Narcolepsy-cataplexy, characterized by sleepiness and rapid onset into REM sleep, affects 1 in 2,000 individuals1,2. Narcolepsy was first shown to be tightly associated with HLA-DR23, and later sublocalized to DQB1*06024. Following studies in dogs5 and mice6, a 95% loss of hypocretin-producing cells in human postmortem hypothalami was reported7,8, Using Genome Wide Association (GWA) in Caucasians with replication in three ethnic groups, we found association with polymorphisms in the T-Cell receptor alpha (TCRA) locus, with highest significance at rs1154155 (average allelic odds ratio 1.69, genotype odds ratios 1.94 and 2.55, p<10−21, 1830 cases, 2164 controls). This is the first documented genetic involvement of the TCRA locus, the major receptor for HLA-peptide presentation, in any disease. It is still unclear how specific HLA alleles confer susceptibility to over 100 HLA-associated disorders9, thus narcolepsy will provide new insights on how HLA-TCR interactions contribute to organ specific autoimmune targeting.
An autoimmune etiology has been suggested for narcolepsy but never proven despite decades of intensive research10,11. Narcolepsy is recognized to be familial and despite the association with DQB1*0602 not fully explained by the HLA locus1. To identify additional susceptibility loci for narcolepsy, we undertook a GWA study. We selected Caucasian cases from Europe and the United States, together with geographically and ethnically matched controls. All cases were HLA-DQB*0602 positive and all had clear-cut cataplexy. Among the 23% on whom we had hypocretin-1 levels, all were found to be hypocretin deficient. Potential controls were typed using Sequence Specific PCR, and only those who were also DQB1*0602 positive were included. The sample was comprised of 807 cases and 1074 controls of mixed European ancestry; 415 cases and 753 controls were recruited from the US and Canada; 392 cases and 321 controls were recruited from European centers. For the GWA study, subjects were genotyped using the Affymetrix Mapping 500K array set or Genome-Wide SNP Array 6.0. Ethnic homogeneity and case/control matching was verified by cluster and principal component analysis12. In addition we compared the allele frequency of 107 of 400 Single Nucleotide Polymorphisms (SNPs) known to predict European substructure and found no significant differences after Bonferroni correction13.
We conducted allele-based association tests in SNPs with allele frequency above 5% in controls using the Mantel-Haenszel (MH) test14 in 3 groups of subjects defined by platform (Affymetrix 500K versus 6.0 typed at UCSF) and location of typing (Affymetrix 6.0 at Institut für Humangenetik, Munich, Germany). The χ2 Quantile-Quantile plot showed a slight deviation from the expected chi-square distribution, and an inflation factor λ of 1.11 was estimated (Supplementary Fig. 1). However, the plot also showed the presence of 3 extreme outlier χ2 values of 47.7, 54.1 and 60.4 (Supplementary Fig. 1, Table 1). These 3 SNPs, all on chromosome 14, clearly exceeded the genome-wide significance level of 9.1×10−8. Other nominally significant associations (p<1×10−6) are reported in Supplementary Table 1.
Table 1.
SNP markers of interest from the association study
| SNP | CHR | Position (bp) | Minor Allele |
Freq Controls (n) |
Freq Cases (n) |
χ2(MH) | P (MH) | OR (95% CI) | χ2 (BD) | P (BD) |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1154155 | 14 | 22072524 | C | 0.14 (1067) | 0.24 (796) | 54.11 | 1.90 × 10−13 | 1.87 (1.58-2.21) | 2.49 | 0.29 |
| rs12587781 | 14 | 22069457 | C | 0.15 (917) | 0.25 (622) | 53.19 | 3.03 × 10−13 | 1.96 (1.63-2.35) | 1.66 | 0.20 |
| 0.14 (1066)* | 0.24 (794)* | 60.42* | 7.65 × 10−15* | 1.93 (1.63-2.28)* | 1.61* | 0.45* | ||||
| rs1263646 | 14 | 22087370 | G | 0.16 (1069) | 0.26 (797) | 47.74 | 4.86 × 10−12 | 1.77 (1.50-2.09) | 0.40 | 0.82 |
| rs5770917† | 22 | 49364219 | G | 0.05 (1063) | 0.046 (796) | 1.068 | 0.30 | 0.84 (0.61-1.16) | 0.39 | n.a. |
The top three genome-wide significant markers after bonferoni correction are listed, together with data obtained with the previously published rs5770917 marker, previously found to be associated in Japanese narcolepsy17. A total of 1074 Controls and 807 Narcolepsy cases were genotyped using SNP Affymetrix Array platforms (500K and 6.0).
Affymetrix 6.0K marker after genotypes were completed using TaqMan (see text). MH: Mantel-Haenszel; BD: Breslow Day heterogeneity test; OR: Odds Ratio.
Note that 388 of the 796 narcolepsy genotypes were previously reported for this marker by Miyagawa et al.17.
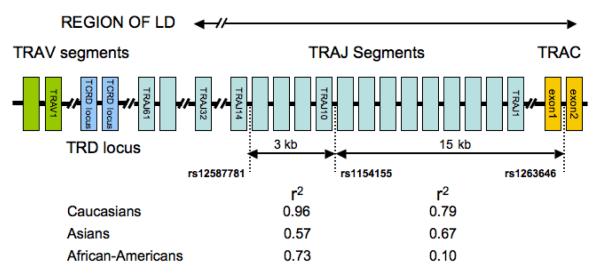
The 3 top markers were in high linkage disequilibrium (LD) and are located within an 18kb segment of the TCRA locus containing the TRA Joining (J) segment subregion (14q11.2, see Fig. 1). One of the nominally significant markers, rs17231, is located within the V segment region of the T-Cell Receptor Beta (TCRB) locus (7q34). Genome wide significant SNPs were genotyped using TaqMan assays (Applied Biosystems, Foster City, CA, USA) in an independent sample of 1057 cases (using the same diagnostic criteria), and 1104 controls (matched by ethnicity) as a replication study. The Caucasian replication sample contained 718 individuals, of whom 542 were recruited from the US and Canada (259 cases, 283 controls), and 176 from Europe (104 cases 72 controls). The Asian sample included 866 Japanese (433 cases, 433 controls) and 300 Koreans (128 cases, 172 controls). Finally, 277 African Americans were studied (133 cases, 144 controls). All subjects had given written informed consent approval.
Figure 1.
Schematic representation of the TCRA locus and of SNPs associated with narcolepsy. The TCRA locus consists of clusters of V and J segments and exons of the C region. The T-Cell Receptor delta locus (TRD) resides within the TCRA locus. A 40kb region of LD encompasses half of the TRAJ segments and is flanked by TRAJ32 and the second exon of the TRAC gene. Within this region, 3 SNPs are highly associated with narcolepsy, separated by 3 and 15 kb successively. In Caucasians, the association is equivalent with rs12587781 and rs1154155 (Tables 1 and 2), and LD is extremely high (r2=0.97 and 0,94 n=1154 cases, n=1425 controls, correlations calculated using Haploview). In contrast, the association is stronger with rs1154155 than rs12587781 in Asians (Table 2), a phenomenon explained by the lower LD in this ethnic group (r2=0.62 and 0.52, n=553 cases, n=603 controls). Intermediate LD was seen in African-American individuals (r2=0.74 and 0.71, n=124 cases, n=142 controls). The association with rs1263646 is weaker across all ethnic groups, most notably Asians and African Americans (Table 2). These results, depicted as values for cases and controls combined in this figure, illustrate the value of trans-ethnic mapping.
As shown in Table 2, the 3 SNPs located within the TCRA locus replicated with high significance across the 3 major ethnic groups combined, and showed significant effects individually in the Caucasians and Asians. In the African Americans, although the Odds Ratios (ORs) trended in the same direction, formal significance was not reached due to small sample size and low allele frequencies (Table 2).
Table 2.
Replication of SNP markers discovered in the GWA study
| Ethnicity | rs12587781 | rs1154155 | rs1263646 |
|---|---|---|---|
| Caucasians | C | C | G |
| Freq Controls (n) | 0.14 (352) | 0.14 (348) | 0.16 (351) |
| Freq Cases (n) | 0.22 (353) | 0.22 (343) | 0.24 (353) |
| χ 2 | 17.08 | 17.04 | 13.66 |
| P | 3.58 × 10−5 | 3.67 × 10−4 | 2.19 × 10−4 |
| OR (95% CI) | 1.79 (1.36-2.37) | 1.80 (1.36-2.39) | 1.65 (1.26-2.15) |
| Asians | C | C | G |
| Freq Controls (n) | 0.61 (601) | 0.47 (599) | 0.45 (600) |
| Freq Cases (n) | 0.68 (552) | 0.57 (549) | 0.51 (553) |
| χ 2 | 11.09 | 26.76 | 9.81 |
| P | 8.70 × 10−4 | 2.30 × 10−7 | 1.73 × 10−3 |
| OR (95% CI) | 1.34 (1.13-1.59) | 1.54 (1.31-1.82) | 1.30 (1.10-1.53) |
| African Americans | C | C | G |
| Freq Controls (n) | 0.11 (142) | 0.08 (138) | 0.13 (139) |
| Freq Cases (n) | 0.13 (124) | 0.10 (113) | 0.17 (124) |
| χ 2 | 0.70 | 0.74 | 1.08 |
| P | 0.40 | 0.39 | 0.30 |
| OR (95% CI) | 1.25 (0.74-2.13) | 1.31 (0.71-2.42) | 1.29 (0.80-2.09) |
Note: Frequencies at the 3 SNPs did not differ between DQB1*0602 positive (n=81) versus DQB1*0602 negative (n=271) controls within the subset of Caucasians with that information. In addition, allele frequency of these three SNPs did not differ between DQB1*0602 positive (n=470) and negative (n=1375) Caucasian controls.
Based on HapMap data (http://www.hapmap.org/)15, the 3 SNPs are located within a 37kb region of increased LD across ethnic groups (CEU, YRI, CHB-JPT). The localized haplotype block structure among these populations differs, with highest LD with rs12587781/rs1154155 extending in opposite directions in Europeans versus Asians. In all ethnic groups, rs1263646, a SNP located closer to the TRAC gene, showed a smaller OR, suggesting that the association peaks in the TRAJ segment region (Fig. 1). Further, ORs differed significantly for rs12587781 but not rs1154155 between Caucasians and Asians (Table 2). This was likely explained by the difference in LD patterns across the two ethnicities. Whereas rs1154155 and rs12587781 are in almost complete LD in Caucasians (r2=0.96), LD is substantially weaker in Asians (r2=0.57, Fig. 1). In Asians, rs1154155 had a stronger impact on risk (OR=1.54) than did rs12587781 (OR=1.34).
To further evaluate this, we estimated the frequency of haplotypes rs12587781-rs1154155 AA, AC, CA, CC in Asian cases and controls. For cases, the frequencies were 0.318, 0.003, 0.109 and 0.571, respectively. For controls, the frequencies were 0.381, 0.005, 0.154 and 0.460, respectively. We note that the OR is increased for haplotype CC (1.49, 95% CI 1.24-1.79) but not for haplotype CA (0.85, 95% CI 0.64-1.12). Thus, SNP rs12587781 appears to have no effect after controlling for SNP rs1154155, suggesting SNP rs1154155 may have functional significance, or is in high LD with another causative SNP nearby; SNPs with r2 >0.8 with rs1154155 are known to exist from HapMap data. This SNP is located 176bp 3′ to TRAJ10, a J segment without known coding polymorphisms. Genotype analysis suggested a dosage effect (CC vs. AA MH OR=2.55, 95% CI 1.92-3.38; AC vs. AA MH OR=1.94, 95% CI 1.68-2.25) (Table 3).
Table 3.
Analysis of rs1154155 Genotypes in Three Replication Cohorts and Combined
| Ethnicity | AA Case/Ctrl |
AC Case/Ctrl |
CC Case/Ctrl |
ORAC | ORCC | ORC |
|---|---|---|---|---|---|---|
| African Americans |
90/117 | 23/20 | 0/1 | 1.50 (0.74,3.04) |
0.00 (0.00,22.90) |
1.31 (0.68,2.52) |
| Asians | 86/161 | 296/318 | 167/120 | 1.74 (1.27,2.39) |
2.61 (1.81,3.76) |
1.54 (1.30,1.83) |
| Caucasians | 201/259 | 132/83 | 10/6 | 2.05 (1.45,2.89) |
2.15 (0.70,6.77) |
1.80 (1.35,2.41) |
| 3 Replication Samples (MH) |
1.83 (1.48,2.27) |
2.50 (1.80,3.48) |
1.59 (1.38,1.83)* |
|||
| All Samples (MH) |
1.94 (1.68,2.25) |
2.55 (1.92,3.38) |
1.69 (1.52,1.88)** |
χ2 = 42.9, P=5.9×10−11
χ2 = 94.2, P=2.8×10−22
Note: ORAC is the odds ratio for genotype AC versus AA; ORCC is the odds ratio for genotype CC versus AA; ORC is the odds ratio for allele C versus A.
Population attributable risks16 for TCRA rs1154155C in Caucasians and Asians were 20% and 42%, respectively. The increased frequency of rs1154155C in Asians likely contributes to the reported increased prevalence in Japan1 despite lower DQB1*0602 frequency4. Our identified TCRA rs1154155C polymorphism showed no interaction with the nominally significant TCRB rs17231T polymorphism of the GWA data (OR interaction=1.0). In our much larger sample, we also did not replicate a previously published rs5770917 association in Japanese narcolepsy (Table 1), suggesting an ethnic specific effect17. Further, interactions between rs5770917 and rs1154155 were non-significant in Caucasians, Asians, and African Americans (OR interaction=1.0 in all samples).
The TCRA locus encodes the of the TCRαβ-heterodimer, a protein expressed by T lymphocytes18. The T-cell receptor is a unique protein which interacts with both HLA class I (CD8 in cytotoxic T-cells) and HLA Class II (CD4 in helper T-cells), including the DQαβ heterodimer denoted DQ0602, encoded by DQB1*0602 and the closely linked DQA1*0102 allele. The TCRA locus, like the TCRB and the Immunoglobulin variable heavy and light chain loci, is unusual in undergoing somatic cell recombination. TCRA and TCRB recombination occur in the thymus, resulting, after deletion of auto reactive clones and positive selection, in the generation of T-cell clones with unique TCRA and TCRB recombined loci. In the TCRA locus, recombination occurs between the 5′ area of one of the 46 functional Variable (V) segments19and the 3′ area of one of the 49 functional J segments20,21,22, with additional amino acid junctional diversity generated by N- and P-additions in the V-J border region. In the TCRB locus, diversity is even more complex and generated by recombination of 48V, 2D and 13J segments22. This mechanism produces a diverse repertoire of distinct TCRαβ idiotype bearing T-cells21, which can be called upon to recognize antigens presented by HLA class I or class II molecules23.
Unlike most other autoimmune diseases9, narcolepsy is almost completely associated with a single HLA allele, DQB1*0602, across Caucasians, Asians and African Americans4. Considering the tight DQB1*0602 association in narcolepsy, it is logical to hypothesize that the DQB0602 heterodimer should interact with a specific TCRαβ receptor subtype whose occurrence is marked by rs1154155C, and less strongly by rs17231T at both TCR loci. This TCR idiotype would bear specific VJα and VDJβ recombinants, with recognition of a peptide that also binds DQ0602, mediating further immune reaction leading to the destruction of hypocretin-producing cells. Precisely how a J segment region polymorphism such as rs1154155C could increase the risk of occurrence of this narcolepsy associated T-cell clone is unknown, but could involve non-random VJα choices in recombination21, as previously reported. Similarly, a polymorphism in the TCRB V region could influence VDJ recombination for the complementary TCRβ chain. Less probably, the TCR-DQ association could also occur without the need for peptide binding, through superantigen-like bridging of TCR and DQ, although most known superantigens interact with TCRβ rather than chains24. Further, superantigen bridging typically results in stimulation of large systemic lymphocyte populations carrying specific TCRB segments such as that seen in toxic shock syndrome.
Surprisingly, of over 10 HLA associated autoimmune diseases that have been subjected to genome-wide analyses and candidate gene studies, none has shown consistent association with either TCR locus25. Further studies of the TCR loci in narcolepsy may for the first time reveal a role for a specific TCR receptor idiotype in the pathophysiology of an autoimmune disorder.
Methods
Cases and Controls
Narcolepsy patients were selected as described, 98% of whom are predicted to be hypocretin deficient. The initial Caucasian sample was comprised of 807 cases and 1074 controls of mixed European ancestry; 415 cases and 753 controls were recruited from the US and Canada; 392 cases and 321 controls were recruited from European centers.
The Caucasian replication sample contained 718 individuals of whom 542 were recruited from the US and Canada (259 cases, 283 controls), and 176 from Europe (104 cases 72 controls). The Asian sample included 866 Japanese (433 cases, 433 controls) and 300 Koreans (128 cases, 172 controls). Finally, 277 African Americans were studied (133 cases, 144 controls). All subjects had given written informed consent approval.
HLA-DQB1*0602 typing
The presence or absence of DQB1*0602 was determined using DQB1 exon 2 sequence-specific primers (see Supplementary Table 2). These primers amplify DQB1*0602 and a few exceptionally rare DQB1*06 alleles (allele frequency<0.5%) as a 218 bp PCR product. The assay includes a DRB1 internal positive control.
Analysis of Affymetrix Data
We obtained Cel file data for all samples and performed genotyping using the birdseed-dev algorithm for Affy 6.0 (Affymetrix Power Tools \apt-1.8.5) (1544 samples) (http://www.affymetrix.com/products/software/specific/birdseed_algorithm.affx), and BRLMM for Affy 500K array set chips (337 samples) (http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf). In each genotype-calling group, individual chips with outlier low call rates (typically <97%) or high heterozygosity were excluded from further analysis. For each Birdseed calling run, SNPs with call rates <0.9, or Hardy Weinberg P<0.01 in controls were excluded. A total of 549,596 SNPs passed all quality control filters and were included in the final analysis. Genotype data was maintained in our database (Progeny Lab 7, http://www.progenygenetics.com), and analyses were performed using the PLINK software package (v1.04 26/Aug/2008, http://pngu.mgh.harvard.edu/purcell/plink/ 14). Interaction studies were performed in the initial set and in replication sets (cases and controls) using Plink epistasis, which performs a logistic regression including main genotype effects plus an interaction term.
Supplementary Material
Acknowledgments
We are most indebted to all the participants of the study, most notably narcoleptic patients. This study was supported primarily by the National Institutes of Neurological Disease and Stroke grant P50 NS2372. Additional funding included National Institutes of Mental Health R01 MH080957 to E. Mignot, 5U01 MH079470 to D. Levinson, 5U01 MH079469-02 to P. Gejman, R01 HL62252 to T. Young, Czech Ministry of Education MSM0021620849 and MZO 0002373601 to S. Nevsimalova, National institutes of Allergic and Infectious diseases 5U19 AI063603 to D. Salomon. National Institute of Neurological Disorders and Stroke R01 NS38523 to W Longstreth, MIUR PRIN Grant 2005065029 to G. Plazzi, a grant from Grants-in-Aid for Scientific Research on Comprehensive Genomics from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K. Tokunaga. G Rouleau is supported by the Canadian Institutes of Health Research. Grant MGC 77493 to J. Montplaisir. E. Mignot is an HHMI supported investigator. We are also grateful to GAIN (the Genetic Association Information Network, NIH) and KORA (Kooperative Gesundheitsforschung in der Region Augsburg, Germany). The KORA research platform was initiated and financed by the German Federal Ministry of Education and Research and by the State of Bavaria. The authors extend their thanks to Eunice Wan, Catherine Chu, Connie Ha, Jing Zhang, and Anna Voros for technical assistance, and Carl Grumet for brainstorming and constant support.
References
- 1.Mignot E. Genetic and familial aspects of narcolepsy. Neurology. 1998;50:S16–22. doi: 10.1212/wnl.50.2_suppl_1.s16. [DOI] [PubMed] [Google Scholar]
- 2.Longstreth WT, Jr., Koepsell TD, Ton TG, Hendrickson AF, van Belle G. The epidemiology of narcolepsy. Sleep. 2007;30:13–26. doi: 10.1093/sleep/30.1.13. [DOI] [PubMed] [Google Scholar]
- 3.Juji T, Satake M, Honda Y, Doi Y. HLA antigens in Japanese patients with narcolepsy. All the patients were DR2 positive. Tissue Antigens. 1984;24:316–9. doi: 10.1111/j.1399-0039.1984.tb02144.x. [DOI] [PubMed] [Google Scholar]
- 4.Mignot E, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–99. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin L, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–76. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- 6.Chemelli RM, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–51. doi: 10.1016/s0092-8674(00)81973-x. [DOI] [PubMed] [Google Scholar]
- 7.Peyron C, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–7. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 8.Thannickal TC, et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27:469–74. doi: 10.1016/s0896-6273(00)00058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens. 2004;64:631–49. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- 10.Scammell TE. The frustrating and mostly fruitless search for an autoimmune cause of narcolepsy. Sleep. 2006;29:601–2. doi: 10.1093/sleep/29.5.601. [DOI] [PubMed] [Google Scholar]
- 11.Overeem S, Black JL, 3rd, Lammers GJ. Narcolepsy: immunological aspects. Sleep Med Rev. 2008;12:95–107. doi: 10.1016/j.smrv.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Seldin MF, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng MC, et al. Replication and identification of novel variants at TCF7L2 associated with type 2 diabetes in Hong Kong Chinese. J Clin Endocrinol Metab. 2007;92:3733–7. doi: 10.1210/jc.2007-0849. [DOI] [PubMed] [Google Scholar]
- 17.Miyagawa T, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet. 2008;40:1324–8. doi: 10.1038/ng.231. [DOI] [PubMed] [Google Scholar]
- 18.Garcia KC, Teyton L. T-cell receptor peptide-MHC interactions: biological lessons from structural studies. Curr Opin Biotechnol. 1998;9:338–43. doi: 10.1016/s0958-1669(98)80004-9. [DOI] [PubMed] [Google Scholar]
- 19.Haynes MR, Wu GE. Evolution of the variable gene segments and recombination signal sequences of the human T-cell receptor alpha/delta locus. Immunogenetics. 2004;56:470–9. doi: 10.1007/s00251-004-0706-x. [DOI] [PubMed] [Google Scholar]
- 20.Koop BF, et al. The human T-cell receptor TCRAC/TCRDC (C alpha/C delta) region: organization, sequence, and evolution of 97.6 kb of DNA. Genomics. 1994;19:478–93. doi: 10.1006/geno.1994.1097. [DOI] [PubMed] [Google Scholar]
- 21.Fuschiotti P, et al. Analysis of the TCR alpha-chain rearrangement profile in human T lymphocytes. Mol Immunol. 2007;44:3380–8. doi: 10.1016/j.molimm.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–12. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–32. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 24.Sundberg EJ, Deng L, Mariuzza RA. TCR recognition of peptide/MHC class II complexes and superantigens. Semin Immunol. 2007;19:262–71. doi: 10.1016/j.smim.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116–21. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.