Abstract
The 3′ untranslated region (3′ UTR) of turnip crinkle virus (TCV) genomic RNA contains a cap-independent translation element (CITE), which includes a ribosome-binding structural element (RBSE) that participates in recruitment of the large ribosomal subunit. In addition, a large symmetric loop in the RBSE plays a key role in coordinating the incompatible processes of viral translation and replication, which require enzyme progression in opposite directions on the viral template. To understand the structural basis for the large ribosomal subunit recruitment and the intricate interplay among different parts of the molecule, we determined the global structure of the 102-nt RBSE RNA using solution NMR and small-angle x-ray scattering. This RNA has many structural features that resemble those of a tRNA in solution. The hairpins H1 and H2, linked by a 7-nucleotide linker, form the upper part of RBSE and hairpin H3 is relatively independent from the rest of the structure and is accessible to interactions. This global structure provides insights into the three-dimensional layout for ribosome binding, which may serve as a structural basis for its involvement in recruitment of the large ribosomal subunit and the switch between viral translation and replication. The experimentally determined three-dimensional structure of a functional element in the 3′ UTR of an RNA from any organism has not been previously reported. The RBSE structure represents a prototype structure of a new class of RNA structural elements involved in viral translation/replication processes.
Keywords: new method, NMR, SAXS, 3' UTR RNA, RNA structure
Structural elements in mRNAs such as the 5′ cap, internal ribosome entry site in the 5′ UTR, and the 3′ poly(A) tail in the 3′ UTR are important determinants for efficient translation initiation (1). These structural elements can function synergistically to attract ribosomes and translation factors to enhance translation initiation (2, 3). In both cap and poly(A) tail-dependent translation in eukaryotes this enhancement is realized by the binding factor elF4G associating with both the polyA binding protein (Pab1p) and elF4E, resulting in a circularized mRNA template (4), which has been visualized under atomic force microscopy (5). In contrast, high level initiation of cap-independent translation in many plant viruses involves 3′ UTR RNA elements known as CITE (6). TCV lacks a 5′ cap and poly(A) tail. Instead the virus uses a structural element in its 3′ UTR that synergistically enhances translation when associated with its 5′ UTR (7) (Fig. 1 A). A model for 3′ UTR involvement in the recruitment of the large ribosomal subunit has been proposed to account for the enhancement (7) but the experimental structural basis for such an involvement has not been demonstrated. Moreover, mechanisms are required to temporally coordinate viral replication and translation because these processes are mutually exclusive due to the opposing directions of protein and RNA synthesis. The infecting genomic RNA must first be translated to produce viral replication proteins before RNA synthesis can initiate. An element critical to these activities is located in approximately 140 nt sequence within the 3′ UTR (7). A subfragment of this RNA, the 118 nt F1 fragment, represents a RNA structural domain based on the ability of this segment derived from the related virus Cardamine chlorotic fleck virus (CCFV), but not most subfragments of CCFV, to support efficient TCV accumulation (McCormack et al., 2008). A recent report using in-line probing indicated that the domain also serves as a highly stable scaffold for canonical basepairing interactions with cis-sequences through the H3 large internal symmetrical loop (LSL) (Fig. 1 B), which are important for the translation/replication switch (8 –11) (Fig. 1 B). These interactions imply that both sides of the LSL are accessible for tertiary interactions.
Fig. 1.
The TCV RBSE. (A) Model for cap-independent translation initiation in TCV. The model suggests that the RBSE functions to recruit or recycle 60S ribosomal subunits to the template that then accesses the 5′ end possibly through interaction with prebound 40S subunits. (B) Schematic drawing of the genome organization of TCV and the secondary structure of the RBSE. The secondary structure of RBSE was verified by imino-NOE walks of this construct and a number of mutants (see SI Text). The hairpins are labeled as H1, H2 and H3, which consists of H3a, H3b and an internal loop (the nomenclature is simplified from previous nomenclature [(7) for this report]. The pseudoknot formed between the residues in the H2 loop and the residues at the 3′ end was previously determined (10). The cis-acting sequences, external to the RBSE on both the 5′ and 3′ ends of the TCV 3′ UTR are also shown. The italic numbers are those of the genome and smaller numbers are those for RBSE starting from position 1. (C) RBSE ribosome binding competition experiments. Deacylated tRNAphe can compete with binding of RBSE to the P site of yeast ribosomes (Left), and RBSE does not compete with acylated Phe-tRNAphe for binding to the A site of ribosomes (Right).
A previous computational study suggested that a 100 nt RNA within the F1 fragment folds into a structure that resembles a tRNA-like shape (10). The F1 fragment binds to 60S ribosomal subunits with an affinity of about 400 nM and competes with N-acetylated phe-tRNA for the P-site of the ribosome (7). Mutations that disrupt H1, which is positionally equivalent to the amino-acceptor arm on a tRNA in the structure, repressed ribosome binding (7). In addition RNA-dependent RNA polymerase (RdRp) binding to the region causes a substantial conformational switch that disrupts the H1 region and likely promotes transcription of complementary strands while suppressing translation (7, 10). To understand the mechanism of 3′ UTR participation in translation and replication, it is important to determine the global structure that outlines the spatial arrangements of the three hairpins, H1, H2 and H3. A three-dimensional global structure of RBSE will also address in part the structural basis for the accessibility of the LSL for interaction with surrounding sequences.
Currently, despite significant advances in x-ray crystallography and solution NMR, structure determination of mid- to large-size RNA molecules with complex folds remain a daunting task. In the meantime, structures of a number of regulatory riboswitches in 5′ UTR (12) have been reported but no structure of any functional structural element from within the 3′ UTR is available. We report here the application of the global structures from global measurements (G2G) method. (13) to determine the global structure of the 102 nt RBSE and provide insight into the structural aspects of this RNA.
Results
Binding of 102 nt RBSE to the P site of 80S Yeast Ribosomes.
The 102 nt RBSE used for this report has activities similar to that of the F1 fragment reported previously (7). The RBSE has two additional Gs at the 5′ end (Fig. 1 B) that stabilize the shorter fragment by extending a previously proposed pseudoknot stem that is important for ribosome binding at the expense of a short hairpin (7, 10). The 102 nt RBSE fragment contains 100 nt from position 3909–4009 in the 3′ UTR of the TCV genomic RNA. This construct is slightly smaller than the F1 fragment, a 118 nt RNA (nt 3900–4017), studied by Stupina et al. for its ribosome-binding activities (7). The published work identified four important biochemical characteristics of the F1 fragment. First, the F1 fragment bound 80S ribosomes with a K d of 450 nM. Second, binding of the F1 fragment to nonprogrammed ribosomes could be blocked by deacylated tRNA (which binds to the P site). Third, the F1 fragment could compete with the binding of N-acetylated phe-tRNAphe to the P site of the ribosome. Finally, the F1 fragment did not prevent acylated phe-tRNAphe from binding to the A site. These observations led to conclusion that the F1 fragment binds to the P site of ribosomes with significant affinity (450 nM) (7).
The RBSE was tested for ribosome binding using nitrocellulose filter binding experiments similar to those published for the F1 fragment (7). The K d of the interaction between the 102 nt RBSE and 80S yeast ribosomes was determined to be 300 ± 90 nM, which agrees with the published value for the F1 fragment of 450 nM. The 102 nt RBSE construct also exhibited the same characteristics as those published for the F1 fragment (Fig. 1 C). Because these experiments duplicate the published results for the nearly identical F1 fragment, we can conclude that the 102 nt RBSE construct also binds to the P site of the ribosome with significant affinity (7).
RBSE has a Twisted “T”-Shape.
The secondary structure of the RBSE consists of hairpins H1, H2, and H3 (H3a + H3b) as shown in Fig. 1 B and the bairpairing scheme was verified by the conventional NOE-walk method aided with spectra and spectra of the mutants (SI Text). The molecular envelope of the RBSE (Fig. 2 A) was derived from the small-angle x-ray scattering (SAXS) data and shows that the structure of this RNA adopts a twisted “T” shape in solution. The approximate angles and dimensions are depicted in Fig. 2 A. The location of hairpin H3 was identified by comparing the envelope shape of the RBSE to that of hairpin H3 and a number of other constructs (SI Text). In particular, the long arm of the RBSE envelope matches remarkably well with that of the hairpin H3 construct (SI Text). We also assigned the left side of the short arm to hairpin H1 and the right side to hairpin H2 whose hairpin loop residues are complementary to those at the 3′ end of RBSE to form a pseudoknot (7) making it considerably larger than a simple hairpin based on dimension measurements and shapes (Fig. 2 A).
Fig. 2.
The molecular envelope of RBSE, RDC waves of RBSE and the TCV3M mutant. (A) The low-resolution molecular envelope of RBSE with dimension measurements. The envelope is an average of 16 bead models calculated from SAXS data using DAMMIN (34). The top short arm on the left of the envelope has a cylindrical shape, about 20 Å in diameter which is comparable in diameter to an A-form RNA duplex. The long arm of the envelope is bent with an angle of approximately 140° and is twisted outward. (B). The RDC waves of the three duplexes in RBSE with the solid curves calculated using parameters D a, R, orientation (Θ,Φ), phase ρ 0 from the simultaneous fit using the program ORIENT and the structural parameters of an A-form duplex. The experimental RDCs are shown as circles. Interestingly, duplex H3a is interrupted at the mismatch A61-G94 but the remaining residues whose imino RDCs are available are still in the A-form conformation as indicated by their imino RDC fits. (C). On the left is the secondary structural drawing of the TCV3M mutant that has an extended H3b (highlighted in red) and a point mutation at A61 to C61. This mutant allowed for a better determination of the angle between H3a and H3b. On the right are the RDC waves of the H3a and H3b. From the simultaneous fit, we obtained the angle between the axes of the H3a and H3b duplexes.
Shape-Assisted Duplex Orientation Determination.
The relative orientation and phase for each duplex were calculated using the program ORIENT in the G2G toolkit package (13). The degenerate combinations of orientations that were not consistent with the molecular envelope were filtered out in the calculation by using the angle restrictions between the duplexes with a ± 30° error range. The dipolar waves of the RBSE duplexes are shown in Fig. 2 B. The average orientations and phases (Θ,Φ,ρ 0) with standard deviations, produced from the top fits with a RDC RMSD cutoff of 1.2 Hz, are (150 ± 12°, 88 ± 4°, 260 ± 9°), (44 ± 2°, 290 ± 5°, 66 ± 3°), and (36 ± 1°, 35 ± 4°, 222 ± 3°) for H1, H2 and H3a, respectively, and with the average D a = -21.0 ± 1.0 and R = 0.35 ± 0.03, where the standard deviations are used as the error ranges. It is interesting to note that even though imino signals for U60, G94 and U93 in hairpin H3a were not detected possibly due to fast hydrogen exchanges with solvent introduced by the A61-G94 mismatch (Fig. 1 B), this mismatch may introduce very limited perturbation detectable neither in the periodicity curve fitting (Fig. 2 B) nor in the SAXS-derived molecular envelope (Fig. 2 A).
The bending Angle in H3.
The bending angle between H3a and the segment involving H3b in hairpin H3 was determined using a construct, TCV3M, in which the A61-G94 mismatch was mutated to a C–G Watson–Crick pair in H3a and a stretch of four basepairs was inserted between the triple CGs and the GAAA tetraloop to extend H3b (Fig. 2 C) to give 7–9 imino RDCs for H3a or H3b, allowing for a more accurate orientation and phase determination. The single mutation in H3a and the insertion of a stretch of four basepairs after the triple CGs in H3b have little impact on the original angle between H3a and H3b as seen in the low-resolution envelope (SI Text). The angle between H3a and H3b in TCV3M is similar, about 140 ± 30°, to that in the intact RBSE or in the hairpin H3 construct. The top simultaneous RDC fits for tcv3M with a RMSD cutoff of 1.0 Hz give an average angle between H3a and H3b in TCV3M of 159° ± 2°, and was taken as the approximate angle between H3a and H3b to generate a starting structure (Fig. 3).
Fig. 3.
A two-dimensional topology drawing (Left) of RBSE, the initial structure (Center) generated with the G2G toolkit, and the structure after regularization that fixes the bond breaks (Right). The regularization was accomplished in Xplor-NIH (35). The orientations and phases in terms of (Θ,Φ,ρ 0) of H1, H2 and H3a, obtained from the best simultaneous fit, are given in the figure. The angles between hairpins labeled on the left figure were calculated from the hairpin orientations (Θ,Φ) on the figure. The linker residues are represented with broken lines in the topology drawing (Left) and residue numbers are drawn on the regularized structure (Right). The angle between H3a and H3b was determined using the mutant TCV3M (Fig. 2 C). Angles between all pairs of duplexes are given in the text.
The Global Structure of RBSE in Solution.
The topological arrangements of the three hairpins is shown on the left side and the three-dimensional structure, generated using programs ORIENT and BLOCK(13), is shown in the middle of Fig. 3, respectively. In addition, the pseudoknot formed between the residues in the terminal loop of H2 and those at the 3′ end of the RNA was restrained by Watson– Crick pairings and loosely restrained A-form duplex torsion angles. Structural calculation statistics are provided in SI Text. We then used the structure determination program Xplor-NIH to regularize the structure. During the regularization the duplexes were allowed to translate but not rotate and the nucleotide chain was connected with optimized geometry at joints. The correlation coefficient between the experimental RDCs and the back-calculated RDCs based on the starting structure prior to the refinement (Fig. 3, Right) is approximately 0.97 for the imino RDCs in duplex regions, as expected because the orientations of the duplexes has been predetermined using the RDC-structural periodicity correlation (Fig. 2 B).
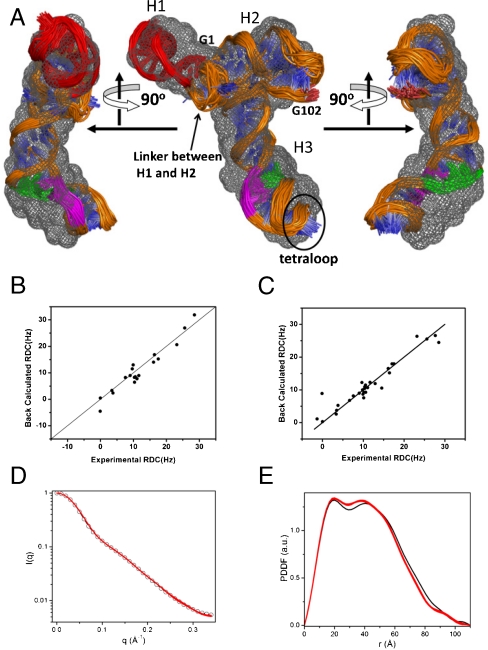
We then refined the structure (Fig. 3, Right) restrained with all RDCs that include those in the duplex and nonduplex regions and the SAXS data. In addition, we also included distance and torsion angle restraints to maintain the A-form duplex geometry, generic distance restraints for maintaining base stacking. The refined global structure of the RBSE is shown in Fig. 4 A. The “correctness” of the global structure in terms of global orientations of duplexes and the overall shape is simultaneously benchmarked by the correlation coefficients of RDCs before the simulated annealing (SA) refinement (Fig. 4 B) and after the refinement (Fig. 4 C) and by the SAXS (Fig. 4 D) and the pair distance distribution function (PDDF) (Fig. 4 E) curves. These correlation coefficients before and after the non-rigid-body SA refinement remain similar (Fig. 4 B and C), suggesting the orientations of the duplexes is consistent with the SAXS data that restrains the overall shape of the molecule and indirectly restrains the duplex orientations. The comparison of the back-calculated SAXS curves based on the refined top 10% structures with the experimental SAXS data is displayed in Fig. 4 D and the RMSD between the two is about 0.20 ± 0.01. The comparison of pair distance distribution function curves of the corresponding SAXS profiles is shown in Fig. 4 E. Thus, the structure has a correct overall shape, with the duplexes in their proper global orientations, phases, and positions that are consistent with the global measurements in solution. The accuracy of the backbone structure is comparable to that of the riboA structure that we determined using the G2G method (13). The linkers between the hairpins were set free without any restraint during the calculation and their possible structures are only indirectly restrained by the orientations, phases, and positions of duplexes and directly by the covalent linkages between the duplexes and linkers.
Fig. 4.
The ensemble of global structures of the RBSE determined using the G2G “top-down” method and SAXS and PDDF curves comparison. (a) The front (Center) and side (Left and Right) views of the superimposed RBSE backbone structures (top 50% lowest energy) overlaid with the molecular envelope in gray mesh (top) or itself (bottom). The linker between the H1 and H2 is located at a position that is similar to the variable-loop found in canonical tRNA. The region important for large subunit ribosome and RdRp binding is highlighted in red; sequences in LSL involved in canonical basepairing with 5′- and 3′ends are colored in green and magenta, respectively. (B) The correlation plot of the back-calculated RDCs based on the starting structure (Fig. 3, Right), where the orientation of the three duplexes were determined using the RDC-structural periodicity correlation. Only RDCs in the duplex regions (the same experimental RDC data as shown in Fig. 2 B) were used the correlation coefficient calculation. The correlation coefficient is approximately 0.97. (C) The correlation plot of the back-calculated RDCs based on the top 10% lowest G2G structures vs. the experimental RDCs. The correlation is near unit. (D) The comparison of experimental (circle) and back-calculated SAXS curves (red) based on the top 10% ensemble. The RMSD between experimental data and the back-calculated curves is 0.20 ± 0.01. RMSD is calculated based on the logarithm of the normalized [i.e., I(q = 0) = 1.0] SAXS intensities. (E) The comparison of PDDFs of the corresponding experimental SAXS (black) and back-calculated SAXS curves (red) in (C).
Discussion
We show that the RBSE folds into a structure that resembles a tRNA not only in overall shape but also in some details. This is in spite of the fact that the secondary structure representation of the element does not resemble a typical tRNA cloverleaf. H1, H2, and the linker between them form the upper part of the RBSE structure. The linker between H1 and H2 is in a position similar to that of the D-loop in a canonical tRNA and part of the linker sequence, 5′-AGCU (Fig. 1 B) is identical to the first part of the sequence of the D-loop in tRNAphe. The biological implication of this loop in RBSE is not known. Furthermore, hairpins H1 and H2 are coaxially stacked similar to that of the acceptor and T-stems in a tRNA. Coaxial stacking is a common packing scheme found in RNA folding and it has been seen in other RNAs besides tRNA (14 –19). H1 is involved in binding to the large ribosomal subunit and to the RdRp (10, 11) (Fig. 4 A). H1 is at the equivalent position of the tRNA amino-acceptor stem, which is the site of large ribosomal subunit binding (20). The structural equivalence between these RNAs of two different classes is significant because the H1 in RBSE and the acceptor stem in tRNA are both involved in binding to the large ribosomal subunit (10, 20, 21) and may provide structural basis for the TCV 3′ UTR’s ability to recruit the large ribosomal subunit for initiation. It is noteworthy that there have been abundant studies reported in literature about how the small ribosomal subunit is recruited (for a recent review, see ref. 22) but little has been discussed as how the large ribosomal subunit is recruited for the initiation. The lower part of RBSE consists of H3, which is linked to H2 in the upper part of RBSE by a short linker. The linker consists of triple uridylates (U54-U56 in RBSE numbering in Fig. 1 B, or U3961-U3963 in the TCV genomic numbering) and is exposed, which is consistent with the in-line probing results that suggest the linker is highly flexible and thus strongly susceptible to in-line cleavage (10) (Fig. 1 B). This linker has also been proposed to be part of the RdRp binding site together with the H1 region (11). Moreover, extensive mutagenesis studies suggest that H3 is the focal point of the viral translation/replication switch (7, 10, 11). On one hand, 5′-G84GGC in the LSL interacts with 3′-CCCG at the 3′-terminus of the TCV 3′ UTR to form a phylogentically conserved pseudoknot (8, 9, 23) (Fig. 1 B). This interaction is important for viral accumulation in vitro (24). On the other hand, residues 5′-A68AAA in the 5′ side of the LSL interact with the UUUU sequence in the terminal loop of an upstream hairpin that is also central to the switch between replication and translation (10, 11) (Fig. 1 B). The simultaneous canonical pairing to both sides of a similar LSL has been reported in the case of the H/AC small nucleolar RNA and 28S rRNA that leads to pseudouridylation (25). The global orientation and position of H3 in the RBSE structure, also shown by molecular dynamics calculations (10), indicate that it is relatively independent, is accessible without significant alteration of the rest of the scaffold, and serves as a structural basis to explain mutagenesis and in-line probing results where mutations in the H3 LSL do not affect cleavage patterns in the remainder of the tRNA-like structure (7).
It is noteworthy that the GAAA tetraloop is located in a position similar to the anticodon loop in a tRNA. The GAAA tetraloop consists of a “U-turn” with three stacked but unpaired adenine bases. In parallel, the anticodon loop of tRNA contains a U-turn followed by three stacked but unpaired anticodon bases. A GAAA can be a very stable structural element that could serve as a nucleation site for RNA folding (26). On the other hand, the GAAA tetraloop is one of the most common motifs that interact with the A-form minor groove via “A-minor” motifs (27 –30). The remaining sequence in the D-loop is UUU, whose counterpart in RBSE is U54-U55-U56 in close proximity. The current available structural data that allows determination of the global fold of this RNA, is not sufficient to determine the details of possible intricate networks of interactions in this junction involving the linkers H2 and H3a.
The shape of the RBSE is also reminiscent of a known class of tRNA-like structures (TLS) (31, 32). Both RBSE and TLS elements are part of 3′ UTRs that lack poly(A) tails. Nevertheless, RBSE differs from TLS (32) in at least two aspects. First, RBSE is internal and is a CITE (6), whereas TLS is located at the 3′ terminus of some 5′ cap-containing plant virus RNAs. Second, unlike TLS, RBSE lacks a 3′ CCA terminus and is therefore not expected to be a substrate of tRNA synthetase.
Furthermore, the tRNA-like structure of the TCV RBSE may not be unique, as CCFV has a similar secondary structural element in its 3′ UTR (10). RBSE plays an important role in mediating viral translation and replication by adopting a tRNA-like shape that allows interplays among different regions that are not proximal in the primary sequence of the 3′ UTR. The RBSE structure represents a prototype for a unique class of structural elements involved in viral gene expression and is a previously undescribed experimentally determined structure of a functional element from the 3′ UTR of any organism. This structure sheds insight into structural basis for the large ribosomal subunit recruitment in viral translation and for the regulation of the viral translation and replication processes. We speculate that more of these types of structural elements will be discovered as more attention is paid to the untranslated, but not unimportant, region of genomic RNAs.
Materials and Methods
Activity Assay of the 102 nt RBSE.
All binding experiments were carried out essentially as described previously (7). In brief the procedures were as follows. For the K D measurements, substoichiometric 3′ end-labeled RBSE was incubated with ribosomes at concentrations from 0–1500 nM and bound to nitrocellulose filters after incubation. For the competition between RBSE and deacylated tRNA, a 20-fold excess of deacylated tRNA was incubated with ribosomes prior to the addition of substoichiometric end-labeled RBSE and after incubation the samples were bound to nitrocellulose filters. For the competition between RBSE and Phe-tRNAPhe, ribosomes were incubated with a 7.5-fold excess of unlabeled RBSE prior to the addition of 14[C]-Phe-tRNAPhe, and after incubation samples were bound to nitrocellulose filters. The N-Ac-Phe-tRNAPhe competition experiments were conducted identically to the Phe-tRNAPhe competition experiments.
SAXS Experiments, Data Analysis, and Bead Model Reconstruction.
Both SAXS and wide-angle x-ray scattering (WAXS) were performed at beamlines 12-ID and 18-ID of Advanced Photon Sources at Argonne National Laboratory. A detailed description about experimental procedure, processing, and analysis of scattering data are presented in SI Text. The experimental SAXS data is presented in Molecular envelope reconstruction from SAXS data in SI Text.
A detailed description about the bead model reconstruction is presented in SI Text. The ± 30° range was estimated conservatively based on the size of a bead that was used in bead model calculation. The diameter of the bead was 7.2 Å. This size is chosen based on the DMAX, the maximum distance determined from the PDDF of RBSE, and the total number of beads used in a specific mode in the DAMMIN calculation. The 5,400 initial number of beads and jagged mode of DAMMIN were used in calculations for RBSE. We tested the error range of a duplex by using a bead model that was calculated in the same way as that for RBSE but using simulated SAXS data, calculated based on an L-shape model RNA similar to that of RBSE. The error range for the helical axis orientation is less than ± 30° (SI Text).
In the fitting of the RDC data, the error range for finding a unique orientation is actually not so important so long that the angle range allows one to discriminate the possible correct relative orientation from other three degenerate orientations. The angular relationships among the four discrete orientations are: (ϑ,Φ), (ϑ, Φ + π), (π - ϑ, π - Φ), (π - ϑ, 2π - Φ), where (ϑ,Φ) are polar angles of a duplex axis (33). We call this approach “SAXS-aided removal of degeneracy in orientations”. If one can discriminate single solution, one can determine the orientation of the duplex with RDC fitting with much higher precision as well as accuracy.
Supplementary Information Available
A more detailed description of the experimental methods and materials is presented in SI Text. The calculation protocols has been deposited in the Protein Data Bank together with the coordinates and restraint files (accession number 2krl), and can also be downloaded from the author’s web site: http://ccr.cancer.gov/staff/links.asp?profileid=5546.
Supplementary Material
Acknowledgments.
We thank Professors R. Green and Samuel E. Butcher for helpful discussions.We thank Drs. L. Guo (BioCAT, sector 18-ID) and S. Seifert (BESSRC, sector 12-ID) at Argonne National Laboratory for their support for synchrotron experiments. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research to B.A.S. and Y-X.W.; by the Intramural Research Program of the NIH, the CIT Intramural Research Program to C.D.S, and U.S. Public Health Service (GM 061515-05A2/G120CD) to A.E.S.. This publication has been funded in whole or in part with federal funds from the National Cancer Institute, NIH, under grant HHSN261200800001E to W.K. and P.Y. Work at Argonne National Laboratory (DMT) and the Advanced Photon Source was supported by the Office of Basic Energy Sciences, Department of Energy under contract DE-AC02-06CH11357. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a National Institutes of Health-supported Research Center RR-08630.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908140107/DCSupplemental.
References
- 1.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 2.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 3.Tarun SZ, Jr, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 4.Tarun SZ, Jr, Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 6.Miller WA, Wang Z, Treder K. The amazing diversity of capindependent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stupina VA, et al. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, Zhang J, Simon AE. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J Virol. 2004;78:7619–7633. doi: 10.1128/JVI.78.14.7619-7633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Zhang G, Guo R, Shapiro BA, Simon AE. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus-strand synthesis. J Virol. 2006;80:9181–9191. doi: 10.1128/JVI.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCormack JC, et al. Structural domains within the 3′ untranslated region of turnip crinkle virus. J Virol. 2008;82:8706–8720. doi: 10.1128/JVI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan X, Kerong Shi K, Meskauskas A, Simon AE. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. in press. [DOI] [PMC free article] [PubMed]
- 12.Montange RK, Batey RT. Riboswitches: Emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, et al. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J Mol Biol. 2009;393:717–734. doi: 10.1016/j.jmb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puglisi JD, Wyatt JR, Tinoco I., Jr. Conformation of an RNA pseudoknot. J Mol Biol. 1990;214:437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazantsev AV, et al. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D’Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 19.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 20.Schafer MA, et al. Codon-anticodon interaction at the P site is a prerequisite for tRNA interaction with the small ribosomal subunit. J Biol Chem. 2002;277:19095–19105. doi: 10.1074/jbc.M108902200. [DOI] [PubMed] [Google Scholar]
- 21.Yuan X, Shi K, Meskauskas A, Simon AE. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. 2009;15:1849–1864. doi: 10.1261/rna.1708709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na H, White KA. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology. 2006;345:305–316. doi: 10.1016/j.virol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Zhang G, McCormack JC, Simon AE. Evolution of virus-derived sequences for high-level replication of a subviral RNA. Virology. 2006;351:476–488. doi: 10.1016/j.virol.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Feigon J. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proc Natl Acad Sci USA. 2007;104:6655–6660. doi: 10.1073/pnas.0701534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin, and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 29.Tanner MA, Cech TR. An important RNA tertiary interaction of group I and group II introns is implicated in gram-positive RNase P RNAs. RNA. 1995;1:349–350. [PMC free article] [PubMed] [Google Scholar]
- 30.Costa M, Michel F. Frequent use of the same tertiary motif by selffolding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond JA, Rambo RP, Filbin ME, Kieft JS. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA. 2009;15:294–307. doi: 10.1261/rna.1360709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fechter P, Rudinger-Thirion J, Florentz C, Giege R. Novel features in the tRNA-like world of plant viral RNAs. Cell Mol Life Sci. 2001;58:1547–1561. doi: 10.1007/PL00000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh JD, Cabello-Villegas J, Wang YX. Periodicity in residual dipolar couplings and nucleic acid structures. J Am Chem Soc. 2004;126:1938–1939. doi: 10.1021/ja039446g. [DOI] [PubMed] [Google Scholar]
- 34.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.