SUMMARY
MicroRNAs (miRNAs) inhibit mRNA expression in general by base pairing to the 3′UTR of target mRNAs and consequently inhibiting translation and/or initiating poly(A) tail deadenylation and mRNA destabilization. Here we examine the mechanism and kinetics of miRNA-mediated deadenylation in mouse Krebs-2 ascites extract. We demonstrate that miRNA-mediated mRNA deadenylation occurs subsequent to initial translational inhibition, indicating a two-step mechanism of miRNA action, which serves to consolidate repression. We show that a let-7 miRNA-loaded RNA-induced silencing complex (miRISC) interacts with the poly(A)-binding protein (PABP) and the CAF1 and CCR4 deadenylases. In addition, we demonstrate that miRNA-mediated deadenylation is dependent upon CAF1 activity and PABP, which serves as a bona fide miRNA coactivator. Importantly, we present evidence that GW182, a core component of the miRISC, directly interacts with PABP via its C-terminal region and that this interaction is required for miRNA-mediated deadenylation.
INTRODUCTION
MicroRNAs (miRNAs) are short single-stranded RNAs (~21 nt in length) encoded within the genome of species ranging from protozoans to plants to mammals (Bartel, 2004; Molnar et al., 2007). miRNAs play key roles in a broad range of biological processes including hematopoiesis, insulin secretion, apoptosis, and organogenesis (Bartel, 2004). When assembled together with Argonaute (Ago) proteins into the miRNA-induced silencing complex (miRISC), miRNAs base pair with and repress mRNA expression through mechanisms that are not fully understood (Eulalio et al., 2008a; Filipowicz et al., 2008).
miRNAs were reported to employ different mechanisms to inhibit expression of targeted mRNAs (Eulalio et al., 2008a; Filipowicz et al., 2008). Some data indicate that miRNAs interfere with mRNA translation at the initiation step (Chendrimada et al., 2007; Ding and Grosshans, 2009; Humphreys et al., 2005; Mathonnet et al., 2007; Pillai et al., 2005; Thermann and Hentze, 2007; Wang et al., 2008), whereas other studies concluded that the miRNA machinery represses translation at postinitiation steps (Gu et al., 2009; Lytle et al., 2007; Maroney et al., 2006; Nottrott et al., 2006; Olsen and Ambros, 1999; Petersen et al., 2006). miRNAs have been observed, although not in every study, to mediate deadenylation and/or decay of targeted mRNAs (Behm-Ansmant et al., 2006; Giraldez et al., 2006; Wakiyama et al., 2007; Wu et al., 2006).
In addition to Ago proteins, GW182 proteins also play key roles in miRNA-mediated repression. One GW182 protein (Gawky) exists in Drosophila, and three GW182 paralogs (TNRC6A, TNRC6B, and TNRC6C) are present in mammals. Direct interaction of GW182 with Ago proteins is critical for miRNA-mediated translation repression and mRNA decay (Eulalio et al., 2008b). Studies conducted with either TNRC6C or Gawky artificially tethered to a reporter mRNA demonstrated that a region within their C termini is required for repression of translation (Chekulaeva et al., 2009; Eulalio et al., 2009b; Zipprich et al., 2009).
Cell culture-based assays invariably measure miRNA effects hours or days after the initial mRNA target site recognition, making it difficult to ascertain the temporal order and contribution of the different proposed mechanisms to mRNA repression. Moreover, RNAi-based approaches for identifying miRNA-associated factors may perturb cellular transcriptional programs in such a way that it becomes difficult to determine direct contributions. Thus, developing an in vitro system that faithfully recapitulates all aspects of miRNA-mediated repression is necessary to elucidate the biochemistry of miRNA mechanisms of action, especially at early time points. Such systems have recently been reported (Mathonnet et al., 2007; Thermann and Hentze, 2007; Wakiyama et al., 2007; Wang et al., 2006).
To explore the mechanisms that miRNAs utilize to repress mRNA expression in mammals, we utilized an in vitro translation extract from mouse Krebs-2 ascites cells (referred to throughout as Krebs extract). We showed before that the earliest detectable effect of miRNA action is the inhibition of cap-dependent translation initiation (Mathonnet et al., 2007). We demonstrate here that miRNA-mediated deadenylation follows the initial inhibition of cap-dependent translation. We further show that Ago2 interacts with the CNOT7/CAF1 (hereafter referred to as CAF1) deadenylase and poly(A)-binding protein (PABP) in an RNA-independent manner, and that both proteins are required to facilitate miRNA-mediated deadenylation. Importantly, we show that PABP physically interacts with the miRISC by directly binding the C terminus of GW182 and is required for deadenylation.
RESULTS
miRNA-Mediated Deadenylation Follows Initial Translation Inhibition
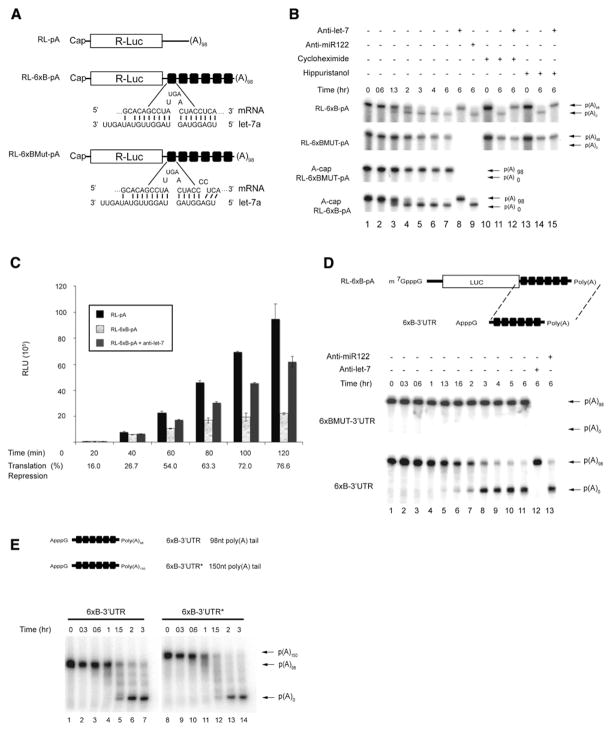
We previously described an in vitro translation extract derived from Krebs-2 ascites cells that contains high levels (~150 pM) of the let-7a (referred throughout as let-7) miRNA and displays a faithful let-7 miRNA response (Mathonnet et al., 2007). The Krebs extract manifests reduced translation initiation of in vitro-transcribed let-7-targeted mRNAs starting within the first 15 min of incubation without detectable mRNA degradation (Mathonnet et al., 2007). Since miRNAs were also reported to induce mRNA deadenylation (Eulalio et al., 2009a; Giraldez et al., 2006; Wakiyama et al., 2007; Wu et al., 2006), and since deadenylation generally results in translational repression (Wormington, 1993), we wished to determine whether miRNA-mediated deadenylation can be recapitulated in a Krebs extract and study the temporal relationship between translation inhibition and deadenylation. A polyadenylated RL-6xB mRNA (Figure 1A), labeled uniformly with 32P-UTP, was incubated in a Krebs extract, and its integrity was analyzed by denaturing polyacryl-amide-gel electrophoresis (PAGE) followed by autoradiography. A new RNA band migrating faster than the full-length mRNA was detected after ~1 hr of incubation (Figure 1B, lanes 3–7, and see Figure S1 available online). Formation of the new RNA species was dependent on let-7 miRNA as (1) inclusion of anti-let-7 2′-O-methylated oligonucleotide (2′-O-Me), but not anti-miR122 2′-O-Me, in the Krebs extract blocked the generation of this product (lanes 8 and 9, respectively); and (2) a reporter containing mutations in nucleotides complementary to the let-7 “seed” sequence (RL-6xBMut-pA; see Figure 1A and Figure 1B, lanes 1–7), and a reporter devoid of let-7 sites (RL-pA; Figure S1) failed to give rise to this band. Cloning and sequencing of the new RNA species using an oligonucleotide-ligation RT-PCR strategy (Figure S2) demonstrated that it represents a deadenylation product of the RL-6xB-pA mRNA. Thus, let-7 miRNA mediates deadenylation of the targeted mRNA in the Krebs extract, but with the earliest detection only after 1.3 hr of incubation. As translational inhibition (~55%) occurs within the first hour of incubation in the same Krebs extract in which deadenylation has been monitored (Figure 1C; see also Mathonnet et al. [2007]), it appears that miRNA-mediated inhibition of cap-dependent translation precedes mRNA deadenylation. When translation of RL-6xB-pA mRNA was allowed to proceed for longer times, the degree of translation repression increased from ~55% at 1 hr to ~77% at 2 hr (Figure 1C; three different experiments). These data indicate that deadenylation may consolidate the initial inhibition of cap-dependent translation.
Figure 1. Deadenylation Mediated by let-7 miRNA in a Krebs Extract.
(A) Schematic representation of the Renilla luciferase (Rluc) reporter mRNAs. Sequences of the let-7-binding sites (RL-6xB) and mutated seed sites (RL-6xBMut) are shown below the drawings.
(B) Time course of RL-6xB-pA and RL-6xBMUT-pA mRNA deadenylation as determined by autoradiography. Reporter mRNAs were incubated in the presence or absence of 10 μM cycloheximide, 1 mM hippuristanol, or 10 nM 2′-O-Me oligonucleotide (either anti-let-7a or anti-miR122).
(C) A time course of translation of RL-pA, RL-6xB-pA, and RL-6xB-pA in the presence of anti-let-7 2′-O-Me. Average percentage repression is labeled below each time point. Error bars represent the standard deviation of three independent experiments.
(D) Schematic representation of the 6xB-3′UTR reporter RNA and time course of 6xB-3′UTR and 6xBMUT-3′UTR RNA deadenylation in a Krebs extract as determined by autoradiography. Reporters were incubated in the presence or absence of 10 nM 2′-O-Me oligonucleotide (either anti-let-7a or anti-miR122), and their stability was monitored by autoradiography.
(E) Schematic representation of the 6xB-3′UTR reporter RNAs with either 98As or 150As (*). Time course of 6xB-3′UTR and 6xB-3′UTR* deadenylation in a Krebs extract as determined by autoradiography. Polyadenylated and deadenylated mRNAs are marked on the right of the figure.
Next, we asked whether deadenylation is dependent on translation. To this end, translation was inhibited in the Krebs extract by the addition of either cycloheximide (Figure 1B, lanes 10–12), which blocks translation elongation, or hippuristanol (lanes 13–15), which inhibits translation initiation (Bordeleau et al., 2006). Inhibiting either step of translation failed to block let-7-induced deadenylation of RL-6xB-pA mRNA. We then examined whether the m7GpppG-cap structure is required for miRNA-mediated deadenylation. Deadenylation assays were conducted with RL-6xB-pA and RL-6xBMUT-pA mRNAs possessing an ApppG-cap, which cannot be bound by eIF4E but protects the RNA against degradation by 5′-3′ exonucleases. Neither the time course nor the extent of deadenylation of A-capped RL-6xB-pA significantly differed from RL-6xB-pA bearing an m7GpppG structure (Figure 1B). Since miRNA-mediated deadenylation is an m7GpppG-cap- and translation-independent event, we examined whether any RNA element upstream of the RL-6xB-pA 3′UTR is required for miRNA-mediated deadenylation. ApppG-capped 3′UTR transcripts were generated that lack an open reading frame and contain six either functional (6xB-3′UTR) or mutated (6xBMUT-3′UTR) let-7 sites and a 98 nt poly(A) tail (Figure 1D). The 6xB-3′UTR RNA recapitulated both the time course and the deadenylation pattern observed for the full-length RL-6xB-pA mRNA (Figure 1D). Deadenylation was dependent on let-7 miRNA as (1) addition of anti-let-7a 2′O-Me oligonucleotide, but not a nonspecific anti-miR122 2′-O-Me oligonucleotide (Figure 1D, lanes 12 and 13, respectively), abrogated the deadenylation of 6xB-3′UTR RNA; and (2) the 6xBMUT-3′UTR RNA was not deadenylated (Figure 1D). A 6xB-3′UTR RNA with a longer poly(A) tail (150 nt, 6xB-3′UTR*) behaved similarly to the 6xB-3′UTR RNA vis-a-vis the time course and the deadenylation pattern (Figure 1E). Taken together, our findings demonstrate that no RNA determinant other than the let-7 target sites is required for miRNA-mediated deadenylation.
Argonaute Proteins Interact with CAF1 and CCR4 Deadenylases
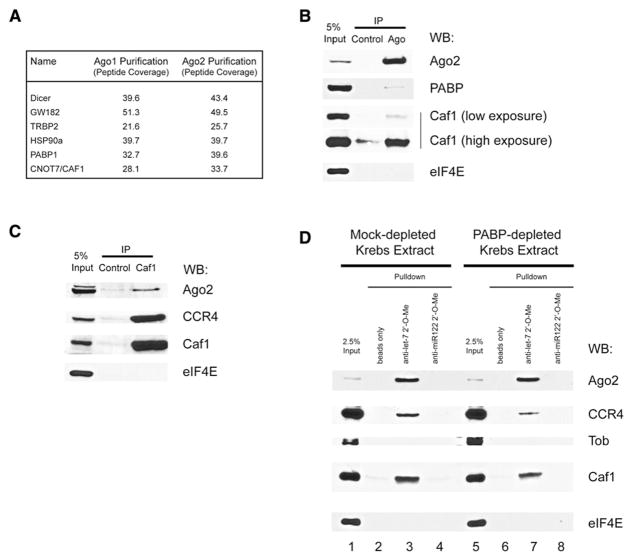
We used several approaches to identify the deadenylase(s) involved in the miRNA-mediated deadenylation. In one approach, Myc-tagged Ago1 and Ago2 were stably transfected into HEK293 cells. Tagged Ago proteins were immunopurified, and the associated proteins were identified by using multidimensional protein identification technology (MuDPIT) (Washburn et al., 2001; Wolters et al., 2001). This method was validated by the identification of known Ago2-interacting proteins such as HSP90, DICER, TRBP, and GW182 (Figure 2A) (Chendrimada et al., 2005; Landthaler et al., 2008; Liu et al., 2005; Meister et al., 2005). In addition, PABP was identified in both Ago1 and Ago2 immunopurifications (Figure 2A) (Hock et al., 2007; Landthaler et al., 2008). One identified protein that was not reported before to interact with Ago proteins was CAF1 deadenylase. To validate this interaction, we performed coimmunoprecipitation experiments using a micrococcal nuclease-treated Krebs extract. When endogenous Ago2 was immunoprecipitated from the Krebs extract, the precipitated fraction contained Ago2 and CAF1, but not eIF4E (Figure 2B). When endogenous CAF1 was immunoprecipitated from Krebs extracts, the precipitated fraction contained CAF1, CCR4 (a CAF1-associated deadenylase [Tucker et al., 2001]), and Ago2, but not eIF4E (Figure 2C).
Figure 2. Ago Proteins Interact with PABP and the CAF1/CCR4 Deadenylase Complex.
(A) MuDPIT analysis of Ago1- and Ago2-interacting proteins. Identified proteins are listed along with corresponding peptide coverage for Ago1 and Ago2 coimmunoprecipitations.
(B) Immunoprecipitation of endogenous Ago2 protein from micrococcal nuclease-treated Krebs extract using anti-Ago2 antibody. Immunoprecipitated complexes were subjected to SDS-PAGE and probed with anti-Ago2 antibody, anti-CAF1 antibody, anti-PABP antibody, or anti-eIF4E antibody.
(C) Immunoprecipitation of endogenous CAF1 protein from micrococcal nuclease-treated Krebs extract using anti-CAF1 antibody. Immunoprecipitated complexes were subjected to SDS-PAGE and probed with anti-Ago2 antibody, anti-CAF1 antibody, anti-CCR4 antibody, or anti-eIF4E antibody.
(D) Pulldown of Ago2, CCR4, and CAF1 from micrococcal nuclease-treated Krebs extracts using biotin-conjugated anti-let-7 2′-O-Me oligo-nucleotide and streptavidin Dynabeads. Isolated complexes were subjected to SDS-PAGE and probed with anti-Ago2 antibody, anti-CAF1 antibody, anti-CCR4 antibody, anti-Tob antibody, or anti-eIF4E antibody.
To determine whether the CAF1 and CCR4 deadenylases can be recruited by the let-7-loaded Ago2, we used a 2′-O-Me RNA target “bait” pulldown assay (Hutvagner et al., 2004). Biotinylated 2′-O-Me oligonucleotides, which mimic partially complementary mRNA target sites for let-7 or miR122 (a liver-specific miRNA [Lagos-Quintana et al., 2002] that can pull down Ago2 from lysates derived from Huh7 liver cells [Figure S3]), were incubated in Krebs extract and pulled down using streptavidin beads. The associated proteins were eluted and analyzed by western blotting. Ago2 bound specifically to the anti-let-7 2′-O-Me beads and failed to bind to control beads or anti-miR122 2′-O-Me beads (Figure 2D, lanes 2–4). Importantly, in these pulldown experiments a similar pattern of enrichment was observed for CAF1 and CCR4, but not for eIF4E or Tob (a protein that can associate with CAF1 to enhance deadenylation [Ezzeddine et al., 2007; Mauxion et al., 2008]). These results demonstrate that CAF1 and CCR4 can be specifically recruited to the target-bound let-7-loaded Ago2.
miRNAs Require CAF1 Activity to Promote Deadenylation
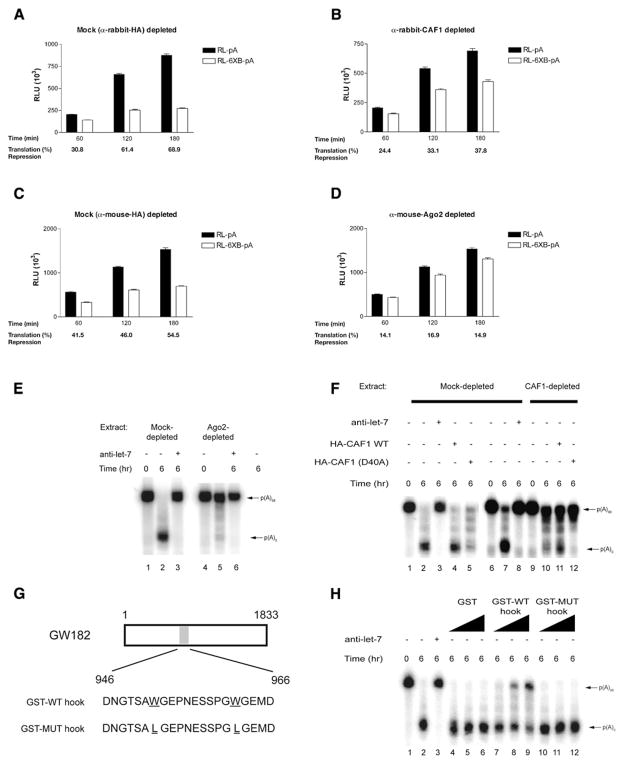
To determine whether CAF1 is required for miRNA-mediated deadenylation, it was immunodepleted (~80%) from the Krebs extract using an affinity-purified CAF1 antibody (Figure S4). Analysis of the depleted extract (Figure 3) demonstrated that miRNA-mediated translation inhibition is partially relieved in both CAF1- and Ago2-depleted extracts (37.8% [Figure 3B] and 14.9% repression [Figure 3D], respectively, after 3 hr incubation) when compared to the corresponding control-depleted extracts (68.9% [Figure 3A] and 54.5% [Figure 3C] repression after 3 hr incubation). The Ago2-depleted extract was dramatically impaired in its ability to deadenylate the 6xB-3′UTR RNA, inasmuch as deadenylation was barely detectable even after 6 hr of incubation (Figure 3E, lane 5). A similar decrease in deadenylation was detected in a Krebs extract depleted of CAF1 (Figure 3F, lane 10). These deadenylation defects were specific, because in a mock-depleted extract, 6xB-3′UTR RNA was deadenylated in a let-7-dependent manner (Figures 3E and 3F, lanes 1–3). miRNA-mediated deadenylation was modestly restored (2.2-fold increase; from ~5% to ~12% deadenylation) by the addition of affinity-purified wild-type HA-CAF1 to the CAF1-depleted extract (Figure 3F, lane 11), while wild-type HA-CAF1 had no noticeable effect on mock-depleted extract (lane 4). Modest restoration was most likely due to a small fraction of affinity-purified wild-type HA-CAF1 being bound to let-7-loaded miRISC. In contrast, addition of affinity-purified catalytically inactive HA-CAF1 mutant (D40A) (Zheng et al., 2008) decreased miRNA-induced deadenylation in both mock- and CAF1-depleted extracts (Figure 3F, lanes 5 and 12, respectively). This is likely due to HA-CAF1(D40A) acting as a dominant-negative mutant in both mock- and CAF1-depleted extracts. Taken together, these results show that miRNA-induced deadenylation is executed, at least in part, by the CAF1 deadenylase.
Figure 3. let-7-Mediated Deadenylation Requires CAF1, Ago2, and GW182.
(A–D) Time course of RL-pA and RL-6xB-pA translation in rabbit anti-HA- (A), rabbit anti-CAF1- (B), mouse anti-HA- (C), and mouse anti-Ago2-depleted Krebs extracts (D). Average percentage repression is labeled below each time point. Error bars represent the standard deviation of three independent experiments.
(E) 6xB-3′UTR RNA deadenylation in the presence or absence of 10 nM anti-let-7a 2′-O-Me in control (mouse anti-HA) or anti-Ago2-depleted Krebs extract. 6xB-3′UTR RNA deadenylation was followed by autoradiography. Polyadenylated and deadenylated mRNAs are marked on the right of the figure.
(F) 6xB-3′UTR RNA deadenylation in control (rabbit anti-HA) or anti-CAF1-depleted extract in the presence or absence of either 10 nM anti-let-7a 2′-O-Me oligonucleotide, or WT or D40A HA-CAF1 protein.
(G) Wild-type and mutant hook peptides derived from GW182.
(H) 6xB-3′UTR RNA deadenylation in Krebs extract in the presence or absence of either GST or GST hook peptides at concentrations ranging from 0.1 to 2.0 μg per reaction, respectively.
Ago2-GW182 Interaction Is Essential for miRNA-Mediated Deadenylation
The Ago-binding protein GW182 is required for efficient miRNA-mediated silencing in C. elegans and in Drosophila S2 cells (Behm-Ansmant et al., 2006; Ding and Han, 2007; Eulalio et al., 2008b). GW182 is required for the assembly of P bodies, protein-RNA assemblies thought to contribute to translation inhibition and mRNA destabilization (Behm-Ansmant et al., 2006; Ding and Han, 2007; Jakymiw et al., 2007; Liu et al., 2005; Pillai et al., 2005; Rehwinkel et al., 2005). CAF1 also localizes to P bodies in mammalian cells (Zheng et al., 2008). We therefore investigated whether the GW182 interaction with Ago2 plays a role in miRNA-mediated deadenylation in vitro. To this end, we used a 22 amino acid fragment of GW182 (called “Ago hook”) (Figure 3G) that competes with GW182 for Ago binding and inhibits miRNA-mediated repression in vivo (Till et al., 2007). A Krebs extract was incubated with either GST alone, GST fused to Ago hook (GST-WT hook), or GST fused to a mutant hook (GST-MUT hook) containing two Trp to Leu mutations that abrogate the ability of the hook to bind to Ago (Till et al., 2007) (Figure S5). Addition of a recombinant GST-WT hook, but not GST alone or GST-MUT hook to the Krebs extract, impaired the deadenylation of 6xB-3′UTR RNA in a concentration-dependent manner (Figure 3H, lanes 7–9 compared to lanes 4–6 and 10–12, respectively). These findings indicate that miRNA-mediated deadenylation in vitro requires GW182 contact with Ago2 at the hook site.
PABP Is Required for miRNA-Mediated Deadenylation
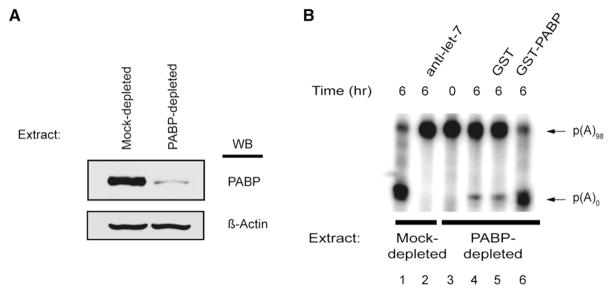
Since the MuDPIT analysis identified PABP as an Ago-interacting protein, it was pertinent to determine whether PABP is necessary for miRNA-induced deadenylation. A Krebs extract was depleted (>95%) of endogenous PABP using a GST-tagged PABP-interacting protein 2 (Paip2) affinity matrix (Figure 4A). Paip2 is a strong translational inhibitor and acts by sequestering PABP and blocking PABP-poly(A) tail and PABP-eIF4G interactions in vitro (Karim et al., 2006; Khaleghpour et al., 2001). GST-Paip2 coupled to a resin was previously used to efficiently deplete PABP from a Krebs extract, resulting in reduced translation (Kahvejian et al., 2005). Strikingly, the PABP-depleted extract was severely impaired in its ability to deadenylate the 6xB-3′UTR RNA (Figure 4B, compare lane 4 to lane 1). This was a specific consequence of PABP depletion as a mock-depleted extract still deadenylated the reporter RNA and was responsive to the addition of anti-let-7 2′-O-Me oligonucleotide (Figure 4B, lanes 1 and 2, respectively). Moreover, addition of recombinant GST-PABP (50% of endogenous PABP levels in a Krebs extract [Figure S6]) to the PABP-depleted extract (lane 6), but not GST alone (lane 5), completely rescued miRNA-mediated deadenylation of 6xB-3′UTR RNA. The rescue was prevented by the addition of anti-let-7 2′-O-Me oligonucleotide (Figure S7, lane 8). These findings clearly show that PABP is essential for miRNA-mediated deadenylation in vitro.
Figure 4. let-7-Dependent Deadenylation Requires PABP.
(A) Western blot analysis of Krebs-2 extracts depleted with either GST (Control Extract) or GST-Paip2 (PABP-depleted Extract) probed with anti-PABP antibody and anti-β-actin antibody.
(B) A-capped 6xB-3′UTR RNA incubated in either mock-depleted (lanes 1–2) or PABP-depleted extract (lanes 3–6). PABP-depleted extract was supplemented with recombinant GST, GST-PABP (100 ng, which is the equivalent of roughly 50% of endogenous PABP present in an in vitro reaction), and RNA stability was monitored by autoradiography. Polyadenylated and deadenylated mRNAs are marked on the right of the panel.
PABP Function in miRNA-Mediated Deadenylation Is Antagonized by eIF4G
How does PABP facilitate miRNA-mediated deadenylation? PABP is probably not required for miRISC target site recognition, as the let-7-loaded Ago2 can be pulled down with anti-let-7 2′-O-Me oligonucleotide from a PABP-depleted Krebs extract almost as well as from a nondepleted extract (Figure 2D, compare lanes 3 and 7). Moreover, PABP is required for recruiting neither CAF1 nor CCR4 as they are pulled down in similar amounts from PABP-depleted extracts with anti-let-7 2′-O-Me oligonucleotide (Figure 2D, compare lanes 3 and 7). It is unlikely that PABP’s role is to compete with other proteins for poly(A) tail binding, as adding free poly(A) to PABP-depleted extracts (Figure S8) does not rescue miRNA-mediated deadenylation.
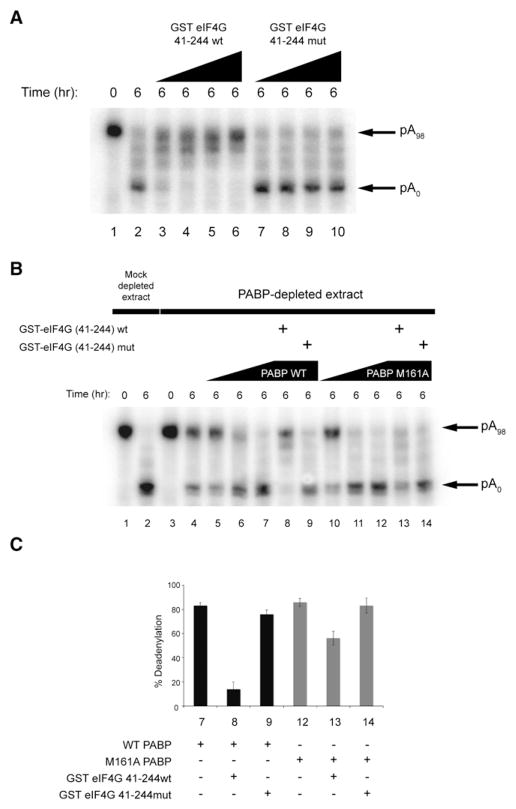
The N-terminal region of PABP can interact with the translation initiation factor eIF4G, and this interaction stimulates translation (Imataka et al., 1998; Wakiyama et al., 2000). To determine whether this interaction might antagonize deadenylation, Krebs extract was incubated with increasing concentrations of either an N-terminal eIF4G fragment (GST-eIF4G 41-244wt) that binds the N terminus of PABP or a mutant eIF4G fragment (GST-eIF4G 41-244mut) that does not bind to PABP (Kahvejian et al., 2005) (Figures 5A and S9). Addition of a wild-type (lanes 3–6), but not the mutant eIF4G fragment (lanes 7–10), impaired the deadenylation of 6xB-3′UTR RNA in a concentration-dependent manner.
Figure 5. eIF4G Contact with PABP Antagonizes miRNA-Mediated Deadenylation.
(A) 6xB-3′UTR RNA deadenylation in Krebs extract in the presence or absence of increasing concentrations (0.15, 0.5, 1.0, and 3.0 μg per reaction) of wild-type or mutant GST-eIF4G 41-244.
(B) 6xB-3′UTR RNA deadenylation in mock- or PABP-depleted Krebs extract in the presence or absence of increasing concentrations (25, 50, or 100 ng per reaction, respectively) of either wild-type (lanes 5–7) or M161A PABP (lanes 10–12) and/or wild-type (lanes 8 and 13) or mutant (lanes 9 and 14) GST-eIF4G (41-244).
(C) Quantification of deadenylated bands as a percentage of total RNA in (B) is shown in bar graphs (with standard deviations).
We next examined whether the effect of GST-eIF4G 41-244wt on miRNA-mediated deadenylation was a result of its binding to PABP. PABP-depleted extracts were supplemented with either wild-type or PABP M161A that cannot bind eIF4G (Groft and Burley, 2002) (Figures 5B, 5C, and S9). miRNA-mediated deadenylation in PABP-depleted extracts can be rescued equally well with PABP M161A as compared to wild-type PABP (Figure 5B, compare lanes 5–7 with lanes 10–12). Addition of GST-eIF4G 41-244wt blocked deadenylation in a PABP-depleted extract supplemented with wild-type PABP (lane 8) but decreased it only minimally when supplemented with PABP M161A (lane 13). These findings suggest that the eIF4G-PABP interaction is not required for, but rather interferes with, miRNA-mediated deadenylation.
PABP Interacts with the C-Terminal Region of GW182
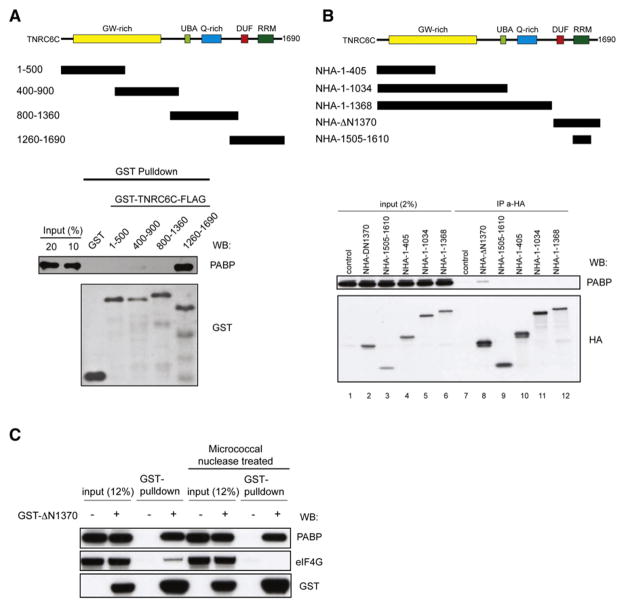
GW182 is a core component of miRISC, and its contact with Ago is required for miRNA-mediated repression (Eulalio et al., 2008b; Till et al., 2007). Mammalian and C. elegans GW182 protein orthologs were previously shown to coimmunoprecipitate with PABP (Landthaler et al., 2008; Zhang et al., 2007), but whether these associations were direct has not been determined. To test for a direct interaction between GW182 and PABP, we performed GST pulldown experiments using recombinant His-tagged PABP and four GST- and FLAG-tagged partially overlapping fragments (covering amino acids 1–500, 400–900, 800–1360, and 1260–1690) of the human GW182 protein TNRC6C (Figure 6A). GST on its own and fusions with TNRC6C fragments 1–500, 400–900, and 800–1360 did not interact with PABP. In contrast, the C-terminal 1260–1690 of TNRC6C, which harbors both the domain of unknown function (DUF; Zipprich et al., 2009) and RRM domains of TNRC6C pulled down PABP very efficiently (20% of input; Figure 6A). GST pulldown experiments using overlapping fragments of another human GW182 paralog (TNRC6A) and PABP yielded similar results (data not shown).
Figure 6. The C Terminus of TNRC6C Directly Binds PABP.
(A) Schematic representation of human TNRC6C and GST- and FLAG-tagged recombinant protein fragments. Western blot analysis of GST pull-downs of PABP incubated with GST or various fragments of GST-TNRC6C-FLAG and probed with anti-PABP and anti-GST antibodies.
(B) Schematic representation of human TNRC6C HA-tagged fragments transfected into HEK293 cells. Cell extracts of HEK293 cells, transiently expressing the indicated fusion proteins, were incubated with Anti-HA Affinity Matrix (Roche), and immunoprecipitated proteins were analyzed by western blotting using the indicated antibodies. Inputs represent 1% of the cell extract used for IP. Nontransfected cells served as a control.
(C) Cell extracts of HEK293 cells transiently expressing GST-ΔN1370 were pulled down using glutathione Sepharose resin in the presence or absence of micrococcal nuclease. GST pulldowns were analyzed by western blotting using anti-PABP, anti-eIF4G, and anti-GST antibodies. Non-transfected cells served as a control.
We next investigated whether the C-terminal region of TNRC6C interacts with PABP in transfected HEK293 cells. Of the HA-tagged fragments spanning different regions of TNRC6C (Figure 6B), only the C-terminal fragment, ΔN1370, encompassing residues 1370–1690, pulled down endogenous PABP (Figure 6B, lane 8). In additional experiments, lysates from cells expressing GST-ΔN1370 were used for GST pulldowns. In the absence of micrococcal nuclease treatment, GST-ΔN1370 pulled down both PABP and eIF4G. However, in nuclease-treated lysates GST-ΔN1370 pulled down only PABP (Figure 6C), demonstrating the RNA independence of the interaction between TNRC6C and PABP. Taken together, these data indicate that the C-terminal region of the GW182 protein TNRC6C interacts directly with PABP in an RNA-independent manner.
GW182 Contact with the PABP C-Terminal Domain Is Required for Maximal miRNA-Mediated Deadenylation
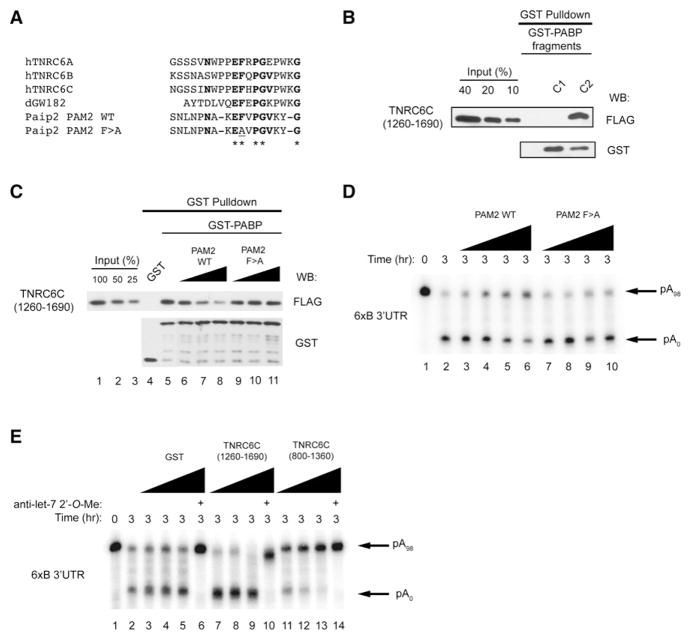
We next performed a sequence analysis of the C terminus of GW182 proteins to identify any potential PABP-interacting motifs. We observed a short sequence within the DUF that shows similarity to the Paip2 PAM2 motif (Figure 7A) that is required for Paip2 to bind the second half of the PABP C terminus (C2) (Khaleghpour et al., 2001; Kozlov et al., 2004). GST pulldown experiments were subsequently carried out using recombinant GST-tagged C-terminal PABP fragments (GST-C1 and GST-C2) and the FLAG-tagged TNRC6C C terminus (covering amino acids 1260–1690 [Figure 7B]). The PABP GST-C1 fusion did not pull down the GW182 1260–1690 fusion. In contrast, GST-C2 pulled down the TNRC6C C-terminal fragment very efficiently (~40% of input).
Figure 7. GW182 Binding to PABP Is Required for miRNA-Mediated Deadenylation.
(A) Alignment of GW182 DUF sequences with Paip2 PAM2 motif.
(B) Western blot analysis of GST pulldowns of TNRC6C (1260-1690)-FLAG incubated with various C-terminal (C1 and C2) fragments of GST-PABP and probed with anti-FLAG and anti-GST antibodies.
(C) Western blot analysis of GST pulldowns of GST-PABP incubated with TNRC6C (1260-1690)-FLAG and/or wild-type or mutant PAM2 peptide and probed with anti-FLAG and anti-GST antibodies.
(D) 6xB-3′UTR RNA deadenylation in Krebs extract in the presence of increasing concentrations (1, 10, 50, and 100 μM) of wild-type or mutant (F > A) Paip2 PAM2 peptides.
(E) 6xB-3′UTR RNA deadenylation in Krebs extract, as determined by autoradiography, in the presence of increasing concentrations (0.5, 1.0, and 2.0 μg per reaction) of GST, TNRC6C (1260-1690), or TNRC6C (800-1360).
To determine whether miRNA-mediated deadenylation requires GW182 contact with the PABP C2 domain, a Paip2-derived PAM2 peptide that specifically binds the C2 domain (Figure 7C) was used. Addition of increasing concentrations of wild-type, but not mutant PAM2 peptide (F117A [Kozlov et al., 2004]) to GST-PABP incubated with TNRC6C 1260–1690 fragment, blocked, albeit not completely, the binding of the TNRC6C C terminus to PABP (lanes 6–8 compared to lanes 9–11). Consistently, addition of the wild-type, but not the mutant PAM2 peptide to a Krebs extract, interfered with miRNA-mediated deadenylation in vitro in a dose-dependent manner (Figure 7D, lanes 3–6 compared to lanes 7–10). Taken together, these findings demonstrate that maximal miRNA-mediated deadenylation in vitro requires GW182 contact with the PABP C2 domain.
To further assess the function of the GW182 C terminus in miRNA-mediated deadenylation, we added the C-terminal recombinant 1260–1690 fragment to in vitro deadenylation reactions. The fragment dramatically enhanced miRNA-mediated deadenylation in vitro (Figure 7E, lanes 7–10 as compared to lane 2). The enhancement is specific, since adding a TNRC6C fragment 800–1360 that overlaps the 1260–1690 fragment but cannot bind PABP inhibited rather than enhanced the deadenylation in the same assays (lanes 11–14 as compared to lane 2). These data demonstrate the key role that the PABP-GW182 interaction plays in miRNA-mediated deadenylation.
DISCUSSION
In this report we used a mammalian cell-free extract to demonstrate that miRNAs mediate deadenylation of a target mRNA subsequent to initial inhibition of cap-dependent translation. Biochemical methods and functional assays in this in vitro system elucidated some of the protein and RNA requirements for miRNA-mediated mRNA deadenylation.
CAF1 and CCR4 Are Mammalian miRISC-Associated Deadenylases
miRNAs have previously been implicated in the deadenylation of targeted mRNAs in mammalian cells (Wu et al., 2006). One major deadenylase complex in mammals is the multisubunit CCR4-NOT complex, which contains two proteins having deadenylase activity, CCR4 and CAF1 (Yamashita et al., 2005; Zheng et al., 2008). Although CCR4 is the active deadenylase in the yeast CCR4-NOT complex (Tucker et al., 2001, 2002), mammalian CAF1 is also a processive deadenylase that regulates mRNA decay (Bianchin et al., 2005; Funakoshi et al., 2007; Schwede et al., 2008; Viswanathan et al., 2004; Zheng et al., 2008). Previous work carried out in Drosophila S2 cells demonstrated that the CCR4-NOT complex (which contains CAF1) facilitates miRNA-mediated deadenylation (Behm-Ansmant et al., 2006). Our results bolster these findings and show that the association of the miRISC with the deadenylase complex is conserved between Drosophila and mammals. Moreover, we provide biochemical evidence that both deadenylases physically interact with the mammalian miRISC, and that CAF1 activity is responsible, at least in part, for miRNA-mediated deadenylation. As CAF1 interacts with both Ago1 and Ago2 in HEK293 cells, this suggests that both Ago proteins are involved in facilitating miRNA-mediated deadenylation in mammals.
PABP as a Coactivator of miRNA-Mediated Deadenylation
Studies aimed at characterizing miRISC-associated proteins have previously identified PABP by mass spectrometry of immunoprecipitates not subjected to ribonuclease treatment (Hock et al., 2007; Landthaler et al., 2008; Zhang et al., 2007). We show that PABP is required for miRNA-mediated deadenylation and physically interacts with the miRISC via direct contact with GW182. Moreover, our results suggest that PABP-GW182 interaction is required to facilitate miRNA-mediated deadenylation. Previous studies have shown that PABP augments the activity of different deadenylases. PABP helps to recruit the PAN2/3 deadenylase complex to poly(A) tails in both yeast and mammalian systems via a direct interaction between the PAN3 subunit and the PABP C-terminal domain (Lowell et al., 1992; Uchida et al., 2004). The PABP C-terminal domain directly binds to the CAF1-interacting protein Tob, which may contribute to the CCR4-CAF1-mediated deadenylation of some mRNAs (Ezzeddine et al., 2007; Simon and Seraphin, 2007). In contrast to these modes of PABP-dependent deadenylation, our data show that PABP is not required for recruitment of either the miRISC or the miRISC-associated deadenylase complex to miRNA-targeted mRNAs (Figure 2D). Furthermore, while CAF1 is recruited to the miRISC, Tob is not Figure 2D).
PABP-GW182 Interaction and miRNA-Mediated Repression
GW182 is a core component of the miRISC and is critical for miRNA-mediated repression. All three mammalian paralogs of GW182 (TNRC6A, TNRC6B, and TNRC6C) are involved in miRNA-mediated repression (Jakymiw et al., 2007; Lazzaretti al., 2009; Liu et al., 2005; Till et al., 2007; Zipprich et al., 2009). Tethering experiments of different TNRC6C fragments to a reporter mRNA demonstrated that a C-terminal fragment of TNRC6C, harboring both the DUF and RRM domains, represses protein synthesis as effectively (>10-fold) as a full-length TNRC6C protein (Zipprich et al., 2009). Experiments performed with Drosophila GW182 protein in S2 cells also pointed to the importance of the protein C terminus for repression of protein synthesis (Chekulaeva et al., 2009; Eulalio et al., 2009b), implying functional conservation. We demonstrate that the mammalian GW182 C terminus directly binds PABP in an RNA-independent manner. Importantly, we show that GW182-PABP contact through the PABP C2 domain is required for efficient miRNA-mediated deadenylation. Because PABP functions as a bona fide translation initiation factor (Kahvejian et al., 2005), these data provide evidence that the mammalian miRISC directly interacts with a component of the translation initiation machinery. It is possible that PABP binding to GW182 may compete with eIF4G binding, as adding an eIF4G fragment that binds to the N terminus of PABP blocks miRNA-mediated deadenylation in vitro (Figure 5). In addition, it is conceivable that PABP binding to GW182 may function to juxtapose the poly(A) tail against the miRISC-associated deadenylase complex (see model, Figure 8). Although intriguing, these possibilities are still speculative at this point and await future experimental validation.
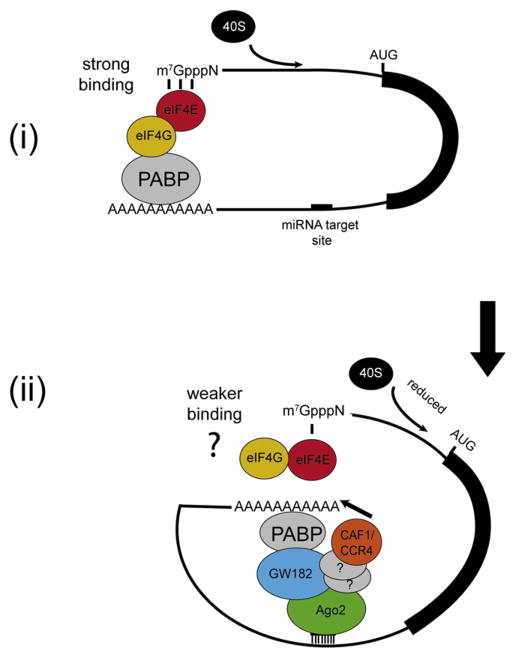
Figure 8. Model for Temporal Stepwise miRNA-Mediated Gene Silencing.
(i) mRNA circularization via eIF4G-PABP interaction stimulates cap-dependent translation by enhancing eIF4E’s binding to the mRNA 5′ cap structure (strong binding [Kahvejian et al., 2005]).
(ii) miRISC binds to its target site in the 3′UTR. GW182 binds to PABP, hypothetically inhibiting its interaction with eIF4G, thereby repressing cap-dependent translation by decreasing eIF4E’s binding to the 5′ cap structure (weaker binding), and sequestering the poly(A) tail into the vicinity of CAF1 and CCR4 deadenylases (illustrated by an arrow) to facilitate deadenylation of the mRNA. The interaction between CAF1/CCR4 and Ago2 is probably indirect through other proteins (depicted as question marks).
Temporal Mode of miRNA Action
miRNAs inhibit translation and/or mediate deadenylation and decay of target mRNAs (Filipowicz et al., 2008). In previous studies, mostly carried out in cultured cells, it was impossible to determine the earliest events leading to the miRNA-mediated repression (Behm-Ansmant et al., 2006; Giraldez et al., 2006; Humphreys et al., 2005; Petersen et al., 2006; Pillai et al., 2005). We have demonstrated that miRNAs inhibit translation initiation as early as 15–40 min after addition of mRNA to the Krebs extract (Mathonnet et al., 2007, and this study). As shown in this work, the miRNA-induced deadenylation of targeted mRNAs in vitro is a slower event, which follows the miRISC-mediated repression of translation initiation. These results indicate that miRNAs can function by two complementary and likely sequential mechanisms, first by inhibiting initiation of cap-dependent translation, which is then followed by the deadenylation of the target mRNA. As miRNA-mediated repression in Krebs extract further increases between 1 and 2 hr of incubation and miRNA-mediated translation repression is partially inhibited in CAF1-depleted extract, it is possible that deadenylation has an additional repressive effect supplementary to the initial inhibition of cap-dependent translation.
EXPERIMENTAL PROCEDURES
DNA Constructs and Protein Purification
Myc-Ago1 and Ago2 DNA constructs have been described (Liu et al., 2005). HA-CAF1 wild-type and HA-CAF1 D40A constructs have been described (Zheng et al., 2008). pGST-Paip2 and pGST-PABP full-length and fragments C1 and C2 have been described (Khaleghpour et al., 2001). Plasmids encoding wild-type and mutant HA-fused CAF1 proteins were transfected into HeLa cells and proteins were eluted with HA peptide (Anaspec). Eluted proteins were analyzed by western blot analysis using CAF1 and Ago2 antibodies. The plasmids pCI-NHA-1-405, pCI-NHA-1-1034, pCI-NHA-1-1368, pCI-NHA-ΔN1370, and pCI-NHA-1505-1610 were previously described (Zipprich et al., 2009). To generate the plasmid pEBG-ΔN1370, the sequence encoding a C-terminal part of TNRC6C was PCR amplified using CCCGTCG GATCCCGTGCCAAATCTGACAG TGA and AACCCTCACTAAAGGGAAGC oligonucleotides as primers and pCI-NHA-ΔN1370 as template. The fragment was digested with BamHI and NotI and inserted into pEBG-Piwi (Tahbaz et al., 2004; Zipprich et al., 2009) precut with BamHI and NotI.
To generate the plasmids used for bacterial expression of GST-FLAG-TNRC6C fragments (pGST-TNRC6C1-500, pGST-TNRC6C400-900, pGST-TNRC6C800-1360, and pGST-TNRC6C1260-1690), the appropriate DNA was amplified by PCR using pCI-NHA-TNRC6C as template and the following primer pairs: GGCCGGCCGTCGACTCATGGC TACAGGGAGTGCCCAGGG CAAC and CTTGTCATCGTCGTCCTTGTAGTCAGCA CTGTTCATGATGGAC CCATCGTTCTTC (1–500), GGCCGGCCGTCGACTCAGTG ATGGTTCTGGC AACCACAATGAAG and CTTGTCATCGTCGTCCTTGTAGTCAG CCACGTC CCCTTCTTCATCCTCCCACTG (400–900), GGCCGGCCGTCGACTCTC ATC AGGCTGGGGAGAAATGCCTAATG and CTTGTCATCGTCGTCCTTGTAGTC AGCGGGAGGACTGGCTGGTGACTCACTGTTC (800–1360), and GGCCGGC CGTC GACTCAACACCTTTGCTCCTTACCCTCTCGCTG and CTTGTCATCG TCGTCCTT GTAGTCAGCCAGGGACTCCCCGCTGAGCAGGTCCCC (1260–1690). These PCR products were subjected to a second round of PCR amplification using the original forward primer and a new reverse primer (CCGGC CGCGGCCGCTCACTTGTCATCGT CGTCCTTGTAGTCAGC). The product of these PCR reactions was then gel purified, digested with SalI and NotI restriction enzymes, and ligated into similarly digested pGEX-6P-1 expression vector (GE Healthcare). This strategy resulted in constructs that express the appropriate fragment of TNRC6C carrying N-terminal GST and C-terminal FLAG epitopes. TNRC6C fragments were expressed in Rosetta-2(DE3) E. coli cells (EMD Biosciences) and purified by two sequential affinity chromatography steps, first over glutathione Sepharose 4B resin (GE), followed by M2-FLAG affinity resin (Sigma).
In Vitro Transcription
Plasmids that lack or contain six let-7 target sites (RL and RL-6xB, respectively) were described (Pillai et al., 2005). A 98 base pair poly(A) sequence was added to the 3′UTR of both constructs. RL-6xBMut was constructed as previously published (Mathonnet et al., 2007; Pillai et al., 2005). A 150 base pair poly(A) sequence was synthesized (IDT) and added to the 3′UTR of RL-6xB (RL-6xB-pA*). For in vitro transcription, plasmids were linearized with ApaI and filled in using the Klenow fragment. Transcription reactions were performed using MAXIscript In Vitro Transcription Kit (Ambion) in 20 μl at 37°C according to the manufacturer’s protocol in the presence of the cap analog m7(3′-O-methyl)(5′)Gppp(5′)G (anti-reverse cap analog, ARCA; New England Biolabs). ApppG-capped mRNAs were synthesized using ApppG (New England Biolabs) instead of ARCA. 6xB-3′UTR and 6xBMUT-3′UTR transcripts were generated from PCR products derived from RL-6xB-pA and RL-6xBMUT-pA templates and T7-3′UTR (GGCGCCTAATACGACTCACTATAGGGGTAAGTACATCAAGAGCTTCG) and Oligo 3R(−) (GGTGACACTATAGAATAGGGCCC) primers. PCR products were digested with ApaI and filled in using the Klenow fragment. To synthesize radiolabeled mRNAs, UTP was substituted with [α-32P]UTP (800 Ci/mmol, 10 mCi/ml; PerkinElmer) according to the manufacturer’s protocol. The mRNA was loaded on a mini Quick Spin RNA Column (Roche) to remove unincorporated nucleotides.
In Vitro Translation Assays
Krebs-2 ascites cell extract was prepared as previously described (Svitkin and Sonenberg, 2004). Translation reactions were performed in a total volume of 10 μl at 30°C. A typical reaction mixture contained 7 μl extract, 1 μl mRNA, and, where indicated, 2′-O-Me oligonucleotide complementary to let-7a or miR-122a or poly(A)30 oligonucleotide (Dharmacon) in water. The mixture was preincubated for 20 min at 16°C and then at 30°C for 120 min. When the 2′-O-Me oligonucleotide was added, the extract was first incubated at 30°C for 20 min in the absence of mRNA to allow for the annealing of the oligonucleotide with its target miRNA. The reaction was stopped by addition of 20 μl cold 1 × PBS. For time course experiments, the reaction was scaled up to 80 μl, and 10 μl was withdrawn at each time point. Luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
mRNA Stability Assay
Radiolabeled RNA (0.1 ng) was incubated in Krebs-2 ascites in a total volume of 10 μl in the absence or presence of 10 nM let-7 2′-O-Me oligonucleotide. Aliquots of the reaction mixture were withdrawn at specific intervals, and the RNA was extracted using TRIzol reagent (Invitrogen) and loaded on a 4% or 4.5% polyacrylamide/urea gel. The gel was dried and analyzed using a Typhoon PhosphorImager (GE Healthcare).
Oligonucleotide Ligation-Mediated Cloning of RNA
Radiolabeled RNA from Krebs extract was extracted with TRIzol and loaded on a 4% polyacrylamide/urea gel. Specific RNA bands were cut from the gel and eluted in 2× proteinase K buffer (100 mM Tris-HCl, pH 8.3; 25 mM EDTA, pH 8.0; 300 mM NaCl; 2% (w/v) SDS), purified and ligated to a miRNA universal linker (NEB) using T4 RNA ligase 1 in the absence of ATP. Ligation products were purified and reverse transcribed with Superscipt III (Invitrogen), and amplified using Titanium DNA polymerase (Clontech). PCR products were cloned and sequenced.
Immunodepletion Assay, GST Pulldown Assay, Western Blotting, and Antibodies
protein G-Sepharose (GE Healthcare) (20 μl) was washed and incubated in 100 μl of Krebs extract with 6 μg of either mouse monoclonal anti-HA (Covance), rabbit anti-HA (Sigma), mouse monoclonal anti-Ago2 (Wako Chemicals), or affinity-purified rabbit polyclonal anti-CAF1 with gentle agitation for 2 hr at 4°C. The resin was then centrifuged at 500 × g, and the supernatant was collected. GST pulldown assays of Krebs extract have been described (Kahvejian et al., 2005). Antibodies and their working dilutions for western blotting were as follows: rabbit polyclonal anti-Ago2, 1:1000; rabbit polyclonal anti-PABP, 1:1000 (Cell Signaling Technologies); mouse monoclonal anti-Actin, 1:5000 (Sigma); mouse monoclonal anti-FLAG, 1:5000 (Sigma); mouse monoclonal anti-CAF1, 1:1000; mouse monoclonal anti-Tob 4B1, 1:1000 (Sigma); and mouse monoclonal anti-CCR4, 1:1000. For the GST pulldown assay with HEK293 cell extracts, cells were lysed with 50 mM Tris-HCl (pH 7.5) containing 150 mM KCl, 0.5% Triton X-100, 2 mM DTT, and complete EDTA-free protease inhibitor cocktail (Roche). The cleared lysate was incubated with glutathione Sepharose 4B (GE Healthcare) followed by washing with 50 mM Tris-HCl (pH 7.5), containing 150 mM KCl, 0.1% Triton X-100, 2 mM DTT, and complete protease inhibitor cocktail (Roche). Proteins associated with glutathione Sepharose beads were eluted with 50 mM glutathione in the same buffer as used for washing the beads and analyzed by western blotting using anti-PABP1 antibody (Cell Signaling Technology), anti-eIF4GI antibody (Gradi et al., 1998), and anti-GST antibody (GE Healthcare). To examine RNA dependence of protein-protein interactions, cleared lysates were treated with micrococcal nuclease (Roche) (10 μg/ml) for 25 min at room temperature in the presence of 1 mM CaCl2 before incubation with glutathione Sepharose 4B beads.
Anti-let-7 2′-O-Me Oligonucleotide Biotin Pulldown Assay
M-280 streptavidin magnetic Dynabeads (Invitrogen) were washed three times in buffer D (25 mM HEPES-KOH [pH 7.3], 2 mM MgCl2, 50 mM KCl, 75 mM KOAc) and resuspended in buffer D with 2 mM DTT and 1 M NaCl and incubated with biotin-labeled anti-let-7 2′-O-Me, anti-miR122 2′-O-Me, or anti-miR35 2′-O-Me oligonucleotide (Integrated DNA Technologies) for 60 min at 4°C. 2′-O-Me-bound beads were washed three times in buffer D and then incubated in aliquots of Krebs extract containing protease inhibitors at 30°C for 60 min. Beads were washed three times in buffer D with 0.5% NP-40 and boiled in SDS sample buffer and analyzed by SDS-PAGE and western blotting.
Cell Lines
HEK293 cells were transfected with Myc-Ago1 and Myc-Ago2 DNA constructs using LT-1 transfection reagent according to the manufacturer’s instructions (Mirus). All constructs contain a G418 resistance cassette. Stable transfectants were selected with 500 μg/ml G418 (Roche) for at least 2 weeks prior to being used in experiments.
Other HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-BRL) supplemented with 2 mM L-glutamine and 10% heat-inactivated fetal calf serum (FCS). Transfections were performed in 10 cm cell culture dishes with ~60% confluent cells using Nanofectin (PAA Laboratories), following the manufacturer’s instructions. For mass spectrometry analysis, cells in one 10 cm cell culture dish were transfected with 6 μg of the plasmid pEBG-DN1370. For IP experiments, cells in 10 cm cell culture dishes were transfected with 6 μg of the plasmids pCI-NHA-1505-1610 and pCI-NHA-ΔN1370 and 20 μg of the plasmids pCI-NHA-1-405, pCI-NHA-1-1034, and pCI-NHA-1-1368. For the GST pulldown experiment cells, 10 cm cell culture dishes were transfected with 4 μg of the plasmid pEBG-ΔN1370.
Mass Spectrometry Analysis
Cells were lysed with 50 mM Tris-HCl (pH 7.5) containing 150 mM KCl, 5 mM MgCl2, 1 mM CaCl2, 0.5% Triton X-100, 2 mM DTT, and EDTA-free protease inhibitor cocktail (Roche). The cleared lysate was incubated with glutathione Sepharose 4B (GE Healthcare) followed by washing with 50 mM Tris-HCl (pH 7.5) containing 150 mM KCl, 5 mM MgCl2, 1 mM CaCl2, 0.5% Triton X-100, 2 mM DTT, and EDTA-free protease inhibitor cocktail (Roche). Cysteine residues of proteins associated with the beads were reduced and alkylated prior to gel separation. The Coomassie-stained bands were digested with trypsin, and tryptic peptides were analyzed by nano-HPLC (Agilent 1100 nanoLC system, Agilent Technologies, Santa Clara, CA) coupled to a 4000 Q TRAP mass spectrometer (Applied Biosystems, Foster City, CA). Peptides were identified searching UniProt database (version 13.8) restricted to human using Mascot (version 2.1, Matrix Science, London).
MuDPIT and Coimmunoprecipitation Analysis
Samples were prepared as follows: HEK293 cells were harvested and washed with phosphate-buffered saline (PBS). Cells were washed once in hypotonic lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 5 mM DTT, and EDTA-free protease inhibitor cocktail [Roche]) and allowed to swell for 20 min on ice prior to homogenization. Cell extracts were centrifuged in a tabletop centrifuge at 10,000 rpm for 30 min at 4°C to clarify the lysate. The salt concentration in the extract was raised to 100 mM KCl. To immuno-precipitate Ago and Ago-interacting proteins, Myc-agarose beads (Sigma) were added to the extract and allowed to incubate for 6 hr with gentle rotation. Immunoprecipitates were washed (wash buffer, 10 mM Tris-HCl [pH 7.5], 100 mM KCl, 1.5 mM MgCl2, 5 mM DTT, and EDTA-free protease inhibitor cocktail [Roche]) four times for 30 min each. Immunocomplexes were eluted from Myc-agarose beads by two serial washes in elution buffer (100 mM Tris-HCl [pH 8.0], 8M urea). Proteins in eluates were precipitated with trichloroacetic acid and submitted for MuDPIT analysis (Washburn et al., 2001). Samples analyzed for coimmunoprecipitation of Ago and Ago-interacting proteins from HEK293 cells were prepared as above. In cases in which immunoprecipitates were subjected to RNase A treatment, immunoprecipitation was performed as described, but the next to last washing step was done in the presence of RNase A (10 units/ml in wash buffer). Samples were washed an additional two times prior to SDS-PAGE and western blot analysis. Samples analyzed for coimmunoprecipitation of Ago2- and CAF1-interacting proteins from Krebs extracts were prepared as follows: Krebs extracts were treated with micrococcal nuclease (Roche) in the presence of CaCl2 for 30 min at 20°C and subsequently with EGTA as previously described (Svitkin and Sonenberg, 2004). Krebs extracts were then mixed with protein G Dynabeads (Invitrogen) already bound to either mouse monoclonal anti-HA (Covance), rabbit anti-HA (Sigma), mouse monoclonal anti-Ago2 (Wako Chemicals), or affinity-purified rabbit anti-CAF1 and gently mixed at 30°C for 60 min. Immunoprecipitates were washed five times with buffer D containing 0.5% NP-40 prior to SDS-PAGE and western blot analysis. For HA epitope IP reactions, cells were lysed with 50 mM Tris-HCl (pH 7.5) containing 150 mM KCl, 0.5% Triton X-100, 2 mM DTT, and protease inhibitor cocktail (Roche). The cleared lysate was incubated with Anti-HA Affinity Matrix (Roche). After washing with 10 mM Tris-HCl (pH 7.5) containing 200 mM KCl, proteins associated with the beads were analyzed by western blotting using anti-HA 3F10 antibody (Roche) and PABP1 antibody (Cell Signaling Technology) (Polacek et al., 2009).
Supplementary Material
Acknowledgments
We thank Bertrand Seraphin and Mauxion Fabienne for providing recombinant mCAF1 protein; Tadashi Yamamoto for providing mouse monoclonal CAF1 and CCR4 antibodies; Kalle Gehring for providing PAM2 peptides; Maria Ferraiuolo, Ler Lian Wee, and Mark Livingstone for helpful comments; and Ragna Sack and Daniel Hess from the FMI Protein Analysis Facility for mass spectrometry analysis. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and a Chercheur-Boursier Junior1 Award to T.F.D.; a CIHR grant to N.S., who is a Howard Hughes Medical Institute (HHMI) International Scholar; and National Institutes of Health (NIH) grants (R01 GM46454 and P41 RR011823) to A-B.S. and J.R.Y, respectively. Postdoctoral support was by the Natural Science and Engineering Research Council of Canada, the Richard H. Tomlinson Foundation, and the Terry Fox Foundation of the Canadian Cancer Society to M.R.F.; and by Fonds de Recherche en Sante du Quebec to G.M. The Friedrich Meischer Institute is supported by the Novartis Research Foundation. The work there was also supported by EC FP6 Program “Sirocco.”
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include ten figures and can be found with this article online at http://www.cell.com/molecular-cell/supplemental/S1097-2765(09)00550-4.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA . 2009;15:794–803. doi: 10.1261/rna.1364909. Published online March 20, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009a;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Buttner R, Weichenrieder O, Izaurralde E, Truffault V. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucleic Acids Res. 2009b;37:2974–2983. doi: 10.1093/nar/gkp173. Published online March 18, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CYA, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft CM, Burley SK. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T. Micro-RNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EK. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci USA. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O’Connor-McCourt M, Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, De Crescenzo G, Lim NS, Siddiqui N, Fantus D, Kahvejian A, Trempe JF, Elias D, Ekiel I, Sonenberg N, et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 2004;23:272–281. doi: 10.1038/sj.emboj.7600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell JE, Rudner DZ, Sachs AB. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that micro-RNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Polacek C, Friebe P, Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol. 2009;90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E, Seraphin B. A specific role for the C-terminal region of the poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 2007;35:6017–6028. doi: 10.1093/nar/gkm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Sonenberg N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudopolysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/3′–5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Imataka H, Sonenberg N. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol. 2000;10:1147–1150. doi: 10.1016/s0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc Natl Acad Sci USA. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- Wormington M. Poly(A) and translation: development control. Curr Opin Cell Biol. 1993;5:950–954. doi: 10.1016/0955-0674(93)90075-2. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. Published online March 20, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.