SUMMARY
Cell-based screening can facilitate rapid identification of compounds inducing complex cellular phenotypes. Advancing a compound toward the clinic, however, generally requires identification of precise mechanisms of action. We previously found that epidermal growth factor receptor (EGFR) inhibitors induce acute myeloid leukemia (AML) differentiation via a non-EGFR mechanism. In this report, we integrated proteomic and RNAi-based strategies to identify their off-target anti-AML mechanism. These orthogonal approaches identified Syk as a target in AML. Genetic and pharmacological inactivation of Syk with a drug in clinical trial for other indications promoted differentiation of AML cells and attenuated leukemia growth in vivo. These results demonstrate the power of integrating diverse chemical, proteomic, and genomic screening approaches to identify therapeutic strategies for cancer.
INTRODUCTION
We previously discovered that EGFR inhibitors induce AML differentiation and inhibit cell viability with case reports now of clinical responses, including two complete remissions (Boehrer et al., 2008; Chan and Pilichowska, 2007; Pitini et al., 2008; Stegmaier et al., 2005; Stegmaier et al., 2004). However, the mechanism by which these molecules induce phenotypic alterations in AML has remained a mystery because EGFR is not expressed in AML (Lindhagen et al., 2008; Stegmaier et al., 2005). This specific example speaks to a more general problem encountered in cell-based, phenotypic screening. While these screens are quite powerful in their ability to identify compounds that modulate complex biological states, the identification of the direct binding target of, or cellular pathway modulated by, a discovered chemical hit can be a serious limitation.
Target identification is critical for optimization of drug specificity and potency, minimization of off-target effects, and monitoring of pharmacodynamic studies in clinical testing. Accordingly, in the absence of a known target, translation of a compound to clinical trial or optimal use of a compound with demonstrated efficacy, can be stymied, even for FDA-approved drugs. Indeed, there are many compounds for which the mechanism of action is unknown even when clinical efficacy has been demonstrated. For example, the thalidomide derivative lenalidomide has been approved by the FDA to treat patients with low or intermediate-1 risk myelodysplastic syndrome (MDS) associated with a deletion 5q cytogenetic abnormality (List et al., 2006). However, the majority of patients with MDS lack the 5q deletion and of these patients, a minority respond to lenalidomide (List et al., 2005). Similarly, sorafenib, initially developed as a RAF inhibitor, has been approved for patients with advanced renal cell carcinoma (RCC) where the relevant target is likely to be a non-RAF tyrosine kinase (Escudier et al., 2007; Ratain et al., 2006). Further optimization in the treatment of RCC ideally would be directed at the relevant target. Because the precise mechanism of action of these drugs is not clear, it has been difficult to identify a priori which patients will benefit from the drug. Similarly, in the absence of target knowledge, our understanding of the development of resistant disease is limited at best.
Although target identification has been a significant roadblock to cell-based screening, this problem now warrants revisiting. With advances such as the application of high-throughput shRNA screening to mammalian systems and emerging abilities to evaluate the phosphorylated tyrosine kinome, interdisciplinary solutions to this challenge are now feasible. We decided to undertake this challenge of target identification by using integrative orthogonal proteomic and genetic approaches. We explored this strategy in the context of the gene expression-based screen that identified gefitinib as an inducer of AML differentiation (Stegmaier et al., 2005; Stegmaier et al., 2004). As in the case of phenotype-based screening, expression-based screening faces the challenge of identifying the protein target of confirmed compound hits. Here, we integrated immunoaffinity profiling of tyrosine phosphorylation by mass spectrometry and RNAi-based signature screening to identify candidate gefitinib targets and validated one of these kinases as a target for AML therapy.
RESULTS
Proteomic and genetic approaches identify Syk as a candidate gefitinib target
Because multiple EGFR inhibitors induce the AML differentiation phenotype, we hypothesized that a shared off-target kinase was the target in AML differentiation. In order to identify candidate targets, we integrated peptide immunoprecipitation-HPLC-mass spectrometry and RNAi-based signature screening. Specifically, we employed a proteomics approach that involved immunoprecipitation (IP) with a cocktail of three phospho-tyrosine antibodies after enzymatic digestion to enrich for phospho-tyrosine peptides (Figure S1) (Rush et al., 2005). We treated the AML cell line HL-60 with gefitinib or vehicle for 10 minutes and then identified peptide phosphorylation sites that were lost after gefitinib treatment by LC-MS/MS (Figure 1A and Table S1). Syk was identified as one of the few tyrosine kinases that exhibited loss of phosphorylation post-treatment. Syk is a nonreceptor tyrosine kinase, previously shown to play an important role in normal B-cell differentiation and hematopoietic signaling, and implicated in hematological malignancies such as myelodysplastic syndrome and lymphoma (Chen et al., 2008; Cheng et al., 1995; Chu et al., 1998; Feldman et al., 2008; Kanie et al., 2004; Kuno et al., 2001; Rinaldi et al., 2006; Sada et al., 2001; Turner et al., 1995; Young et al., 2008). Two other direct targets of Syk kinase activity, the adaptor protein Vav1, and the ubiquitin ligase Cbl, were also differentially phosphorylated (Deckert et al., 1996; Lupher et al., 1998).
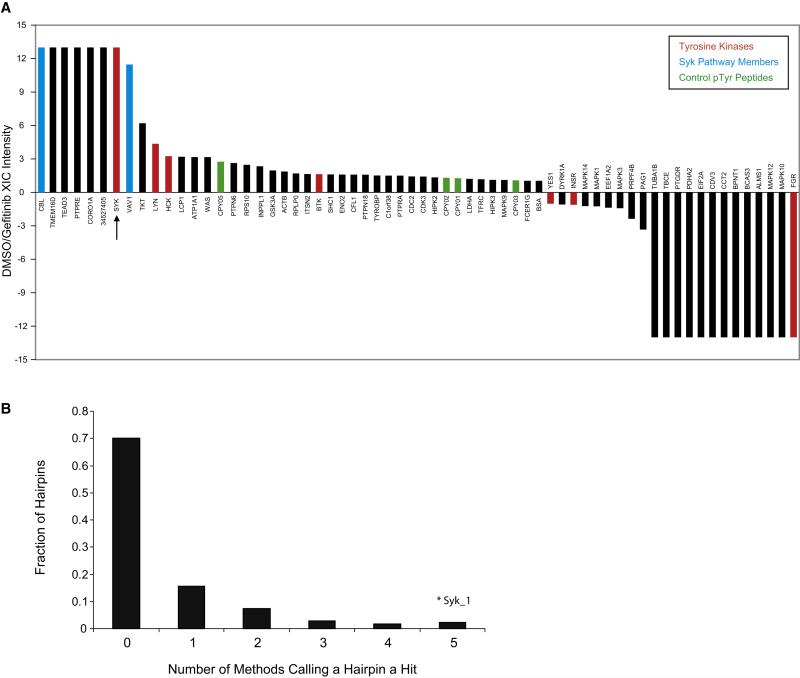
Figure 1. Proteomic and RNAi-based approaches identify Syk as a target for AML differentiation.
(A) Peptide-IP/LC-MS/MS was used to identify proteins dephosphorylated upon gefitinib treatment. HL-60 cells were treated with vehicle (DMSO) or gefitinib at 10 μM for 10 minutes. Phosphotyrosine peptides were identified in Syk, Cbl, and Vav1 that were dephosphorylated with gefitinib. The LC-MS/MS extracted ion currents (XIC) for all identified pTyr peptide precursor ions belonging to an individual protein were summed and used to calculate differential expression ratios in DMSO- and gefitinib-treated cells. The specific phosphopeptides and pTyr sites observed are shown in Table S1. CPY01,2,3,5 are control pTyr peptides spiked into each sample at the same concentration. Positive ratios indicate higher expression for DMSO treatment, negative ratios indicate higher expression for gefitinib treatment. Ratios of 13 and −13 indicate that phosphopeptides for a protein were only detected in one of the treatments.
(B) High-throughput RNAi Screen Performance. Five scoring methods were used to identify hairpins inducing a myeloid differentiation signature (Summed Score, Weighted Summed Score, Naïve Bayes, K-Nearest Neighbor, and Support Vector Machine). The majority of shRNAs did not score by any method. The SYK-specific shRNA (Syk_1) scored in all methods.
In order to increase our confidence in hits identified by this proteomics screen, we performed an orthogonal shRNA screen using a lentivirally delivered short hairpin RNA (shRNA) library targeting the human kinome (Moffat et al., 2006). We screened 5,036 shRNAs for those that induced a complex 19-gene signature of myeloid differentiation (Table S2). Gene expression was measured by the previously described approach: Gene Expression-based High-throughput Screening (GE-HTS) (Stegmaier et al., 2004). GE-HTS is based on the premise that gene expression signatures can be used for high-throughput screening. GE-HTS overcomes some of the challenges inherent in traditional target- and phenotype-based screening in that it is generic, does not require unique assay customization, and does not depend on a priori target knowledge. Briefly, mRNAs are captured on 384-well plates coated in oligo-dT and reverse transcribed to create cDNA. Then, ligation-mediated amplification (LMA) is performed to amplify marker genes with universal primers (one set biotinylated). Molecular barcodes are incorporated into the flanking regions of each amplicon and detected by fluorescent beads, each coupled to a capture oligonucleotide complementary to one of the barcodes. The use of beads of different colors (each corresponding to a different barcode capture probe) facilitates quantitation of the LMA products by dual color flow cytometry. Bead color denotes the signature transcript identity, and the phycoerythrin intensity denotes the transcript’s abundance (Peck et al., 2006). In this integrative approach to target identification, we combined high-throughput RNAi-based screening with high complexity signature-based readouts. In the primary RNAi screen, the total hit rate was 2% for shRNAs scoring across all scoring metrics. One of these top scoring shRNAs targeted SYK (Figure 1B). This shRNA suppressed SYK to the most significant degree of the five SYK-directed shRNAs included in the screen.
SYK-directed RNAi induces myeloid differentiation in AML cell lines
In a secondary screen performed in two AML cell lines, HL-60 and U937, we tested a collection of additional shRNAs designed to target the top scoring candidate kinases with an expanded 32-gene myeloid differentiation signature detected by the GE-HTS assay (Table S3). Multiple shRNAs specific for SYK scored in both cell lines. We next confirmed that the shRNAs that triggered the 32-gene differentiation signature indeed suppress SYK expression (Figures 2A and 2B and Figure S2). These shRNAs also induce morphological evidence of differentiation, including nuclear condensation and cytoplasmic ruffling (Figure 2C), and expression of the mature myeloid cell surface proteins CD11b and CD14 (Figure 2D and Figure S3A). Expression of shRNAs that induced the greatest degree of gene suppression generally resulted in the most striking phenotypic changes. Taken together, these results suggest that loss of Syk in HL-60 and U937 induces evidence of AML differentiation by multiple measurements.
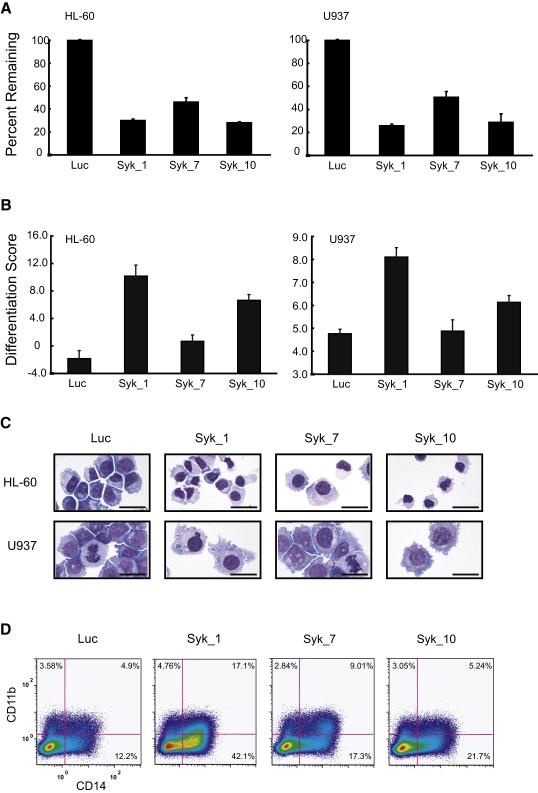
Figure 2. SYK-directed shRNA induces differentiation in AML cell lines.
(A) Knockdown performance of shRNAs targeting SYK in HL-60 and U937 cell lines. Transcript levels were measured at 5 days post-infection using real-time PCR and are reported for each hairpin (Syk_1, Syk_7, and Syk_10) relative to the transcript level of a luciferase shRNA control (Luc). Error bars depict mean ±standard deviation (SD) across three replicates.
(B) HL-60 and U937 cells were infected with shRNAs targeting SYK and luciferase (Luc) and differentiation evaluated 7 days post-infection. A 32-gene differentiation signature was quantified by the LMA/bead-based approach and a Weighted Summed Score (Differentiation Score) calculated for all genes. Error bars denote mean ±SD of 16 replicates. The two SYK-specific shRNAs that induced the greatest degree of gene suppression also induced the highest Differentiation Score.
(C) May Grunwald Giemsa staining of HL-60 and U937 cells 10 and 12 days post-infection, respectively, with shRNAs targeting SYK demonstrates cellular differentiation when compared to a luciferase shRNA control. Images were acquired with an Olympus BX41 microscope, 1000X magnification under oil, and Qcapture software. The scale bar equals 25 μm.
(D) FACS analysis was performed with FITC and PE-labeled antibodies for CD11b and CD14, respectively. Six days post-infection with shRNAs targeting SYK, HL-60 cells were positive for single stained CD11b and CD14 and double staining compared to a luciferase control.
Pharmacological inhibition of Syk induces myeloid differentiation in AML cell lines
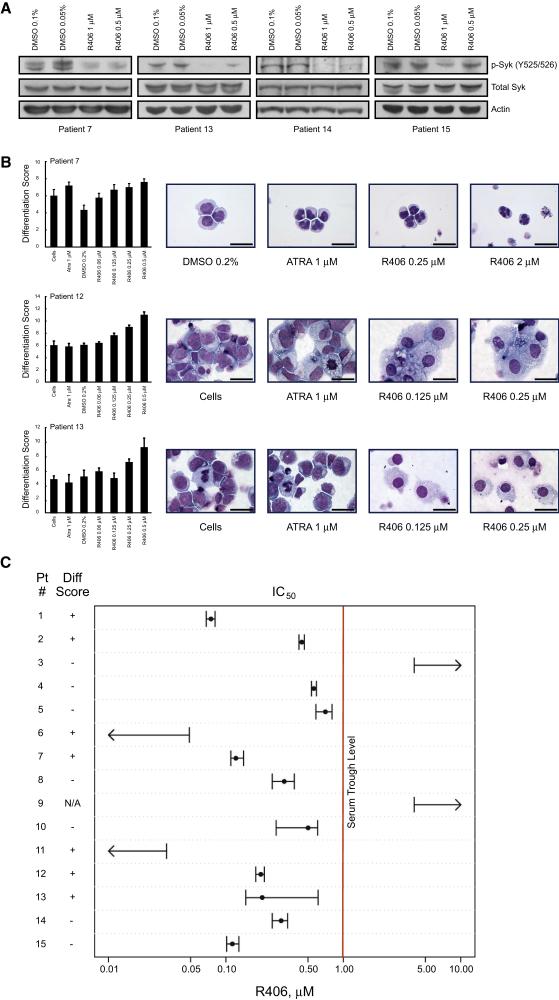
If Syk plays a role in counteracting differentiation in AML cells, specifically designed pharmacological Syk inhibitors should exhibit pro-differentiation activity. To address this hypothesis, we chose to focus on the potent Syk inhibitor already in clinical development, R406, an ATP-competitive pyrimidinediamine with less potency reported for FLT-3, c-Kit, and Lck (Braselmann et al., 2006; Weinblatt et al., 2008). First, we confirmed that treatment with either R406 or gefitinib resulted in inhibition of Syk phosphorylation at Y525/526, the kinase activation site (Figure 3A). As we found when we suppressed SYK expression with shRNAs, pharmacological inhibition of Syk induced differentiation in HL-60 and U937 cells as measured by multiple assays: a 32-gene differentiation signature (Figure 3B), morphological changes including nuclear condensation and cytoplasmic ruffling (Figure 3C), mature cell surface proteins CD11b and CD14 (Figure 3D and Figure S3B) and nitro-blue tetrazolium reduction (Figure 3E), a functional assay of myeloid maturation measuring superoxide anion production. Taken together these genetic and pharmacologic studies implicate Syk as a negative regulator of differentiation in AML.
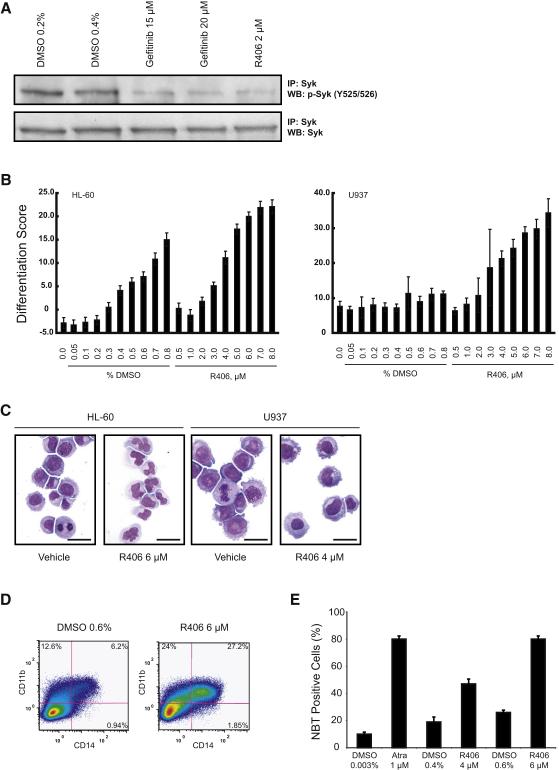
Figure 3. R406 induces differentiation in AML cell lines.
(A) Western immunoblot of HL-60 cells treated with vehicle, gefitinib, or R406 for one hour. Lysates were immunoprecipitated with anti-Syk antibody and blotted with antibody to p-Syk (Y525/526). Treatment with gefitinib and R406 dephosphorylates Syk at Y525/526. Total Syk was evaluated for a loading control.
(B) HL-60 and U937 cells were treated for three days with R406 and DMSO and a Weighted Summed Score (Differentiation Score) determined. Error bars denote the mean ±SD of 16 replicates. A dose response was seen in both cell lines. High-dose DMSO is known to induce differentiation in HL-60; however, the Differentiation Score is higher for each corresponding dose of R406.
(C) May Grunwald Giemsa staining of HL-60 and U937 cells treated for three days with DMSO or R406. R406 induces myeloid maturation in both cell lines as characterized by nuclear condensation and cytoplasmic ruffling. Images were acquired with an Olympus BX41 microscope, 1000X magnification under oil, and Qcapture software. The scale bar equals 25 μm.
(D) FACS analysis was performed with FITC- and PE-labeled antibodies for CD11b and CD14, respectively. HL-60 cells were treated for three days with vehicle or R406. R406 induced both CD11b and CD14 stained double positive cells and single stained positive CD11b consistent with myeloid maturation.
(E) HL-60 cells were treated in triplicate for three days with vehicle, ATRA, or R406. The percentage of NBT-positive cells and mean ±SD are shown. Both ATRA and R406 reduced NBT consistent with the functional maturity of HL-60 cells.
Syk inhibition affects a diverse set of AML cell lines
To determine whether Syk inhibition has broad applicability in AML, we extended testing to additional AML cell lines, evaluating effects of pharmacologic and genetic inhibition on growth and differentiation. When we tested doses up to 10 μM, we found that all of the AML cell lines tested, except for THP-1, exhibited decreased viability (Table S4). We hypothesized that Syk would be differentially phosphorylated across these AML samples and that phosphorylation status would predict response to Syk inhibition. Consistent with this notion, the two cell lines with the greatest degree of Y525/526 phosphorylation, MOLM-14 and KG-1, were amongst the most sensitive to the effects of pharmacological inhibition of Syk on viability, apoptosis, and colony formation in methylcellulose (Figures 4A and 4B and Figure S4). In contrast, R406 had minimal effects on apoptosis in HL-60 cells until a dose of 6 μM (data not shown). However, some AML cell lines that exhibited Y525/526 phosphorylation, such as THP-1, were relatively insensitive to R406, while Kasumi-1 with lower levels of p-Syk were quite sensitive, suggesting that at least in vitro there are factors other than degree of Syk phosphorylation that modify response to R406.
Figure 4. Syk inhibition affects a diverse set of AML cell lines.
(A) Western immunoblot of AML cell lines reveals MOLM-14 and KG-1 cell lines to be highly phosphorylated at the p-Syk (Y525/526) enzymatic site.
(B) MOLM-14 and KG-1 cells were treated with vehicle or R406 for 6 days. Cells were stained with annexin V-FITC and propidium iodine (PI) and evaluated by flow cytometry. R406 treatment induced increased annexin V positive cells in a dose responsive manner consistent with apoptosis.
(C) MOLM-14 and KG-1 cells were infected with shRNAs targeting SYK and luciferase. After 8 days of infection, cell viability was evaluated at days 0, 3, and 6 using an ATP-based assay. Genetic loss of Syk resulted in dramatic decrease in proliferation. Error bars depict mean ±SD across ratios of 6 replicate measurements at each time point relative to the 6 replicate measurements at time zero.
(D) Ectopic expression of TEL-Syk immune to the SYK-directed shRNA Syk_1 in MOLM-14 cells rescues the effects of Syk knockdown on cell viability. Cell viability was evaluated 4 days after infection at days 0, 3, and 6 with an ATP-based assay. Error bars depict mean ±SD across ratios of 8 replicate measurements at each time point relative to the 8 replicates at time zero.
(E) May Grunwald Giemsa staining of KG-1 cells 12 days post infection with shRNAs targeting SYK versus a luciferase shRNA control demonstrate evidence of macrophage-like differentiation. Images were acquired with an Olympus BX41 microscope, 1000X magnification under oil, and Qcapture software. The scale bar equals 25 μm.
(F) KG-1 cells were treated with R406 versus DMSO and the 32-gene differentiation signature measured at 24 hours. R406 induces a myeloid differentiation signature as measured by a Weighted Summed Score (Differentiation Score). Error bars depict mean ±SD across 8 replicates.
We next evaluated genetic inhibition of Syk in MOLM-14 and KG-1. The introduction of SYK-specific shRNA dramatically inhibited their proliferation (Figure 4C). Ectopic expression of a TEL-Syk cDNA immune to the shRNA rescued the phenotypic alteration suggesting that the effects of the shRNA were on-target for Syk (Figure 4D). While both perturbagens induce differentiation in KG-1, as measured by morphological and gene expression changes that precede apoptosis (Figures 4E and 4F and Figure S5), their predominant effects in MOLM-14 were on reduced cell viability. However, the subtle effects of the shRNA on differentiation were also rescued with the expression of TEL-Syk. (Figures S6 and S7). These results suggest that the effects of Syk inhibition on differentiation and apoptosis can be uncoupled. Indeed, this uncoupling was previously demonstrated for the EGFR inhibitor, erlotinib, in AML (Boehrer et al., 2008). Several lines of evidence support the conclusion that the primary consequence of R406 is due to its effects on Syk rather than an off-target kinase of R406. Specifically, we found that SYK-specific shRNAs recapitulate the effects of R406. Second, there is a tight correlation between loss of Syk phosphorylation with R406 at Y525/526 and the differentiation score (Figure S8). Moreover, many of the samples that we used harbor wild type FLT-3, and FLT-3 wild type cell lines did not respond to a FLT-3 inhibitor (Figure S9).
Pharmacological inhibition of Syk has in vivo activity in an orthotopic AML cell line and syngeneic mouse models of AML
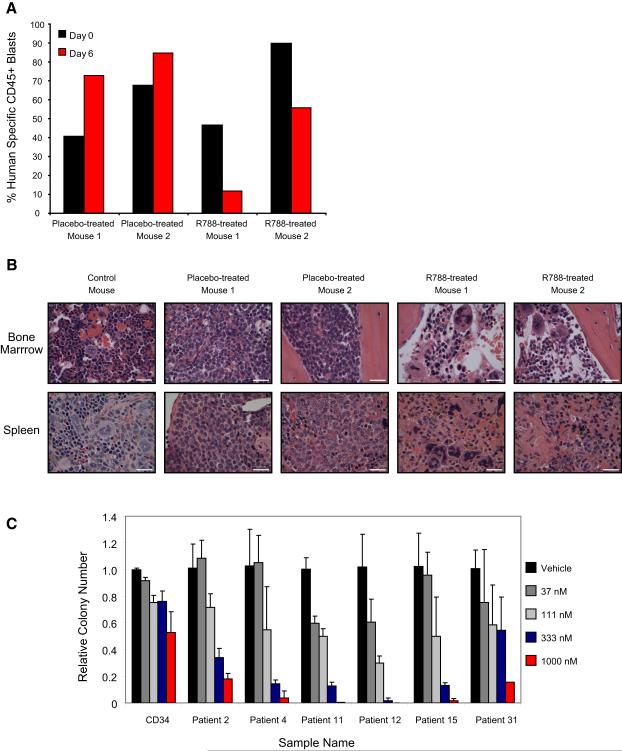
Although evaluation of human cell lines in vitro is invaluable in the preclinical testing of potential anti-leukemia compounds, the ex vivo environment cannot fully recapitulate the bone marrow niche. Thus, in vivo studies are critical. Because KG-1 had the most dramatic level of Syk phosphorylation, we focused our attention on this cell line. First, we evaluated colony formation in methylcellulose. Both chemical and genetic inhibition of Syk abrogated colony forming potential in KG-1 (Figure 5A and Figure S4). Next, we used an orthotopic xenograft model in which KG-1 cells were labeled with luciferase and propagated in NOD-SCID IL2Rγnull (NOG) xenografts. Eight days of treatment with R788, the prodrug of R406, resulted in significantly decreased leukemia burden (p < 0.05) and spleen weight (p < 0.001) compared to control-treated animals (Figure 5B and Figure S10). Moreover, to confirm these findings made in human cancer cell lines in primary murine AML, we extended in vivo testing of Syk inhibition to a syngeneic mouse model of MLL-AF9 AML (Stubbs et al., 2008). Again, eight days of treatment with R788 resulted in significantly decreased leukemia burden (p < 0.005) and spleen weight (p < 0.01) compared to control-treated animals (Figure 5C and Figure S11).
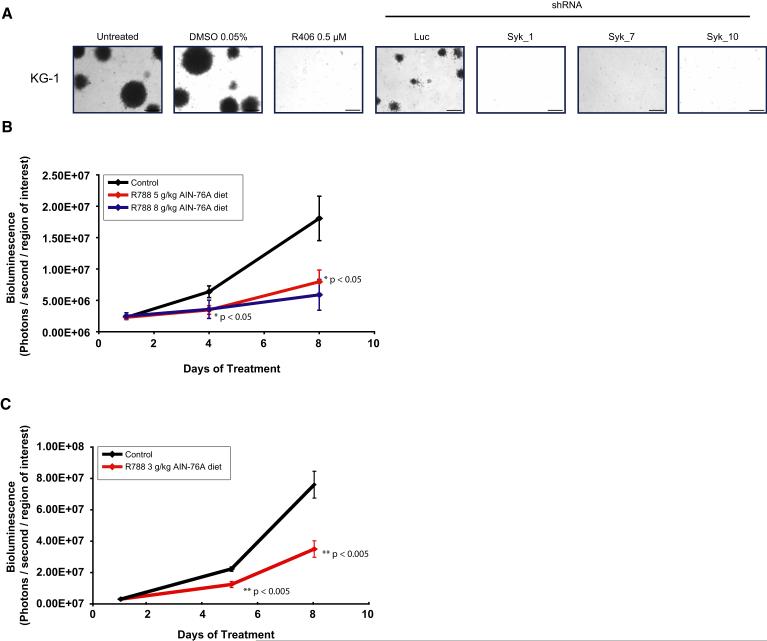
Figure 5. R406 has in vivo activity in AML.
(A) The ability of KG-1 to form colonies in methylcellulose with R406 and multiple shRNAs directed against SYK was assessed. Both chemical and genetic inhibition of Syk abrogate colony formation in KG-1. The scale bar equals 1 mm.
(B) KG-1 luciferase positive xenografts were established in NOG mice. Mice were treated with placebo (n=4), food impregnated with 5g R788 / kg AIN-76A food (n=5), and food impregnated with 8g/kg AIN-76A food (n=3) for 8 days. Mice treated with R788 had a significant difference in tumor burden compared to those treated with placebo. Serial in vivo BLI was used to assess disease burden, and data plotted as the mean ±SEM for each group. One way ANOVA analysis with Tukey post-test was used to determine the significance of all pairwise comparisons.
(C) A syngeneic, luciferase positive mouse model of MLL-AF9 AML was established in C57BL/6:TyrC/C mice. Mice were treated with placebo (n=10) or food impregnated with 3g R788 / kg AIN-76 food (n=10) for 8 days. Serial in vivo BLI was used to assess disease burden and plotted as mean ±SEM. Statistical significance was calculated using Student’s t-test.
Syk is constitutively active in primary AML blasts
Cell lines have been a critical tool for cancer discovery, but extrapolation of response to primary disease can be problematic secondary to selection for, or acquisition of, additional genetic hits ex vivo. Although studies in primary patient AML blasts are notoriously challenging, they are critical in the validation of therapeutic targets and response to small molecule inhibitors. We first determined whether Syk is expressed in primary AML blasts. Syk mRNA had previously been reported to be widely expressed in AML (Tomasson et al., 2008), and we identified Syk protein in over 90% of primary AML samples evaluated by immunoblotting (Figure S12). Moreover, using phosphorylation at Y525/526 as a marker of Syk activation, we identified Syk phosphorylation that was abrogated by R406 in an independent collection of primary patient AML blasts for which we obtained adequate cells (Figure 6A). These observations suggested that Syk was not only expressed but constitutively active in primary AML blasts.
Figure 6. Primary AML cells are responsive to Syk inhibition in vitro.
(A) Fresh primary patient AML cells were treated in vitro with vehicle versus R406 and Syk phosphorylation at Y525/526 was assessed by Western immunoblotting. In all four samples Syk is phosphorylated at the kinase activity site, and this phosphorylation is inhibited by R406.
(B) Primary AML blasts were treated with DMSO, ATRA, or R406 for three days and the 32-gene differentiation signature evaluated. Error bars denote mean ±SD across 8 replicates per condition. R406 induces comparable or greater levels of differentiation to ATRA. May Grunwald Giemsa staining of ATRA- and R406-treated primary AML at four days reveals myeloid differentiation with lobulation of nuclei in the APL sample and macrophage-like differentiation in Patients 12 and 13. Images were acquired with an Olympus BX41 microscope, 1000X magnification under oil, and Qcapture software. The scale bar equals 25 μm.
(C) Primary patient AML cells were treated in quadruplicate with R406 in a two-fold dilution series. At day three, cell viability was evaluated using an ATP-based assay. Values for the IC50 are depicted with error bars denoting the 95% confidence interval for the SEM for the four chemical replicates relative to the four control replicates. At day three the 32-gene differentiation signature was measured. Samples are scored as differentiating if there was a 1.5-fold change in the Differentiation Score across at least 2 doses with p < 0.05 by one-tailed t-test assuming two samples with unequal variance.
The majority of primary AML blasts respond to Syk inhibition in vitro
We next asked whether Syk inhibition would induce differentiation and/or inhibit cell viability in primary AML blasts in vitro. First, we tested the in vitro activity of R406 in primary cells from a patient with acute promyelocytic leukemia (APL), the clinical AML subtype most responsive to differentiation therapy in vivo. Syk inhibition induced differentiation, evaluated by changes in gene expression and morphological appearance with nuclear condensation and lobulation, comparable to the well validated differentiation agent, all-trans retinoic acid (ATRA) (Figure 6B). We next extended testing to 13 additional AML patient samples widely representing the FAB subclasses and one CMML sample. Twelve of these 13 AML patient samples responded with an IC50 less than 1 μM at three days (range 0.05 to 0.7 μM) (Figure 6C and Table 1). We noted that one sample from a patient with relapsed M1-AML and another from a patient with CMML failed to respond at doses up to 4 μM. R406 induced evidence of differentiation in 6 of 13 evaluable patient samples in addition to the APL sample (Figures 6B and C). As in the case of AML cell lines, the effects of R406 on differentiation and viability were sometimes uncoupled. Treatment response was independent of French-American-British (FAB) subtype.
Table 1. Primary Patient AML Characteristics.
| Patient | Diagnosis | Cytogenetic Findings | FLT-3 ITD | FLT-3 D835 Codon change |
|---|---|---|---|---|
| 1 | M1-AML | 89–92, XXXX | No | No |
| 2 | M2-AML | 45 XY, −7, inv(3)(q21q26) | No | No |
| 3 | CMML | Complex karyotype* | N/A | N/A |
| 4 | M4-AML | 46 XY | No | D835H |
| 5 | M4-AML | 46 XX | No | D835E |
| 6 | M2-AML | 46 XX | Yes | No |
| 7 | M3-AML | 47 XX, +8, t(15;17) (q22;q21) | Yes | No |
| 8 | M4-Eo | 45 X,−Y[11] / 46, XY[9]. Nuc ish (CBF x2) [100] |
No | No |
| 9 | M1-AML | 46 XX | No | No |
| 10 | M-5b-AML | 46 XX | No | D835Y |
| 11 | M4-AML | 46 XY | Yes | No |
| 12 | AML** | N/A | Yes | No |
| 13 | AML** | 46 XY | No | No |
| 14 | M4-AML | 46 XY | Yes | No |
| 15 | AML** | 47 XX, +8 [19] / 46, XX [1] | Yes | No |
| 31 | M7-AML | 52,XY,+8,+14,+19,+21,+21,+21c[14]/ 47,XY,+21c[6] |
N/A | N/A |
N/A = not available
46, XY, t (1;19) (p36;q13), del (2) (p2?3), t (3;15) (p21;q22), add (4) (q35), −6, der (7) t (7;14) (q36;q11.2), −14, −21 + mar [cp10]/ 46 XY, del (2) (p2?1), add (7) (q11.2) [cp4]/ 46, XY, add (1) (q42), t (1;14) (p12;q11), del (3) (q1?3), der (7) del (7) (q3?1q3?4) t(3;7) (q13;p22, t(17;21) (q2?1;q22) [2]/ 46, XY, non clonal abnormalities [4]
FAB not reported
Pharmacological inhibition of Syk with R788 has activity in a primary AML orthotopic model and a therapeutic index in AML
There is an ongoing debate about the utility of conventional mouse models for predicting successful therapies in humans. In response, there has been a shift in the experimental approach to in vivo animal studies with the development of mouse models using tumor collected directly from primary human tumors. In order to address this issue, we tested R788 in a primary human AML orthotopic model. A primary patient M4-AML sample responsive in vitro to R406 (Patient 11) was used to generate a human xenograft in NOG mice by tail vein injection. Seventy days post injection, mice developed greater than 40% peripheral AML blasts as measured by human specific CD45+ cells from peripheral blood. Mice were then divided into 2 cohorts with 2 animals per cohort. One group received R788, the second a placebo. A NOG control mouse without human AML was included. Six days post treatment, tail vein blood was sampled and the two treated animals were noted to have a decrement in human specific CD45+ cells (Figure 7A). At day 7, animals were sacrificed. Spleen weights were dramatically different across the groups: control mouse (0.03 g), placebo-treated (0.14 g and 0.16 g) and R788-treated (0.04 g and 0.05 g). Histopathology of the bone marrow and spleen (Figure 7B) revealed a dramatic decrease in the number of infiltrating AML cells. We next explored the therapeutic window of R406 by comparing the effects of R406 on primary AML blast colony formation in methylcellulose compared to normal CD34+ myeloid progenitor cells. All six primary AML samples were more sensitive to the effects of R406 than were the normal CD34 myeloid progenitor cells suggesting that there is a therapeutic index for R406 in AML (Figure 7C).
Figure 7. Primary AML is responsive to Syk inhibition in an orthotopic model.
(A) A primary AML (Patient 11) orthotopic xenograft was established in NOG mice. Six days after initiation of treatment with food containing 5g R788/kg AIN-76 rodent diet, the AML blast percentage, measured by percentage of human specific CD45+ cells in peripheral blood, continued to increase in the placebo-treated mice but decreased in the R788-treated mice.
(B) Histopathology studies of bone marrow in R788-treated versus placebo-treated animals reveal near resolution of AML infiltration with areas of necrosis and areas of recovering marrow identified. Hematoxylin and eosin staining of spleen reveals areas of leukemic infiltration and extramedullary hematopoiesis in placebo-treated mice and near resolution of AML infiltration in the R788-treated animals. Images were acquired with an Olympus BX41 microscope, 1000X magnification under oil, and Qcapture software. The scale bar equals 25 μm.
(C) The ability of normal CD34 myeloid progenitor cells and primary patient AML blasts to form colonies in methylcellulose with R406 was assessed. Colony formation was assessed in duplicate and colonies per 4 cm2 counted and displayed relative to control cells. Error bars depict mean ± SD across 4 ratios of dose response to vehicle. Primary patient AML blasts were more sensitive to the effects of R406 than were normal myeloid progenitors.
DISCUSSION
Cure rates for patients with AML have remained poor despite intensive chemotherapy and stem cell transplantation. For older adults, long-term survival is dismal with many older patients unable to tolerate standard cytotoxic therapy. Although much has been learned about the pathogenesis of AML, many of the potential targets involve abnormalities of transcription factors, a class of proteins considered “undruggable” with standard pharmacological methods. Emerging approaches, such as RNAi-based (Vorhies and Nemunaitis, 2009), gene expression-based (Lamb et al., 2006; Stegmaier et al., 2007), stapled peptide technology (Walensky et al., 2004), or other peptide-based methods (Polo et al., 2004) may ultimately enable targeting of this important protein class. Alternatively, the identification of more pharmacologically tractable targets in AML offers a parallel route to therapies for this disease.
In this work, we build upon the striking observation that multiple EGFR inhibitors had anti-AML activity by a non-EGFR mechanism and hypothesized that this was via a shared off-target effect. We integrate cross-disciplinary proteomic and genetic approaches to meet the difficult challenge of understanding the molecular basis for the biological activity of these EGFR inhibitors in AML. At the intersection of these two approaches, Syk was identified as a tyrosine kinase target in AML. Syk is a cytoplasmic tyrosine kinase widely expressed in hematopoietic cells and critical in B cell differentiation and signal transduction pathways. Syk is a member of the Syk/ZAP-70 family of nonreceptor kinases and is characterized by two N-terminal Src homology 2 domains and a C-terminal kinase domain separated by a flexible linker (Sada et al., 2001). Syk activation has been implicated in a variety of hematopoietic cellular responses (Chu et al., 1998; Sada et al., 2001; Turner et al., 2000), and there is a growing literature supporting the role of Syk in hematological malignancies, particularly the lymphomas (Chen et al., 2008; Feldman et al., 2008; Rinaldi et al., 2006; Streubel et al., 2006). Interestingly, we see variable patterns of p-Syk expression and response to R406 in acute lymphoid leukemia cell lines (Figure S13 and Table S5).
Our demonstration of a role for Syk in the pathogenesis of myeloid malignancies is supported by the case report of the fusion of the TEL to the Syk gene in a patient with MDS with t(9;12)(q22;p12) (Kuno et al., 2001). Importantly, this TEL-Syk fusion transformed the interleukin-3 dependent murine hematopoietic cell line Ba/F3 to growth factor independence (Kanie et al., 2004). Syk mRNA transcript has been reported to be expressed in primary AML blasts (Tomasson et al., 2008) and its expression correlated with response to treatment with gemtuzumab ozogamicin (Balaian and Ball, 2006). In our studies, we demonstrate that Syk protein is not only expressed but is also constitutively activated in nearly all of the samples evaluated. Determining the mechanism of activation of Syk and its critical downstream effectors in AML is under active investigation.
The screening of libraries enriched for FDA-approved compounds is an attractive strategy to repurpose drugs for alternative indications and to more rapidly test clinically relevant hypotheses in patients. However, the potency and selectivity of such drugs are often not optimal for discovered indications. Moreover, there is the desire to optimize on-target activity and minimize off-target side effects. While EGFR inhibitors have had anti-AML effects, including complete responses, these molecules do not potently inhibit Syk and are often not well tolerated at doses anticipated to inhibit Syk in vivo. Therefore, we need to identify more potent and selective Syk inhibitors for this indication. The Syk inhibitor R788 is the orally available prodrug of R406. Phase I testing of R788 has been completed with peak concentrations in excess of 10 μM, steady state concentrations of 1–3 μM, and trough concentrations of 1 μM (Elliott Grossbard, Rigel Pharmaceuticals, Personal Communication). In Phase I testing in patients with rheumatoid arthritis and heavily pretreated patients with lymphoma, the drug has been well tolerated with significant clinical activity demonstrated (Weinblatt et al., 2008 and Margaret Shipp, Personal Communication). In our studies, the majority of primary patient AML samples responded in a clinically achievable dose range, with an IC50 below the serum trough concentration in humans. Thus, these observations suggest that inhibition of Syk can be achieved with doses in current use in clinical trials.
The last decade has seen a marked shift in the approach to cancer-related small molecule library screening performed by the pharmaceutical industry with a transition away from phenotype-based screening and a primary focus now on target-based screening. While target-based screening has been successful for known, tractable protein targets, as with any approach, it does have limitations. The majority of known oncoproteins are not considered easily druggable, and for many malignancies, driver events have not yet been identified. In addition, target-based screening is most commonly performed with ex cellulo assays and may not fully recapitulate the complexity within a cell. Alternative screening approaches, such as traditional phenotype-based and expression-based approaches, create the possibility of screening in the absence of a priori target knowledge. Moreover, expression-based screening holds promise for modulating intractable targets and can be systematized as a screening paradigm. However, the transition from compound discovery to drug development has been stymied for these cell-based screens by the challenge of identifying the protein target of the identified compound. Emerging genomic, genetic, and proteomic approaches have altered the landscape of target identification for cancer. Similarly, these approaches can transform the drug discovery process. Their integration into the compound discovery phase has already begun. Our work suggests that a cross-disciplinary integration of genome-wide expression profiling, proteomics, and high-throughput RNAi-based screening could systematize protein target identification for cell-based chemical screens. Moreover, innovations in synthetic chemistry and proteomics should further facilitate this process in the future. For example, diversity oriented synthesis small molecule collections enable incorporation of chemical handles to facilitate efficient, systematic attachment of affinity resins (Burke et al., 2003). After affinity capture, the specific protein/compound interactions can be identified by differential isotopic labeling of amino acids as recently described (Ong et al., 2009).
In conclusion, we demonstrate that the majority of AML cell lines and primary blasts, and three in vivo AML models had a therapeutic response to Syk inhibition with effects on cell growth and differentiation. With an orally available, well-tolerated Syk inhibitor currently in clinical development for other indications, the results reported here should have immediate relevance for clinical testing of Syk inhibition in patients with AML. Furthermore, these results demonstrate the feasibility of integrating shRNA-based screening and phosphoproteomic studies to identify small molecules and their mechanism of action.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary patient AML blasts were collected from peripheral blood or bone marrow aspirate after obtaining patient informed consent under Dana-Farber Cancer Institute and the University of Pennsylvania Internal Review Board-approved protocols. Mononuclear cells were isolated using Ficoll-Paque Plus (Amersham Biosciences) and red blood cells lysed. Cryopreserved human bone marrow CD34+ cells were obtained from Poietics, and use of these materials is considered exempt as Human Subjects by the Dana-Farber Cancer Institute Internal Review Board.
HL-60, U937, Kasumi-1, and KG-1 were purchased from the American Type Culture Collection. THP-1, MOLM-14, 697, NALM-6, REH, RS4-11, SEMK2, and SUP-B15 were kindly provided by Dr. Scott Armstrong. DND-41, HPBALL, KOPTK1, MOLT-4, and PF-382 were all kindly provided by Dr. Jon Aster. All cell lines and primary patient cells were maintained in RPMI 1640 (Cellgro) supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (Sigma-Aldrich) at 37 °C with 5% CO2.
Chemicals
Gefitinib (WuXi PharmaTech, Shanghai, China) and All-trans retinoic acid (ATRA) (Sigma-Aldrich) were dissolved in dimethyl sulfoxide (DMSO) and stored at −20 °C. R406 (Rigel Pharmaceuticals, Inc., San Francisco, CA) was resuspended in DMSO and stored at −80 °C. R788 was also supplied by Rigel Pharmaceuticals.
Phosphoproteomic Studies
HL-60 cells (1 × 109) were treated in duplicate with 10 μM gefitinib for 10 minutes. Based on a protocol described by Rush et al., protein lysate was extracted with a urea-containing lysis buffer and protease inhibitor (Rush et al., 2005). Upon reduction of the disulfide bonds and alkylation of cysteines with iodoacetamide, the lysate was split into two aliquots and diluted with 20 mM Hepes to 2 M or 0.5 M urea before digestion with trypsin or chymotrypsin, respectively. The total peptide mixtures were then desalted by Sep-Pak cartridge and resuspended in immunoprecipitation (IP) buffer, 50 mM MOPS/NaOH pH 7.2, 10 mM Na2PO4, 50 mM NaCl, along with four exogenous pTyr-containing peptides added as controls. IP was performed with a cocktail of three protein G agarose bead-bound phospho-tyrosine antibodies pY100 (Cell Signaling), 4G10 (UpState), and pY99 (Santa Cruz Biotechnology) to enrich for phosphotyrosine-containing peptides. Peptides captured by phosphotyrosine antibodies were eluted under basic followed by acidic conditions. The IP eluates were analyzed by data-dependent LC/MS/MS using a ThermoFisher LTQ-Orbitrap instrument. All MS and MS/MS data were processed using the Spectrum Mill software package which assigns peptide identity with < 1% false discovery rate using a target-decoy database search approach, maps phosphorylation sites, and quantitates using extracted ion chromatograms of each peptide precursor ion (Table S1).
Differentiation Studies
Morphological Evaluation
Changes in cellular morphology were evaluated by May-Grunwald Giemsa staining (Sigma) under light microscopy under oil at 1000X magnification with an Olympus BX41 microscope and Q-capture software.
Nitro-blue Tetrazolium (NBT) Reduction Assay
NBT reduction assays were performed in triplicate. Compound-treated cells were compared to DMSO-treated controls after three days of treatment. Cells were incubated at 37 °C for 1 hour in a mixture containing total medium, 0.1% NBT (Sigma), and 1 μg/ml TPA (12-O-tetradecanoylphorbol-13-acetate; Sigma). The percentage of blue cells was counted by light microscopy for at least 200 cells per sample. Drug-treated cells were compared with DMSO-treated cells with a one-tailed t-test analysis assuming two samples with unequal variance.
Flow Cytometry
Cells were stained with 1:25 CD11b-FITC (Beckman Coulter IM0530U) and 1:25 CD14-PE (Beckman Coulter IM0650U) for 30 minutes, detected by flow cytometry (Beckman Cytomics FC500), and data analyzed using the FlowJo software package (Tree Star). Fluorescence gating was set based on single-stained mouse Ig κ compensation bead fluorescence intensity (BD Biosciences #552843).
Gene Expression Studies
Marker genes for myeloid differentiation were chosen using previously published Affymetrix AML-related data sets (Stegmaier et al., 2004). We selected an initial collection of 19 genes used in the RNAi primary screen and then an expanded group of 32 genes. (Tables S2 and S3). These genes distinguish AML from either neutrophil or monocyte with p < 0.05 by t-test and distinguish undifferentiated versus differentiated HL-60 with either ATRA, PMA, or VitD with p < 0.05 by t-test. The GE-HTS assay was performed as detailed in the Supplementary Methods. Two primary methods are used to compare signature gene induction. The Summed Score combines expression ratios (marker gene/control gene) by summing them with a sign determined by the expected direction of regulation from gefitinib- or ATRA-treated positive controls. The Weighted Summed Score combines expression ratios by summing them with a weight and sign determined by the signal-to-noise ratio of each expression ratio for the positive control (gefitinib- or ATRA-treated) and negative control (DMSO-treated) samples.
Viability Assay
Viability experiments were performed using the Promega Cell-Titer Glo ATP-based assay per the manufacturer’s instructions. Values for IC50 (drug concentrations that reduced cell viability to 50 percent of the vehicle controls) were calculated by interpolating a natural cubic spline fit to the measured viability data in R (using the spline function).
Apoptosis Studies
AML cell lines were treated in triplicate with DMSO versus R406 for 6 days. Annexin V FITC/PI staining was performed with the Annexin V: FITC Apoptosis Detection Kit I (BD Pharmingen). Cells were analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson) and CELLQuest analytical software.
Methylcellulose Colony Forming Assay
For the RNAi studies, KG-1 and MOLM-14 cells were infected with shRNA directed against a luciferase control or SYK (three unique constructs). After 48 hours of puromycin selection, cells were recounted by trypan blue exclusion and 3×104 were plated at 1:10 (vol/vol) in methylcellulose (ClonaCell-TCS Medium, 03814) with 1% penicillin-streptomycin and appropriate drug treatment. For pharmacological treatment of primary AML, AML cell lines, and normal CD34 myeloid progenitor studies, cells were treated with R406 or matched vehicle control for 48 hours in liquid culture. After 48 hours, an equal number of cells were plated at 1:10 (vol/vol) in MethoCult GF+H4435 methylcellulose (StemCell Technologies, #04445) with 1% penicillin-streptomycin and appropriate drug treatment. All plates were incubated at 37 °C and 5% CO2 and colony numbers counted 8–17 days later.
RNAi Screening
High-throughput screening was performed with the RNAi Platform at the Broad Institute. HL-60 cells were plated in 384-well format in 30 μl of medium at 15,000 cells/well and incubated overnight at 37 °C with 5% CO2. Polybrene (Sigma) was added to a final concentration of 8 μg/ml. The lentivirally delivered sub-library of the RNAi Consortium shRNA library (http://www.broad.mit.edu/rnai/trc/lib) was screened in quadruplicate (Moffat et al., 2006). Virus was added at 2.5 μl per well, plates spun for 30 minutes at 2250 rpm, and incubated for 48 hours. Selection was performed on three replicates with 1 μg/ml puromycin (Sigma) and medium changed on the unselected control plate. Uninfected controls included cells only (8 wells), 1 μM ATRA (8 wells), and 25 μM gefitinib (8 wells). Plates were incubated for 72 hours. ATP-based viability was assessed for one puromycin treated plate and the unselected plate using Cell-Titer Glo (Promega). GE-HTS was carried out on two replicates as previously described using a 19-gene myeloid differentiation signature. (See Supplementary Methods and Table S2) (Peck et al., 2006; Stegmaier et al., 2004).
Screen plates were filtered, scaled and five scoring algorithms applied: Summed Score, Weighted Summed Score, Naïve Bayes, K-Nearest Neighbor (KNN), and Support Vector Machine (SVM). An shRNA was considered a hit if the shRNA was classified as being more like gefitinib-treated controls than untreated HL-60 controls by all five methods. (See Supplementary Methods for details of analysis). A secondary screen was performed as described above with the following exceptions: HL-60 and U937 cells were plated at 15,000 cells/well and 4,500 cells/well, respectively. shRNAs that scored in the primary screen, as well as additional available shRNAs developed against the hits, were screened in five replicates. In addition, an expanded 32-gene myeloid differentiation signature was measured (Table S3).
Lentiviral Vectors and Infection
Oligonucleotides encoding shRNAs were cloned into pLKO.1 as described previously (Moffat et al., 2006). Sequences targeted by each SYK shRNA are listed in Table S6. For large scale infections, 500,000 293T cells were plated in 6 cm plates and transfected 24 hours later with 1 μg DNA from lentiviral backbone vector and packaging plasmids (pCMVdeltaR8.91 and pMD.G) according to FuGENE 6 (Roche) protocol. Medium was changed to RPMI 1640 24 hours post-transfection and viral supernatant harvested and filtered 48 hours post-transfection. Cells were infected for two hours at 37 °C with 2 ml lentivirus and 8 μg/ml polybrene (Sigma). Cells were selected 48 hours later with 1 μg/ml puromycin (Sigma).
To generate the TEL-Syk construct, RT-PCR was used to isolate the sequences encoding amino acids 1–336 of human ETV6 (TEL) and amino acids 266–635 of human Syk. The assembled cDNA was subcloned in place of mCherry in the lentiviral plasmid FUW-Luc-mCherry-puro (Kimbrel et al., 2009) to give the plasmid FUW-Luc-TEL-SYK-puro. VSVG-pseudotyped virus was produced by co-transfection of 293T cells along with the helper plasmids delta8.9 and CMV-VSVG. Infection of MOLM-14 was performed as above.
Real-time PCR
Total RNA was isolated using TRIZOL reagent (Invitrogen), cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) and oligo d(T)16 primers, and cDNA analyzed in the real-time quantitative PCR reactions prepared with TaqMan Universal Master Mix (Applied Biosystems). RPL13A expression was evaluated for each sample as a control for total RNA. Primers and probes for real-time RT-PCR were obtained from Applied Biosystems (RPL13A # Hs01926559_g1, and SYK Hs00895374_m1).
Immunoblotting
Cells were lysed in Cell Signaling Lysis Buffer (Cell Signaling) containing Complete, EDTA-free Protease Inhibitor Cocktail Tablet (Roche Diagnostics) and PhosSTOP Phosphatase Inhibitor Tablet (Roche Diagnostics), resolved by gel electrophoresis, and transferred to nitrocellulose membranes (BioRad Laboratories). Blots were incubated with primary antibody to p-Syk (Y525/526) (Cell Signaling, 2711), total Syk (Santa Cruz Biotechnology, SC-1240), β-Actin (Abcam, ab8227–50), GAPDH (Abcam, ab22556–100) or Vinculin (Abcam, ab18058) followed by secondary antibodies (anti-rabbit-HRP (Amersham #NA9340V) or anti-mouse-HRP (Amersham #NA9310V). Bound antibody was detected by chemiluminescence. For immunoprecipitation, two mg of each protein in lysis buffer were incubated with 2 μg/ml anti-Syk antibody (Santa Cruz, SC-1240) for four hours at 4 °C. The immunocomplex was precipitated using 30 μl UltraLink Immobilized Protein A/G beads (Pierce) overnight at 4 °C. The beads were then washed twice with cold 1x Cell Signaling Lysis Buffer (Cell Signaling) containing protease inhibitor, resuspended in 10 μl of 4X SDS-sample buffer, and boiled for five minutes. The protein was then separated by gel electrophoresis and transferred to a nitrocellulose membrane (BioRad Laboratories). Blots were probed with anti-pSyk (Y525/526) (Cell Signaling #2711) and incubated with anti-rabbit-HRP (Amersham #NA9340V). Bound antibody was detected by SuperSignal West Dura Extended Duration Substrate (Pierce).
In Vivo Studies
All animal studies were performed on Dana-Farber Cancer Institute IACUC approved protocols. For KG-1 xenografts, three hours prior to injection, 6 week old male NOD-SCID IL2Rγnull mice (NOG, Jackson Laboratory) were sublethally irradiated with 200 rad. 2×106 KG-1-LucNeo cells were injected via tail vein, and total body leukemia burden was assessed by bioluminescence imaging (BLI) as previously described (Armstrong et al., 2003). Animals were imaged 5 and 7 days after injection, and mice with established disease were divided into cohorts that were treated with normal feed or feed impregnated with R788 at 5 g/kg or 8 g/kg AIN-76A rodent diet. Serial imaging was used to assess disease burden, and data plotted as the mean ± SEM for each group. After 8 days on treatment, all mice were euthanized, and tissue collected. Spleen weights were expressed as mean ± SEM. One way ANOVA analysis with Tukey post-test was used to determine the significance of all pairwise comparisons.
Murine AML was induced by retroviral transduction of bone marrow with MLL-AF9 as previously described (Stubbs et al., 2008). Briefly, bone marrow was harvested from a transgenic mouse with ubiquitous luciferase expression (C57BL/6:UbC6-Luc) and transduced with a retrovirus encoding MLL-AF9 (MSCV-MLL-AF9-pgkNeo). Development of leukemia was determined by BLI and clinical scoring. Thirty albino co-isogenic mice (C57BL/6:TyrC/C, Jackson Labs) were sub-lethally irradiated with 300 rads, then injected with 106 mononuclear cells isolated from the spleen of a mouse with primary AML. Mice underwent BLI weekly, and at 3 weeks post injection, animals with documented disease (increasing bioluminescence) were divided into 2 cohorts of mice with equal mean bioluminescence. The treatment group (n=10) was started on diet impregnated with R788 at 3 g/kg AIN-76A rodent diet. The control group (n=10) was kept on a normal diet. Mice underwent serial BLI at the indicated days of treatment and were euthanized after the last imaging time point and tissue collected. Total body bioluminescence was quantitated using standardized regions of interest (Living Images, Caliper Life Sciences), and are expressed as mean ± SEM. Statistical significance for BLI data and spleen weights were calculated using Student’s t-test.
For the primary human orthotopic model of AML, NOG mice were sublethally irradiated with 200 rad and then injected by tail vein injection with 4 × 105 viable AML cells (Patient 11). Seventy days post injection, mice developed greater than 40% peripheral AML blasts as measured by FACS analysis of human specific CD45+ cells from peripheral blood. Mice were then divided into 2 cohorts with 2 animals per cohort. One group received 5g R788/kg AIN-76 rodent diet and the second a placebo food. A NOG control mouse without human AML was included. Six days post treatment, tail vein blood was sampled and peripheral AML blasts again measured by FACS analysis of human specific CD45+ cells. At day 7, animals were euthanized and tissue collected.
See Supplementary Methods for the full details of experimental methods.
Microarray Data
Previously published raw microarray data (Stegmaier et al., 2004) is available at http://www.broad.mit.edu/cancer/pub/GE-HTS_leuk or http://www.ncbi.nlm.nih.gov/geo
Supplementary Material
ACKNOWLEDGMENTS
We thank Rigel Pharmaceuticals, particularly Elliott Grossbard and Polly Pine, for supplying R406 and R788 chow. We also thank Jen Grenier for RNAi screening guidance; John Daley, Jill Angelosanto and Kathleen Brosnahan for FACS assistance; Jinyan Du and Shao-En Ong for insight regarding the proteomics experiment; Susan Buchanan for primary sample collection; Jeff Kutock for pathology slide review; Anu Narla and Benjamin Ebert for facilitating testing of normal myeloid progenitor cells; Giovanni Roti, Dorhyun Johng, and Robin Perry for technical assistance, and Curtis Glavin for graphical design. We thank all patients and clinicians who contributed invaluable primary AML samples. This work was supported by the National Cancer Institute (NCI; 5K08 CA098444), Howard Hughes Medical Institute, Sidney Kimmel Cancer Foundation, the Claudia Adams Barr Foundation, and the Gloria Spivak Support Fund (K.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SIGNIFICANCE
Long-term survival for patients with AML remains poor despite dose-intensive chemotherapy regimens. Identification of pharmacologically tractable targets offers an alternative treatment strategy. We build upon our prior observation that multiple EGFR inhibitors possess anti-AML activity by a non-EGFR mechanism. We develop a strategy to identify their off-target activity integrating proteomic and RNAi-based screening approaches. We identify the kinase Syk as a therapeutic target in AML and demonstrate that a Syk inhibitor has activity in AML cell lines, primary patient blasts, and in vivo AML models. With Syk inhibitors already in Phase II trials, these studies can be immediately translated to the clinic and demonstrate the feasibility of a paradigm for target identification in cell-based screens.
REFERENCES
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Balaian L, Ball ED. Cytotoxic activity of gemtuzumab ozogamicin (Mylotarg) in acute myeloid leukemia correlates with the expression of protein kinase Syk. Leukemia. 2006;20:2093–2101. doi: 10.1038/sj.leu.2404437. [DOI] [PubMed] [Google Scholar]
- Boehrer S, Ades L, Braun T, Galluzzi L, Grosjean J, Fabre C, Le Roux G, Gardin C, Martin A, de Botton S, et al. Erlotinib exhibits antineoplastic off-target effects in AML and MDS: a preclinical study. Blood. 2008;111:2170–2180. doi: 10.1182/blood-2007-07-100362. [DOI] [PubMed] [Google Scholar]
- Braselmann S, Taylor V, Zhao H, Wang S, Sylvain C, Baluom M, Qu K, Herlaar E, Lau A, Young C, et al. R406, an orally available spleen tyrosine kinase inhibitor blocks fc receptor signaling and reduces immune complex-mediated inflammation. J Pharmacol Exp Ther. 2006;319:998–1008. doi: 10.1124/jpet.106.109058. [DOI] [PubMed] [Google Scholar]
- Burke MD, Berger EM, Schreiber SL. Generating diverse skeletons of small molecules combinatorially. Science. 2003;302:613–618. doi: 10.1126/science.1089946. [DOI] [PubMed] [Google Scholar]
- Chan G, Pilichowska M. Complete remission in a patient with acute myelogenous leukemia treated with erlotinib for non small-cell lung cancer. Blood. 2007;110:1079–1080. doi: 10.1182/blood-2007-01-069856. [DOI] [PubMed] [Google Scholar]
- Chen L, Monti S, Juszczynski P, Daley J, Chen W, Witzig TE, Habermann TM, Kutok JL, Shipp MA. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111:2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Rowley B, Pao W, Hayday A, Bolen JB, Pawson T. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- Chu DH, Morita CT, Weiss A. The Syk family of protein tyrosine kinases in T-cell activation and development. Immunol Rev. 1998;165:167–180. doi: 10.1111/j.1600-065x.1998.tb01238.x. [DOI] [PubMed] [Google Scholar]
- Deckert M, Tartare-Deckert S, Couture C, Mustelin T, Altman A. Functional and physical interactions of Syk family kinases with the Vav proto-oncogene product. Immunity. 1996;5:591–604. doi: 10.1016/s1074-7613(00)80273-3. [DOI] [PubMed] [Google Scholar]
- Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- Feldman AL, Sun DX, Law ME, Novak AJ, Attygalle AD, Thorland EC, Fink SR, Vrana JA, Caron BL, Morice WG, et al. Overexpression of Syk tyrosine kinase in peripheral T-cell lymphomas. Leukemia. 2008;22:1139–1143. doi: 10.1038/leu.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie T, Abe A, Matsuda T, Kuno Y, Towatari M, Yamamoto T, Saito H, Emi N, Naoe T. TEL-Syk fusion constitutively activates PI3-K/Akt, MAPK and JAK2-independent STAT5 signal pathways. Leukemia. 2004;18:548–555. doi: 10.1038/sj.leu.2403266. [DOI] [PubMed] [Google Scholar]
- Kimbrel EA, Davis TN, Bradner JE, Kung AL. In vivo pharmacodynamic imaging of proteasome inhibition. Molecular Imaging. 2009 In Press. [PubMed] [Google Scholar]
- Kuno Y, Abe A, Emi N, Iida M, Yokozawa T, Towatari M, Tanimoto M, Saito H. Constitutive kinase activation of the TEL-Syk fusion gene in myelodysplastic syndrome with t(9;12)(q22;p12) Blood. 2001;97:1050–1055. doi: 10.1182/blood.v97.4.1050. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lindhagen E, Eriksson A, Wickstrom M, Danielsson K, Grundmark B, Henriksson R, Nygren P, Aleskog A, Larsson R, Hoglund M. Significant cytotoxic activity in vitro of the EGFR tyrosine kinase inhibitor gefitinib in acute myeloblastic leukaemia. Eur J Haematol. 2008;81:344–353. doi: 10.1111/j.1600-0609.2008.01120.x. [DOI] [PubMed] [Google Scholar]
- List A, Dewald G, Bennett J, Giagounidis A, Raza A, Feldman E, Powell B, Greenberg P, Thomas D, Stone R, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355:1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- List A, Kurtin S, Roe DJ, Buresh A, Mahadevan D, Fuchs D, Rimsza L, Heaton R, Knight R, Zeldis JB. Efficacy of lenalidomide in myelodysplastic syndromes. N Engl J Med. 2005;352:549–557. doi: 10.1056/NEJMoa041668. [DOI] [PubMed] [Google Scholar]
- Lupher ML, Jr., Rao N, Lill NL, Andoniou CE, Miyake S, Clark EA, Druker B, Band H. Cbl-mediated negative regulation of the Syk tyrosine kinase. A critical role for Cbl phosphotyrosine-binding domain binding to Syk phosphotyrosine 323. J Biol Chem. 1998;273:35273–35281. doi: 10.1074/jbc.273.52.35273. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. A method for high-throughput gene expression signature analysis. Genome Biol. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitini V, Arrigo C, Altavilla G. Erlotinib in a patient with acute myelogenous leukemia and concomitant non-small-cell lung cancer. J Clin Oncol. 2008;26:3645–3646. doi: 10.1200/JCO.2008.17.0357. [DOI] [PubMed] [Google Scholar]
- Polo JM, Dell’Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD, Melnick A. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- Ratain MJ, Eisen T, Stadler WM, Flaherty KT, Kaye SB, Rosner GL, Gore M, Desai AA, Patnaik A, Xiong HQ, et al. Phase II placebo-controlled randomized discontinuation trial of sorafenib in patients with metastatic renal cell carcinoma. J Clin Oncol. 2006;24:2505–2512. doi: 10.1200/JCO.2005.03.6723. [DOI] [PubMed] [Google Scholar]
- Rinaldi A, Kwee I, Taborelli M, Largo C, Uccella S, Martin V, Poretti G, Gaidano G, Calabrese G, Martinelli G, et al. Genomic and expression profiling identifies the B-cell associated tyrosine kinase Syk as a possible therapeutic target in mantle cell lymphoma. Br J Haematol. 2006;132:303–316. doi: 10.1111/j.1365-2141.2005.05883.x. [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Sada K, Takano T, Yanagi S, Yamamura H. Structure and function of Syk protein-tyrosine kinase. Journal of biochemistry. 2001;130:177–186. doi: 10.1093/oxfordjournals.jbchem.a002970. [DOI] [PubMed] [Google Scholar]
- Stegmaier K, Corsello SM, Ross KN, Wong JS, Deangelo DJ, Golub TR. Gefitinib induces myeloid differentiation of acute myeloid leukemia. Blood. 2005;106:2841–2848. doi: 10.1182/blood-2005-02-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmaier K, Ross KN, Colavito SA, O’Malley S, Stockwell BR, Golub TR. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- Stegmaier K, Wong JS, Ross KN, Chow KT, Peck D, Wright RD, Lessnick SL, Kung AL, Golub TR. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007;4:e122. doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel B, Vinatzer U, Willheim M, Raderer M, Chott A. Novel t(5;9)(q33;q22) fuses ITK to SYK in unspecified peripheral T-cell lymphoma. Leukemia. 2006;20:313–318. doi: 10.1038/sj.leu.2404045. [DOI] [PubMed] [Google Scholar]
- Stubbs MC, Kim YM, Krivtsov AV, Wright RD, Feng Z, Agarwal J, Kung AL, Armstrong SA. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22:66–77. doi: 10.1038/sj.leu.2404951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Mee PJ, Costello PS, Williams O, Price AA, Duddy LP, Furlong MT, Geahlen RL, Tybulewicz VL. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature. 1995;378:298–302. doi: 10.1038/378298a0. [DOI] [PubMed] [Google Scholar]
- Turner M, Schweighoffer E, Colucci F, Di Santo JP, Tybulewicz VL. Tyrosine kinase SYK: essential functions for immunoreceptor signalling. Immunology today. 2000;21:148–154. doi: 10.1016/s0167-5699(99)01574-1. [DOI] [PubMed] [Google Scholar]
- Vorhies JS, Nemunaitis JJ. Synthetic vs. natural/biodegradable polymers for delivery of shRNA-based cancer therapies. Methods Mol Biol. 2009;480:11–29. doi: 10.1007/978-1-59745-429-2_2. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinblatt ME, Kavanaugh A, Burgos-Vargas R, Dikranian AH, Medrano-Ramirez G, Morales-Torres JL, Murphy FT, Musser TK, Straniero N, Vicente-Gonzales AV, et al. Treatment of rheumatoid arthritis with a syk kinase inhibitor: A twelve-week, randomized, placebo-controlled trial. Arthritis Rheum. 2008;58:3309–3318. doi: 10.1002/art.23992. [DOI] [PubMed] [Google Scholar]
- Young RM, Hardy IR, Clarke RL, Lundy N, Pine P, Turner BC, Potter TA, Refaeli Y. Mouse models of non-Hodgkins lymphoma reveal Syk as an important therapeutic target. Blood. 2008 doi: 10.1182/blood-2008-05-158618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.