoplast isolation and transactivation assay
About four-week-old plants were used for protoplast isolation as described previously (http://genetics.mgh.harvard.edu/sheenweb)28. All the chemicals used in protoplast isolation were obtained from Sigma. About 0.5–1 mm width leaf strips cut from the middle part of leaves were vacuum infiltrated with enzyme solution containing 20 mM MES (pH 5.7), 1.5% (wt/vol) cellulase R10 (Yakult Pharmaceutical Ind. Co., Ltd., Japan), 0.4% (wt/vol) macerozyme R10 (Yakult Pharmaceutical Ind. Co., Ltd., Japan), 0.4 M mannitol and 20 mM KCl, 10 mM CaCl2, 1 mM β-mercaptoethanol (optional) and 0.1% BSA. After 30 minutes infiltration, leaf strips in enzyme solution was incubated in dark for 3h. Protoplasts were diluted with W5 (2 mM MES pH 5.7 containing 154 mM NaCl, 125 mM CaCl2 and 5 mM KCl) solution to a final concentration of 2 × 105 cells mL-1. After 30 min resting, W5 solution was removed and protoplasts were resuspended in MMg solution (4 mM MES pH 5.7 containing 0.4 M mannitol and 15 mM MgCl2). For transfection, 100 μL of protoplasts (2 × 104 cells) were mixed with plasmid constructs (in 10 μL) and 110 μl of PEG solution (40%, wt/vol, PEG4000 in ddH2O containing 0.2 M mannitol and 100 mM CaCl2). After 10 minute incubation, transfection was stopped by adding 440 μl of W5 solution. Protoplasts were harvested by centrifugation at 100 × g for 2 min at room temperature and resuspended in about 100 μl of W5 solution. Then, protoplasts were incubated WI solution (4 mM MES pH 5.7 containing 0.5 M mannitol and 20 mM KCl) with 0 or 5 μm (+) ABA for 5 h. Protoplasts were harvested by centrifugation at 100 × g for 2 min, frozen in liquid N2 and store at −80 °C until further use. The frozen protoplasts were resuspended in 100 μl of protoplast lysis buffer (2.5 mM Tris–phosphate pH 7.8 containing 1 mM DTT, 2 mM DACTAA, 10% (vol/vol) glycerol and 1% (vol/vol) Triton X-100). 100 μl LUC mix (Promega) was added in 10 μl of the lysate and LUC activity was measured with a plate reader (Wallac VICTOR2 plate reader). 2 μl of the protoplast lysate was mixed with 10 μl of MUG substrate mix (10 mM Tris–HCl pH 8 containing 1 mM MUG and 2 mM MgCl2) and incubated for 30 min at 37 °C. The reaction was stopped by adding 10 μl of 0.2 M Na2CO3 and the fluorescence of MU was quantified using a plate reader (Wallac VICTOR2 plate readers).
The RD29B promoter fused with LUC coding sequence was used as an ABA-responsive reporter gene (7 μg plasmid per transfection). ZmUBQ::GUS was included in each sample as an internal control (3 μg per transfection). ABF2-HA and its mutant versions, SnRK2.6-Flag and SnRK2.6K50N-Flag, His-PYR/PYLs and HAB1-myc plasmid constructs were used at 3 μg per transfection, while ABI1 was used at 2 μg per transfection.
In vitro phosphatase and kinase assays
MBP-SnRK2.6 (1 μg), MBP (1 μg) or FLAG-SnRK2.6 on beads was incubated with GST-ABI1 (1 μg), GST (1 μg), His-PYR1 (1 μg) and/or His-PYR1P88S (1 μg) as indicated in 40 μl phosphatase buffer (50 mM Tris-HCl, pH 7.0, 60 mM magnesium acetate, 0.1 mM EGTA and 0.1% β-mercapto ethanol) containing indicated concentration of (+)-ABA (Biosynth AG, Switzerland) at 30 °C for 20 min. After removal of the solution, GST-ABF2 fragment (10 μg) was added. The reaction mixture (20 μl) contained 25 mM Tris (pH 7.4), 12 mM MgCl2, 1 μM ATP, 5 μCi [γ-32P] ATP, 1 mM DTT, 1mM Na3VO4, 5 mM NaF. The reaction mixtures were incubated at 30 °C for 30 min. The reaction was stopped by the addition of Laemmli's sample buffer, and the mixtures were then subjected to SDS-polyacrylamide gel electrophoresis. Radioactivity was detected with a Typhoon phosphoimager (GE Healthcare). SnRK2.6 kinase deactivation assays were done by previous incubation with protein phosphatase for 30 minutes at room temperature. Assays to test recovery of SnRK2.6 activity were done by incubation for 10 min of the protein phosphatase together with the receptors in the presence of 1 μM (+) ABA.
To measure phosphate release from synthetic phosphopeptide, the phospho peptide (10 nmol) from Biomatik corporation (DE) was incubated with 1 μg of ABI1 or GST in the phosphatase buffer (40 μl) at 30 °C for 10 min. Released phosphate was measured as described previously29.
Construction of plasmids for yeast two-hybrid and triple-hybrid analysis
The coding sequence of SnRK2.6, SnRK2.2, SnRK2.3 was amplified by PCR using the following pairs of primers: F4g33950 and R4g33950, F3g50500 and R3g50500, F5g66880 and R5g66880, respectively. The PCR product was initially cloned into pCR8/GW/TOPO, doubly digested EcoRI-SalI and cloned into pGBT9, where the kinase coding sequences were fused to the GAL4 DNA-binding domain (GBD). pGBT9 constructs where SnRK2s acted as baits were faced in two-hybrid assays with the following pGADT7 constructs: ABI1, ABI2 and HAB1, where the PP2C coding sequences were fused to the GAL4 activation domain (GAD).
To perform triple-hybrid experiments, the SnRK2.6 EcoRI-SalI fragment described above was cloned into the pBridge vector (Clontech), where the kinase coding sequence is fused to GBD. Next, the coding sequences of PYL8 and PYL5 were cloned into the NotI site of pBridge-OST1. To reproduce the yeast two-hybrid interaction described above, yeast host AH109 was co-transformed with one of the following plasmids, pGADT7-ABI1/pGADT7-ABI2/pGADT7-HAB1, encoding GAD-PP2C fusions and pBridge-OST1, encoding a GBD-OST1 fusion; whereas to test the interference of ABA-receptors on this interaction, pBridge-OST1+PYL8 and pBridge-OST1+PYL5 were employed
BiFC
Experiments were performed as described previously13. Constructs were made in the pSPYNE-35S30 as well as the Gateway vector pYFPC43 (kindly provided by A. Ferrando). ABI1 coding sequence was excised from pCR8/GW/TOPO as a BamHI-SmaI fragment and cloned into pSPYNE-35S. The coding sequences of SnRK2.6, SnRK2.2, SnRK2.3 and ΔCOST1, previously cloned into pCR8/GW/TOPO, were recombined by LR reaction into pYFPC43 destination vector.
Immunoprecipitation
Protein extracts for immunodetection experiments were prepared from N. benthamiana leaves infiltrated with Agrobacterium tumefaciens C58C1 (pGV2260) transformed with the different fusion proteins containing the SnRK2.6/OST1, the C-terminal deletion of SnRK2.6, the N-terminal deletion and full-length ABI1. Protein extracts from the transfected plants were used for immunoprecipitation at 4 °C with super-paramagnetic MACS microbeads coupled to monoclonal anti c-myc antibody (http://www.miltenyibiotec.com). The antigen-antibody complex was eluted, fractionated on an 8% SDS-PAGE gel and transferred onto Immobilon-P membranes (Millipore), and probed either with monoclonal anti-c-myc (clone 9E10, Roche) or anti-GFPC (clone JL-8, Clontech) as primary antibodies and ECL anti-mouse-peroxidase (GE Healthcare) as secondary antibody. Detection was performed using the ECL advance western blotting detection kit (GE Healthcare).
To immunoprecipitate SnRK2.6, three weeks old seedlings of transgenic plants expressing FLAG-SnRK2.6 treated with 100 μM ABA was used. Protein extracts were incubated with anti-FLAG antibody beads (Sigma) for 2 h at 4 °C. The beads were washed six times with the prechilled extraction buffer.
Supplementary Material
Acknowledgements
This work was supported by a National Institutes of Health grant (J-K Z) and MICIIN and CSIC fellowships (SR and RA). We thank Ray Bressan for helpful discussions and editing of the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper on www.nature.com/nature.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant. Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hetherington AM. Guard cell signaling. Cell. 2001;107:711–714. doi: 10.1016/s0092-8674(01)00606-7. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering of drought hardiness in plants. Nature. 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- 4.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002;5:33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 6.Roelfsema MRG, Hedrich R. In the light of stomatal opening – new insights into “the Watergate”. New Phytol. 2005;167:665–691. doi: 10.1111/j.1469-8137.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- 7.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 8.Shen YY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, et al. G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 10.Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 12.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago J, et al. Modulation of drought resistance by the abscisic acid-receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03981.x. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Assmann SM, Albert R. Predicting essential components of signal transduction networks: A dynamic model of guard cell abscisic acid signaling. PLoS Biol. 2006;4(10):e312. doi: 10.1371/journal.pbio.0040312. doi:10.1371/journal.pbio.0040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCourt P, Creelman R. The ABA receptors-we report you decide. Curr. Opin. Plant Biol. 2008;11:474–478. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Pennisi E. Stressed out over a stress hormone. Science. 2009;324:1012–1013. doi: 10.1126/science.324_1012. [DOI] [PubMed] [Google Scholar]
- 17.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida R, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida R, et al. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 20.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii H, Zhu JK. Arabidopsis mutant deficient in three abscisic acid-activated protein kinases reveals critical roles in growth, reproduction and stress. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson RR, Wagner RL, Verhey SD, Walker Simmons MK. The abscisic acid responsive kinase PKABA interacts with a seed-specific abscisic acid response elementbinding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belin C, et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubio S, et al. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn JM, Schroeder JI. Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 2003;6:463–469. doi: 10.1016/s1369-5266(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 27.Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: A versitile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 29.Van Veldhoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]
- 30.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.