Abstract
Different truncated and conformationally constrained analogs of corticotropin-releasing factor (CRF) were synthesized on the basis of the amino acid sequences of human/rat CRF (h/rCRF), ovine CRF (oCRF), rat urocortin (rUcn), or sauvagine (Svg) and tested for their ability to displace [125I-Tyr0]oCRF or [125I-Tyr0]Svg from membrane homogenates of human embryonic kidney (HEK) 293 cells stably transfected with cDNA coding for rat CRF receptor, type 1 (rCRFR1), or mouse CRF receptor, type 2β (mCRFR2β). Furthermore, the potency of CRF antagonists to inhibit oCRF- or Svg-stimulated cAMP production of transfected HEK 293 cells expressing either rCRFR1 (HEK-rCRFR1 cells) or mCRFR2β (HEK-mCRFR2β cells) was determined. In comparison with astressin, which exhibited a similar affinity to rCRFR1 (Kd = 5.7 ± 1.6 nM) and mCRFR2β (Kd = 4.0 ± 2.3 nM), [dPhe11,His12]Svg(11–40), [dLeu11]Svg(11–40), [dPhe11]Svg(11–40), and Svg(11–40) bound, respectively, with a 110-, 80-, 68-, and 54-fold higher affinity to mCRFR2β than to rCRFR1. The truncated analogs of rUcn displayed modest preference (2- to 7-fold) for binding to mCRFR2β. In agreement with the results of these binding experiments, [dPhe11,His12]Svg(11–40), named antisauvagine-30, was the most potent and selective ligand to suppress agonist-induced adenylate cyclase activity in HEK cells expressing mCRFR2β.
Corticotropin-releasing factor (CRF), believed to synchronize the endocrine, autonomic, immunologic, and behavioral responses to stress, was characterized as a 41-residue polypeptide (1) on the basis of its ability to stimulate the secretion of adrenocorticotropic hormone (ACTH) from the anterior pituitary (2).
CRF exhibits its activity through G protein-coupled receptors. CRF receptor, type 1 (CRFR1), mainly found in pituitary and brain, was cloned from human, mouse, rat, chicken, and frog (3–8). cDNAs coding for two splice variants of CRF receptor, type 2, CRFR2α and CRFR2β, were cloned from brain, heart, and skeletal muscle (9–12). In rodents, CRFR2α has been found exclusively in the central nervous system (CNS), whereas CRFR2β is predominantly distributed in the periphery. In humans, both receptor subtypes have been found in the CNS (13). Recently, it has been proposed that urocortin (Ucn), a natural CRF analog, is the endogenous ligand to CRFR2 (14).
CRF is assumed to play a major role in a number of neuropsychiatric diseases including affective disorders, anxiety disorders, anorexia nervosa, and Alzheimer’s disease (15). There is substantial interest in the design and synthesis of CRF antagonists acting selectively at one of the different CRFR forms. After the discovery of potent peptide antagonists based on the N-terminally truncated amino acid sequence of human/rat CRF (h/rCRF) (16–20), several CRFR1-selective nonpeptidic antagonists have been developed (21–23) that attenuate CRF-mediated seizure (24) or interleukin-1β-induced fever or exhibit anxiolytic activity in vivo (25). However, that CRF antagonist α-helical CRF(9–41) exhibits different inhibitory potencies in three different in vivo bioassay systems (26) suggests that distinct physiological functions of endogenous CRF or Ucn are mediated via CRFR1, CRFR2, or both receptor types.
Our objective was to develop CRFR2-specific antagonists to permit discrimination between receptor type-specific functions. To this end, we used truncated and conformationally constrained analogs of CRF based on the amino acid sequences of h/rCRF, ovine CRF (oCRF), rat urocortin (rUcn), and sauvagine (Svg). This strategy was based on the observation that CRFR1 and CRFR2 discriminate between these peptides as indicated by different binding affinities and biologic potencies (27). Therefore, it was expected that CRF antagonists developed on this structural basis may exhibit receptor subtype selectivity. Comparison of the amino acid sequences of oCRF, rUcn, and Svg with the sequence of h/rCRF reveals 45–83% amino acid identity. The CRF ligands mentioned share high amino acid identity at the N terminus (47%) stretching from amino acids 2–20 (h/rCRF and oCRF) and 1–19 (rUcn and Svg), but little at the C terminus (14%) of the peptides stretching from amino acids 21–41 and 20–40, respectively (Fig. 1).
Figure 1.
Comparison of the amino acid sequences of [dPhe11,His12]Svg(11–40) (a Svg-30), astressin, α-helical CRF(9–41), Svg, rUcn, oCRF, and h/rCRF. B, norleucine; f, d-phenylalanine; Z, pyroglutamic acid; lactam bridge is indicated by a bracket. Identical amino acids are shaded.
We assumed that the ligand–receptor interactions of the truncated forms of the CRF peptides ranging from amino acid 11–40 (rUcn and Svg) or 12–41 (h/rCRF and oCRF) acted differently than the full-length CRF peptides on CRFR1 or CRFR2 (8, 14, 28, 29). The CRF analogs were tested in binding studies with [125I-Tyr0]oCRF or [125I-Tyr0]Svg as radioligands and membrane homogenates of human embryonic kidney (HEK) 293 cells stably transfected with cDNA coding for rat CRFR1 (rCRFR1) or mouse CRFR2β (mCRFR2β). The agonistic activity of the peptides to increase second messenger production and their antagonistic activity to suppress oCRF- or Svg-stimulated cAMP accumulation was investigated in whole cells expressing rCRFR1 (HEK-rCRFR1 cells) or mCRFR2β (HEK-CRFR2β cells).
MATERIALS AND METHODS
Synthesis and Analysis of Peptides.
The CRF-like peptides (0.1 mmol scale) were synthesized with fluorenylmethoxycarbonyl (Fmoc) chemistry on TentaGel S RAM resin (Rapp, Tübingen, Germany) with a model ABI 433A peptide synthesizer (Applied Biosystems). For the synthesis of the cyclized CRF analogs, astressin and cyclo(29–32)[dPhe11,Glu29,Lys32]rUcn(11–40), the amino acid derivatives Fmoc-l-Glu(OAll)-OH and Fmoc-l-Lys(Alloc)-OH (PerSeptive Biosystems, Hamburg, Germany) were used. The side-chain-protected peptides were reacted with Pd0[PPh3]4 in HOAc/N-methylaniline/dichloromethane (vol/vol; 2:1:40) for 3 hr and then cyclized with 1-hydroxybenzotriazole/O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate in dimethylformamide and N,N-diisopropylethylamine (DIEA) in N-methylpyrrolidine (NMP) for 8 hr. After removal of the N-terminal Fmoc group with piperidine in NMP, the peptides were cleaved from the resin under standard conditions. The crude peptides were purified by preparative reverse-phase high-performance liquid chromatography (RP-HPLC) performed on a Waters Prep Nova-Pak HR C18 silica gel column (5 × 30 cm, 6-μm particle size, 6-nm pore size) with a mixture of aqueous 0.1% trifluoroacetic acid and acetonitrile. The mass spectra of the purified peptides were confirmed with a plasma desorption mass spectrometer (BioIon 20, Uppsala). Amino acid analysis was performed after hydrolysis of peptides (6 M HCl, 3 hr, 150°C) with a Beckman HPLC Analyzer System 6300 (Beckman).
Binding to rCRFR1.
CRF agonists and antagonists were tested in an in vitro assay for their ability to displace [125I-Tyr0]oCRF or [125I-Tyr0]Svg from membranes of HEK-rCRFR1 cells (30) or HEK-mCRFR2β cells (11). Binding assays were performed in 96-well MultiScreen plates (Millipore) with GF/B filters (pore size, 1.0 μm). Fifty microliters of membrane suspension (25 μg of protein from HEK-rCRFR1 cells; 50 μg of protein from HEK-mCRFR2β cells) was added to a plate containing CRF peptides (0–1 μM) and 50,000 cpm of either [125I-Tyr0]oCRF (specific activity, 81.4 TBq/mmol, 68.25 pM, DuPont/NEN) for the analysis of rCRFR1 or [125I-Tyr0]Svg (specific activity, 81.4 TBq/mmol, 68.25 pM, DuPont/NEN) for the analysis of mCRFR2β in 100 μl of incubation buffer [50 mM Tris⋅Cl/5 mM MgCl2/2 mM EGTA/100,000 kallikrein inhibitor units/liter Trasylol (Bayer, Leverkusen, Germany)/1 mM DTT/1 mg/ml BSA, pH 7.4]. After incubation (60 min, 23°C), the membrane suspension was aspirated through the plate, followed by two washes with assay buffer (0.2 ml, 23°C). Radioactivity of the punched filters was measured with a 1470 Wizard automatic γ counter (Wallac, Turku, Finland). Specific binding of [125I-Tyr0]oCRF or [125I-Tyr0]Svg to membranes of transfected cells was calculated by subtraction of nonspecific binding found in the presence of 1 μM nonlabeled ligand from total binding. Data were analyzed with the nonlinear curve-fitting program ligand. Statistical analysis was performed with ANOVA, and significant differences between groups were determined by post hoc comparison using the Dunn test.
Chemical Cross-Linking Experiments with [125I-Tyr0]oCRF or [125I-Tyr0]Svg.
Chemical cross-linking was carried out in 1.5-ml polypropylene tubes (Sigma) as for the binding assay except that no BSA was used. Samples (50 μg and 100 μg of protein from membrane fractions of HEK-rCRFR1 cells and HEK-mCRFR2β cells, respectively) were reacted with 10 μl of disuccinimidyl suberate (1.5 mM in dimethyl sulfoxide, 23°C, 20 min) after incubation with ligand (300 μl, 100,000 cpm, 1 hr, 23°C). The reaction was terminated by the addition of 1.0 ml of ice-cold buffer (10 mM Tris-Cl/1 mM EDTA, pH 7.0, 4°C). In some experiments, the chemically cross-linked receptor was deglycosylated with peptide N-glycosidase F (PNGase F; New England Biolabs). Samples then were heated (100°C, 5 min) and subjected to SDS/PAGE. Autoradiography was carried out on a BAS-IP NP 2040P imaging plate. Radioactivity was monitored with a Fujix BAS 2000 scanner (Raytest, Straubenhardt, Germany). Gel documentation was accomplished with the program tina (Raytest).
cAMP Stimulation.
HEK-rCRFR1 cells or HEK-mCRFR2β cells were incubated for 30 min at 37°C with different CRF agonists in the presence or absence of 1 μM or 10 nM antagonist, or with 1 μM CRF antagonist alone. After removal of the medium, cells were lysed with aqueous 6% trichloroacetic acid (5 min, 100°C) (29). The cell lysates were stored at −70°C until assayed with an RIA kit (Amersham). Data were analyzed with the sigmoidal dose–response curve-fitting program allfit. Statistical significance was determined across groups with ANOVA, and significant differences between groups were determined by post hoc comparison using the Dunn test.
RESULTS
Displacement of [125I-Tyr0]oCRF or [125I-Tyr0]Svg from Recombinant rCRFR1 or mCRFR2β by CRF Analogs.
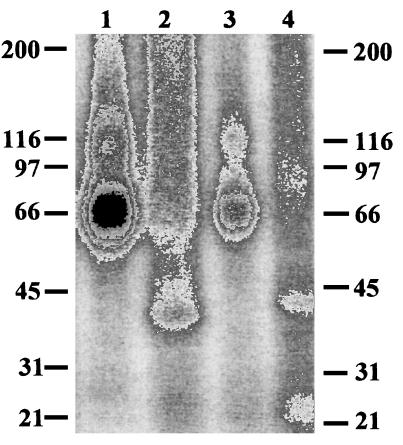
The specific binding of [125I-Tyr0]oCRF to membranes of HEK-rCRFR1 cells was found to be 93% when the radioligand was displaced by oCRF. In analogous displacement experiments with Svg, the specific binding of [125I-Tyr0]Svg to membranes of HEK-mCRFR2β cells was determined to be 94%. No specific binding of the two radioactively labeled CRF analogs to membranes of nontransfected HEK 293 cells could be observed. These data could be confirmed when [125I-Tyr0]oCRF and [125I-Tyr0]Svg were chemically cross-linked to transfected rCRFR1 and mCRFR2β, respectively. The molecular weight of both cross-linked receptors was 66,000. After deglycosylation with peptide N-glycosidase, molecular weights of 43,000 and 41,000 were found for cross-linked mCRF2β and rCRFR1, respectively (Fig. 2). No chemical cross-links could be observed with either radioligand to nontransfected HEK 293 cells (not shown).
Figure 2.
Chemical cross-linking of [125I-Tyr0]oCRF or [125I-Tyr0]Svg to membrane homogenates of HEK 293 cells stably transfected with cDNA coding for rat CRF receptor, type 1 (rCRFR1) (lanes 1 and 2) or mouse CRF receptor, type 2β (mCRFR2β) (lanes 3 and 4), respectively. Fifty micrograms of total membrane protein was labeled with approximately 100,000 cpm of [125I-Tyr0]oCRF (lanes 1 and 2) or [125I-Tyr0]Svg (lanes 3 and 4), and incubated (37°C, 30 min) in the presence (lane 2 and 4) or absence (lane 1 and 2) of 2,000 units of peptide N-glycosidase.
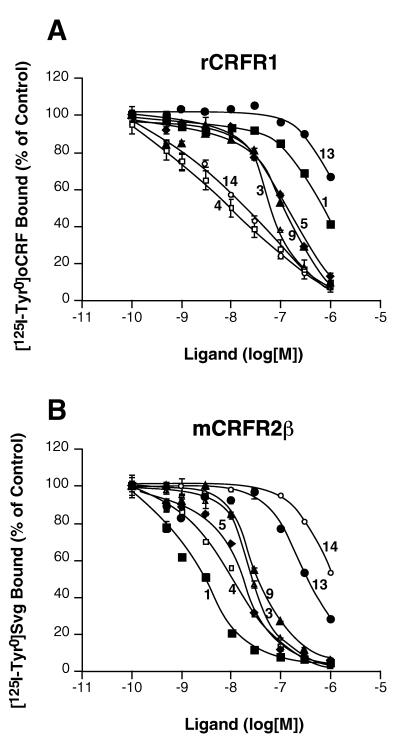
As expected for the CRF peptide agonists (8, 28, 29), oCRF and Svg exhibited similar high-affinity binding to rCRFR1, but differed significantly in their binding to mCRFR2β (Table 1 and Fig. 3).
Table 1.
Binding constants of different CRF agonists and antagonists displacing [125I-Tyr0]oCRF from recombinant rCRFR1 or [125I-Tyr0]Svg from recombinant mCRFR2β
| Compound | Peptide | [125I-Tyr0]Svg Kd(mCRFR2β), nM | [125I-Tyr0]oCRF Kd(rCRFR1), nM | Kd(rCRFR1)/ Kd(mCRFR2β) |
|---|---|---|---|---|
| 1 | [dPhe11,His12]Svg(11-40) | 1.4 ± 0.4 | 153.6 ± 33.5 | 109.71 |
| 2 | [dPhe11]Svg(11-40) | 3.5 ± 0.2 | 237.3 ± 27.7 | 67.80 |
| 3 | Astressin | 4.0 ± 2.3 | 5.7 ± 1.6 | 1.42 |
| 4 | Svg | 4.5 ± 0.6 | 0.7 ± 0.1 | 0.15 |
| 5 | [dPhe11]rUcn(11-40) | 5.2 ± 1.5 | 33.0 ± 5.9 | 6.34 |
| 6 | α-Helical CRF(9-41) | 6.4 ± 0.9 | 60.3 ± 10.6 | 9.42 |
| 7 | [dPhe11,Glu12]rUcn(11-40) | 9.5 ± 2.0 | 68.2 ± 20.5 | 7.18 |
| 8 | Svg(11-40) | 15.4 ± 2.5 | 831.2 ± 668.8 | 53.97 |
| 9 | [dPhe12,Nle21,38]h/rCRF(12-41) | 17.7 ± 2.2 | 46.4 ± 9.4 | 2.62 |
| 10 | [dLeu11]Svg(11-40) | 20.9 ± 4.1 | 1,670.0 ± 500.0 | 79.90 |
| 11 | Cyclo(29-32) [dPhe11,Glu29,Lys32]rUcn(11-40) | 22.4 ± 4.6 | 47.1 ± 8.9 | 2.10 |
| 12 | [dLeu11,Glu12]rUcn(11-40) | 27.9 ± 3.4 | 91.1 ± 21.2 | 3.26 |
| 13 | [dPhe12]oCRF(12-41) | 153.8 ± 26.8 | 290.2 ± 74.7 | 1.88 |
| 14 | oCRF | 162.4 ± 37.6 | 0.6 ± 0.1 | 0.00 |
Figure 3.
Displacement of [125I-Tyr0]oCRF (A) or [125I-Tyr0]Svg (B) bound to membrane homogenates of HEK 293 cells stably transfected with cDNA coding for rat CRF receptor, type 1 (rCRFR1) (A), or mouse CRF receptor, type 2β (mCRFR2 β) (B), by oCRF (compound 14, ○), Svg (compound 4, □), astressin (compound 3, ▵), [dPhe12,Nle21,38]h/rCRF(12–41) (compound 9, ▴), [dPhe12]oCRF(12–41) (compound 13, •), [dPhe11]rUcn(11–40) (compound 5, ♦), and [dPhe11,His12]Svg(11–40) (compound 1, ■). Error bars represent SEM and are not shown when smaller than the symbol size.
The antagonist astressin (19), cyclo(30–33)[dPhe12,Nle21,38,Glu30,Lys33]h/rCRF(12–41), was found to bind nonselectively with similar affinity to rCRFR1 and mCRFR2β, whereas α-helical CRF(9–41) and [dPhe12,Nle21,38]h/rCRF(12–41), described earlier (19), showed moderate selectivity for mCRFR2β (Table 1). [dPhe12]oCRF(12–41), based on the amino acid sequence of oCRF, showed low-affinity binding to rCRFR1 and mCRFR2β (Table 1, Fig. 3).
The truncated Ucn analogs [dPhe11]rUcn(11–40), [dPhe11,Glu12]rUcn(11–40), [dLeu11,Glu12]rUcn(11–40), and cyclo(29–32)[dPhe11,Glu29,Lys32]rUcn(11–40) exhibited moderate binding affinity to rCRFR1 and exhibited low binding preference for mCRFR2β (Table 1, Fig. 3).
The Svg-derived peptides [dLeu11]Svg(11–40), Svg(11–40), [dPhe11]Svg(11–40), and [dPhe11,His12]Svg(11–40) showed low-affinity binding to rCRFR1, but high-affinity binding to mCRFR2β (Table 1).
cAMP Stimulation.
The peptide agonists oCRF and Svg exhibited high potency to increase cAMP accumulation in HEK-rCRFR1 cells with EC50 values of 0.41 ± 0.08 nM and 0.19 ± 0.05 nM, respectively, but differed significantly in their potencies to stimulate cAMP production in HEK-mCRFR2β cells with EC50 values of 11.79 ± 1.96 nM and 0.23 ± 0.05 nM, respectively (not shown).
The CRF antagonists (compounds 1–3 and 5–13, Table 2) mentioned above were tested for their ability to enhance cAMP production in HEK-mCRFR2β and HEK-rCRFR1 cells. This test served as a measure for the intrinsic activity. The agonist activity of the antagonists acting on mCRFR2β-HEK cells ranged from 0.4 to 1.1% of the corresponding values obtained with Svg. In contrast, the agonist activity of the antagonists on rCRFR1-HEK cells was 4–23% of the values found for oCRF (Table 2).
Table 2.
Relative potency of CRFR antagonists
| Compound | Peptides | HEK-mCRFR2β cells
|
HEK-rCRFR1 cells
|
||
|---|---|---|---|---|---|
| cAMP (1 μM antag.)/ cAMP (1 nM Svg)* | cAMP (Svg + antag.)/ cAMP (Svg)† | cAMP (1 μM antag.)/ cAMP (1 nM oCRF)* | cAMP (oCRF + antag.)/ cAMP (oCRF)† | ||
| 1 | [dPhe11,His12]Svg(11-40) | 0.004 ± 0.001 | 0.42 ± 0.02a | 0.04 ± 0.01 | 0.73 ± 0.07b |
| 2 | [dPhe11]Svg(11-40) | 0.005 ± 0.001 | 0.71 ± 0.04 | 0.07 ± 0.01 | 0.75 ± 0.13 |
| 3 | Astressin | 0.004 ± 0.001 | 0.57 ± 0.04 | 0.10 ± 0.02 | 0.11 ± 0.03 |
| 5 | [dPhe11]rUcn(11-40) | 0.011 ± 0.003 | 0.57 ± 0.06 | 0.07 ± 0.01 | 0.18 ± 0.03 |
| 6 | α-Helical CRF(9-41) | 0.007 ± 0.004 | 0.67 ± 0.02 | 0.16 ± 0.02c | 0.58 ± 0.05 |
| 7 | [dPhe11,Glu12]rUcn(11-40) | 0.004 ± 0.001 | 0.61 ± 0.02 | 0.22 ± 0.05c | 0.42 ± 0.11 |
| 8 | Svg(11-40) | 0.005 ± 0.001 | 0.76 ± 0.02 | 0.14 ± 0.02c | 0.76 ± 0.16 |
| 9 | [dPhe12,Nle21,38]h/rCRF(12-41) | 0.007 ± 0.002 | 0.87 ± 0.08 | 0.06 ± 0.01 | 0.33 ± 0.08 |
| 10 | [dLeu11]Svg(11-40) | 0.008 ± 0.002 | 0.75 ± 0.06 | 0.22 ± 0.04c | 0.92 ± 0.16 |
| 11 | Cyclo(29–32)[dPhe11,Glu29,Lys32]rUcn(11-40) | 0.005 ± 0.001 | 0.89 ± 0.01 | 0.10 ± 0.06 | 0.78 ± 0.10 |
| 12 | [dLeu11,Glu12]rUcn(11-40) | 0.009 ± 0.002 | 0.81 ± 0.06 | 0.23 ± 0.04c | 0.56 ± 0.05 |
| 13 | [dPhe12]oCRF(12-41) | 0.008 ± 0.003 | 0.92 ± 0.07 | 0.05 ± 0.01 | 0.89 ± 0.07 |
| 14 | oCRF | — | — | 1.00 | 1.00 |
| 4 | Svg | 1.00 | 1.00 | — | — |
| Control without peptide | 0.004 ± 0.001 | 0.01 ± 0.003 | |||
Statistically significant differences between the relative potencies of the peptides: a, P < 0.001 vs. 6, 9, 10, 11, 12, and 13; b, P ≤ 0.001 vs. 3 and 5. Statistically significant differences between the relative agonist activities of the peptides: c, P > 0.001 vs. control without peptide.
The ratio of cAMP production of transfected HEK cells stimulated by antagonist or Svg or oCRF served as measure of the intrinsic activity.
The relative potency determined by the effect of 10 nM (mCRFR2β) or 1 μM (rCRFR1) CRFR antagonist on the cAMP production stimulated by 1 nM Svg (mCRFR2β) or 1 nM oCRF (rCRFR1).
ANOVA indicated statistically significant differences in relative agonist activity, F(12,50) = 11.68, P < 0.0001, of compounds 1–3 and 5–13 to stimulate cAMP production in HEK-rCRFR1 cells. Post hoc comparison demonstrated a significantly higher activity of compounds 6, 7, 8, 10, and 12 when compared with the basal level of cAMP in the same cells (P < 0.001) (Table 2). ANOVA indicated no statistically significant differences in relative agonist activity of compounds 1–3 and 5–13 to stimulate cAMP production in HEK-mCRFR2β cells. Compound 1 exhibited the lowest relative agonist activity of all tested CRF antagonists in experiments with either recombinant system, HEK-rCRFR1 cells, and HEK-mCRFR2β cells (Table 2).
The antagonist potencies were tested by the inhibition of Svg- or oCRF-stimulated cAMP production of transfected HEK cells (Table 2). The rank order of potencies for the CRF-related peptide antagonists to suppress Svg-stimulated cAMP production in HEK-mCRFR2β cells by CRF antagonists was as follows: [dPhe11,His12]Svg(11–40) (compound 1) > astressin (compound 3), [dPhe11]rUcn(11–40) (compound 5), [dPhe11,Glu12]rUcn(11–40) (compound 7), and α-helical CRF(9–41) (compound 6) > [dPhe11]Svg(11–40) (compound 2), [dLeu11]Svg(11–40) (compound 10), and Svg(11–40) (compound 8) > [dLeu11,Glu12]rUcn(11–40) (compound 12) > [dPhe12,Nle21,38]h/rCRF(12–41) (compound 9), cyclo(29–32)[dPhe11,Glu29,Lys32]rUcn(11–40) (compound 11), and [dPhe12]oCRF(12–41) (compound 13). In contrast, the following pharmacological profile was obtained for the inhibition of oCRF-induced cAMP accumulation in HEK-rCRFR1 cells: astressin (compound 3) and [dPhe11]rUcn(11–40) (compound 5) > [dPhe12,Nle21,38]h/rCRF(12–41) (compound 9) and [dPhe11,Glu12]rUcn(11–40) (compound 7) > [dLeu11,Glu12]rUcn(11–40) (compound 12) and α-helical CRF(9–41) (compound 6) > [dPhe11,His12]Svg(11–40) (compound 1), [dPhe11]Svg(11–40) (compound 2), Svg(11–40) (compound 8), and cyclo(29–32)[dPhe11,Glu29,Lys32]rUcn(11–40) (compound 11) > [dPhe12]oCRF(12–41) (compound 13) and [dLeu11]Svg(11–40) (compound 10).
ANOVA indicated statistically significant differences in potency, F(12,65) = 6.34, P < 0.001, of compounds 1–3 and 5–13 to inhibit Svg-stimulated cAMP production in HEK-mCRFR2β cells. Post hoc comparison demonstrated a significantly higher potency of compound 1 when compared with compounds 6, 9, 10, 11, 12, and 13 (P < 0.001).
ANOVA also indicated statistically significant differences in potency, F(12,39) = 7.93, P < 0.001, of compounds 1–3 and 5–13 to inhibit oCRF-stimulated cAMP production in HEK-rCRFR1 cells. Post hoc comparison demonstrated a significantly lower potency of compound 1 when compared with compounds 3 and 5 (P < 0.001).
DISCUSSION
A highly specific antagonist, [dPhe11,His12]Svg(11–40), directed against mCRFR2β has been developed. It is proposed to call this antagonist antisauvagine-30 (a Svg-30). The test system used for the characterization of this selective peptide antagonist was represented by HEK 293 cells transfected with cDNA coding for rCRFR1 or mCRFR2β.
Two radioactively labeled ligands, [125I-Tyr0]oCRF and [125I-Tyr0]Svg, which bound with high affinity to rCRFR1 (30) or mCRFR2β (31), were displaced by truncated CRF analogs to determine their binding affinity. In chemical cross-linking experiments with the two radioligands, the observed molecular size of the CRFRs after enzymatic deglycosylation was in agreement with the molecular weight predicted on the basis of DNA data (5, 11). The difference of 2,000 between the molecular weights of rCRFR1 and mCRFR2β was probably a result of the additional 16 amino acids found in mCRFR2β in comparison to rCRFR1.
In comparison with astressin, the most potent CRF antagonist described to date (19), the intrinsic activity of antisauvagine-30 was not significantly different in experiments with HEK-rCRFR1 or HEK-mCRFR2β cells. However, the inhibitory potency of antisauvagine-30 toward rCRFR1 was found to be 30% of the potency of astressin. This difference was determined to be significant. In contrast, no significant difference between the inhibitory potencies of astressin and antisauvagine-30 was observed when HEK-mCRF2β cells were tested (Table 2). The difference between astressin and antisauvagine-30 was even more pronounced in binding experiments (Table 1) which demonstrated that antisauvagine-30 exhibited, in contrast to astressin, selective tight binding to mCRFR2β. On the basis of ligand comparisons, antisauvagine-30 thus was demonstrated to be a selective, mCRFR2β-directed CRF antagonist with low intrinsic activities directed toward rCRFR1 and mCRFR2β.
In contrast to CRFR2, mammalian CRFR1 has been reported to be nonselective for CRF and CRF-like peptides including the structurally related 40-aa peptides Svg and Ucn (3, 8, 14, 27, 28). Experimental data available thus far do not show significant pharmacological differences between mammalian CRFR2α and CRFR2β (28). On this basis, it is expected that antisauvagine-30 inhibits CRFR2α similarly as it inhibits CRFR2β.
It is interesting that the most potent and selective antagonist, antisauvagine-30, synthesized here was developed on the basis of the sauvagine and not the urocortin sequence. If urocortin would be the specific ligand of mammalian CRFR2, as suggested (14), it would be expected that a peptide developed on the basis of the urocortin sequence would be more potent than antisauvagine-30. On the basis of the opposite result, it is speculated that a sauvagine-like peptide not yet found in mammals serves as CRFR2 ligand.
Antisauvagine-30, [dPhe11,His12]Svg(11–40), was developed from [dLeu11]Svg(11–40), which bound with high specificity to mCRFR2β. The affinity of this peptide to both receptors was enhanced by consecutive replacement of amino acids dLeu11 by dPhe11 and Glu12 by His12 to produce antisauvagine-30 without significant change of the selectivity of the peptides to either receptor. This dipeptide fragment appears to be a general but important motif responsible for increased binding affinity of CRF peptide antagonists to CRFR1 and CRFR2, as could also be observed in our experiments with truncated Ucn analogs.
It is interesting to note that all nonpeptidic CRFR1 antagonists known to date contain a substituted phenyl ring attached to a five-membered nitrogen heterocycle (23). It is likely that this common feature in nonpeptide CRF antagonists mimics an important and favorable arrangement of the adjacent amino acids phenylalanine and histidine of CRF peptide antagonists in receptor–ligand complexes.
The shortened peptide analogs based on the amino acid sequences of oCRF and h/rCRF did not exhibit the expected selectivity for CRFR1 binding that had been observed for the full-length peptide analogs. It is therefore conceivable that the N termini of the full-length peptides stretching from amino acid 1 to amino acid 11 not only increase high-affinity binding of oCRF and h/rCRF to CRFR1, but also contribute to ligand selectivity. Although [dPhe12]oCRF(12–41) and [dPhe12,Nle21,38]h/rCRF(12–41) share 80% of their amino acid sequence and differ only in 6 amino acids, the oCRF analog in contrast to the h/rCRF analog exhibited relatively low-affinity binding to either receptor. As [dPhe12,Nle21,38]h/rCRF(12–41) or [dPhe11]rUcn(11–40) but not [dPhe12]oCRF(12–41) or [dPhe11,His12]Svg(11–40) showed similar high-affinity binding to CRFR1, it is assumed that the common amino acids Ala22, Arg23 or Ala21, Arg22 in the h/rCRF and Ucn analogs, respectively, are responsible for the increased binding affinity of these peptides to CRFR1.
Intramolecular cyclization of [dPhe12,Nle21,38]h/rCRF(12–41) to astressin or [dPhe11]rUcn(11–40) to cyclo(29–32)- [dPhe11,Glu29,Lys32]rUcn(11–40) enhanced the binding affinity of the h/rCRF analog, but decreased the binding affinity of the Ucn analog to CRFR1 and CRFR2β without changing the selectivity of the peptides for binding to either receptor. Similar results indicating different ligand requirements of CRFR1 for CRF and Ucn also have been observed in a site-directed-mutagenesis approach (32) and single acid replacement studies using chimeric peptides of CRF and Ucn (A.R. and J.S., unpublished results).
In summary, we have designed, synthesized, and characterized a high-affinity mCRFR2β-specific peptide antagonist. Because of the similarity of the pharmacological profile of mammalian CRFR2α and CRFR2β, the new ligand with its amino acid sequence based on Svg should serve as a useful tool to detect CRFR2 and elucidate its functional role in the brain and peripheral organs.
Acknowledgments
Thomas Liepold is acknowledged for the performance of the amino acid analysis. Dr. Bodo Zimmermann is acknowledged for the performance of the mass spectrometric experiments. We thank Drs. Andreas K. E. Köpke and Frank M. Dautzenberg for providing us with the HEK-rCRFR1 cells and Drs. Klaus Eckart and Jelena Radulovic for discussions.
ABBREVIATIONS
- CRF
corticotropin-releasing factor (h, human
- o
ovine
- r
rat)
- CRFR
CRF receptor
- Svg
sauvagine
- Ucn
urocortin
- HEK
human embryonic kidney
Note
After the completion of this manuscript, we determined that antisauvagine-30 did not exhibit detectable specific binding to the rat CRF-binding protein (Olaf Jahn, Klaus Eckart, and J.S., unpublished results).
References
- 1.Spiess J, Rivier J, Rivier C, Vale W. Proc Natl Acad Sci USA. 1981;78:6517–6521. doi: 10.1073/pnas.78.10.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 3.Vita N, Laurent P, Lefort S, Chalon P, Lelias J-M, Kaghad M, Le Fur G, Caput D, Ferrara P. FEBS Lett. 1993;335:1–5. doi: 10.1016/0014-5793(93)80427-v. [DOI] [PubMed] [Google Scholar]
- 4.Chen R, Lewis K A, Perrin M H, Vale W. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrin M H, Donaldson C J, Chen R, Lewis K A, Vale W. Endocrinology. 1993;133:3058–3061. doi: 10.1210/endo.133.6.8243338. [DOI] [PubMed] [Google Scholar]
- 6.Chang C-P, Pearse R V, II, O’Connell S, Rosenfeld M G. Neuron. 1993;11:1187–1195. doi: 10.1016/0896-6273(93)90230-o. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Xie L Y, Abou-Samra A B. Endocrinology. 1996;137:192–197. doi: 10.1210/endo.137.1.8536612. [DOI] [PubMed] [Google Scholar]
- 8.Dautzenberg F M, Dietrich K, Palchaudhuri M R, Spiess J. J Neurochem. 1997;69:1640–1649. doi: 10.1046/j.1471-4159.1997.69041640.x. [DOI] [PubMed] [Google Scholar]
- 9.Lovenberg T W, Liaw C W, Grigoriadis D E, Clevenger W, Chalmers D T, De Souza E B, Oltersdorf T. Proc Natl Acad Sci USA. 1995;92:836–840. doi: 10.1073/pnas.92.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrin M, Donaldson C, Chen R, Blount A, Berggren T, Bilezikjian L, Sawchenko P, Vale W. Proc Natl Acad Sci USA. 1995;92:2969–2973. doi: 10.1073/pnas.92.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishimoto T, Pearse R V, II, Lin C R, Rosenfeld M G. Proc Natl Acad Sci USA. 1995;92:1108–1112. doi: 10.1073/pnas.92.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenzel P, Kesterson R, Yeung W, Cone R D, Rittenberg M B, Stenzel-Poore M P. Mol Endocrinol. 1995;9:637–645. doi: 10.1210/mend.9.5.7565810. [DOI] [PubMed] [Google Scholar]
- 13.Valdenaire O, Giller T, Breu V, Gottowik J, Kilpatrick G. Biochim Biophys Acta. 1997;1352:129–132. doi: 10.1016/s0167-4781(97)00047-x. [DOI] [PubMed] [Google Scholar]
- 14.Vaughan J, Donaldson C, Bittencourt J, Perrin M H, Lewis K, Sutton S, Chan R, Turnbull A V, Lovejoy D, Rivier C, et al. Nature (London) 1995;378:287–292. doi: 10.1038/378287a0. [DOI] [PubMed] [Google Scholar]
- 15.Vale W, Vaughan J, Perrin M. The Endocrinologist. 1997;7:3S–9S. [Google Scholar]
- 16.Rivier J, Rivier C, Galyean R, Miranda A, Miller C, Craig A G, Yamamoto G, Brown M, Vale W. J Med Chem. 1993;36:2851–2859. doi: 10.1021/jm00072a003. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez J F, Kornreich W, Rivier C, Miranda A, Yamamoto G, Andrews J, Tache Y, Vale W, Rivier J. J Med Chem. 1993;36:2860–2867. doi: 10.1021/jm00072a004. [DOI] [PubMed] [Google Scholar]
- 18.Miranda A, Koerber S C, Gulyas J, Lahrichi S L, Craig A G, Corrigan A, Hagler A, Rivier C, Rivier J. J Med Chem. 1994;37:1450–1459. doi: 10.1021/jm00036a010. [DOI] [PubMed] [Google Scholar]
- 19.Gulyas J, Rivier C, Perrin M, Koerber S C, Sutton S, Corrigan A, Lahrichi S L, Craig A G, Vale W, Rivier J. Proc Natl Acad Sci USA. 1995;92:10575–10579. doi: 10.1073/pnas.92.23.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miranda A, Lahrichi S L, Gulyas J, Koerber S C, Craig A G, Corrigan A, Rivier C, Vale W, Rivier J. J Med Chem. 1997;40:3651–3658. doi: 10.1021/jm970311t. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Dagnino R, Jr, De Souza E B, Grigoriades D E, Huang C Q, Kim K-I, Liu Z Y, Moran T, Webb T R, Whitten J P, Xie Y F, McCarthy J R. J Med Chem. 1996;39:4358–4360. doi: 10.1021/jm960149e. [DOI] [PubMed] [Google Scholar]
- 22.Schulz D W, Mansbach R S, Sprouse J, Braselton J P, Collins J, Corman M, Dunaiskis A, Faraci S, Schmidt A W, Seeger T, et al. Proc Natl Acad Sci USA. 1996;93:10477–10482. doi: 10.1073/pnas.93.19.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christos T E, Arvanitis A. Expert Opin Ther Patents. 1998;8:143–152. [Google Scholar]
- 24.Baram T Z, Chalmers D T, Chen C, Koutsoukos Y, De Souza E B. Brain Res. 1997;770:89–95. doi: 10.1016/s0006-8993(97)00759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundkvist J, Chai Z, Teheranian R, Hasanvan H, Bartfai T, Jenck F, Widmer U, Moreau J-L. Eur J Pharmacol. 1996;309:195–200. doi: 10.1016/0014-2999(96)00337-8. [DOI] [PubMed] [Google Scholar]
- 26.Fisher L, Rivier C, Rivier J, Brown M. Endocrinology. 1991;129:1312–1316. doi: 10.1210/endo-129-3-1312. [DOI] [PubMed] [Google Scholar]
- 27.Spiess J, Dautzenberg F M, Sydow S, Hauger R L, Rühmann A, Blank T, Radulovic J. Trends Endocrinol Metab. 1998;9:140–145. doi: 10.1016/s1043-2760(98)00037-x. [DOI] [PubMed] [Google Scholar]
- 28.Donaldson C J, Sutton S W, Perrin M H, Corrigan A Z, Lewis K A, Rivier J E, Vaughan J M, Vale W W. Endocrinology. 1996;137:2167–2170. doi: 10.1210/endo.137.5.8612563. [DOI] [PubMed] [Google Scholar]
- 29.Gottowik J, Goetschy V, Henriot S, Kitas E, Fluhman B, Clerc R G, Moreau J L, Monsma F J, Kilpatrick G J. Neuropharmacology. 1997;36:1439–1446. doi: 10.1016/s0028-3908(97)00098-1. [DOI] [PubMed] [Google Scholar]
- 30.Rühmann A, Köpke A K E, Dautzenberg F M, Spiess J. Proc Natl Acad Sci USA. 1996;93:10609–10613. doi: 10.1073/pnas.93.20.10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoriadis D E, Liu X-J, Vaughan J, Palmer S F, True C D, Vale W W, Ling N, De Souza E B. Mol Pharmacol. 1996;50:679–686. [PubMed] [Google Scholar]
- 32.Liaw C W, Grigoriadis D E, Lorang M T, De Souza E B, Maki R A. Mol Endocrinol. 1997;11:2048–2053. doi: 10.1210/mend.11.13.0034. [DOI] [PubMed] [Google Scholar]