Abstract
In vivo, retroviral integration is mediated by a large nucleoprotein complex, termed the preintegration complex (PIC). PICs isolated from infected cells display in vitro integration activity. Here, we analyze the roles of different host cell factors in the structure and function of HIV type 1 (HIV-1) PICs. PICs purified by size exclusion after treatment with high salt lost their integration activity, and adding back an extract from uninfected cells restored this activity. In parallel, the native protein–DNA intasome structure detected at the ends of HIV-1 by Mu-mediated PCR footprinting was abolished by high salt and restored by the crude cell extract. Various purified proteins previously implicated in retroviral PIC function then were analyzed for their effects on the structure and function of salt-treated HIV-1 PICs. Whereas relatively low amounts (5–20 nM) of human barrier-to-autointegration factor (BAF) protein restored integration activity, substantially more (5–10 μM) human host factor HMG I(Y) was required. Similarly high levels (3–8 μM) of bovine RNase A, a DNA-binding protein used as a nonspecific control, also restored activity. Mu-mediated PCR footprinting revealed that of these three purified proteins, only BAF restored the native structure of the HIV-1 protein–DNA intasome. We suggest that BAF is a natural host cofactor for HIV-1 integration.
Integration of retroviral cDNA is required for efficient virus replication. In infected cells, integration is mediated by a large subviral nucleoprotein complex, termed the preintegration complex (PIC). Retroviral PICs isolated from infected cells can integrate their endogenous cDNA into an exogenously added target DNA in vitro (1–4). The product of this recombination reaction is a gapped intermediate wherein the 3′ ends of the linear viral cDNA are covalently joined to the 5′ phosphates of a double-stranded staggered cut in the target DNA, and the 5′ ends of the viral DNA remain unjoined (5, 6). Repair of the single-stranded gaps in infected cells yields the integrated provirus flanked by the sequence duplication of the double-stranded staggered cut (for a recent review, see ref. 7).
Although purified retroviral integrase proteins display in vitro integration activity, the majority of these recombination products are incomplete in that they result from the integration of only one viral DNA end into just one strand of target DNA (8–12). This single-ended integration would be nonproductive in vivo and differs from the high efficiency of authentic two-ended integration catalyzed in vitro by PICs isolated from infected cells. Thus, analyzing PICs should reveal details of nucleoprotein complex structure and function important for authentic retroviral integration, which are missing from more simplified in vitro integration systems.
The only viral components required for the in vitro integration activity of HIV type 1 (HIV-1) PICs are integrase and cDNA (13). Host-encoded proteins also have been shown to participate in retroviral PIC function in vitro. In these systems, PICs first are treated with high concentrations of salt to remove factors required for integration, the bulk complexes are purified away from the freed proteins, and PIC activity then is reconstituted by adding back extracts from uninfected host cells (14–16). For Moloney murine leukemia virus (Mo-MLV), high salt not only disrupted normal intermolecular integration, but in doing so stimulated autointegration, a suicidal DNA recombination pathway that uses the viral cDNA itself as the target (14). In this case, the host-cell extract both restored normal intermolecular integration and blocked autointegration activity (14). Unlike Mo-MLV, treating HIV-1 PICs with high salt did not detectably induce autointegration (15).
Purified proteins from diverse sources have been shown to stimulate the intermolecular integration activity of retroviral PICs. Whereas a subset of these proteins apparently acts nonspecifically (16), two proteins, the barrier-to-autointegration factor (BAF) and HMG I(Y), have been implicated as specific cofactors. HMG I(Y) restored intermolecular integration activity to salt-stripped HIV-1 (15) and Mo-MLV PICs (16), but had no effect on preventing Mo-MLV autointegration (16). BAF, on the other hand, both restored intermolecular integration activity to salt-stripped Mo-MLV PICs and prevented their autointegration (17).
Here, we use high-salt stripping and functional reconstitution to analyze the roles of various host proteins in HIV-1 PIC function. In addition, we monitor the protein–DNA structure at the ends of HIV-1 by using Mu-mediated PCR (MM-PCR) footprinting (18). We find that relatively low concentrations of recombinant human BAF protein restored both the integration activity of salt-disrupted HIV-1 PICs and the unique protein–DNA structure detected at the ends of HIV-1 by MM-PCR footprinting.
MATERIALS AND METHODS
Oligonucleotides, Proteins, and Reagents.
The Mu and HIV-1 oligonucleotides used for MM-PCR footprinting will be described in detail elsewhere (H.C., S.-Q. Wei, and A.E., unpublished work). Mu A protein was kindly provided by Michiyo Mizuuchi, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD. Purified recombinant human BAF was a generous gift from Robert Craigie, National Institute of Diabetes and Digestive and Kidney Diseases. Human BAF differs from murine BAF (17) at three amino acid positions (R. Zheng and R. Craigie, personal communication). Human HMG I(Y) was purified from Escherichia coli cells as described (15) after expression from plasmid pET7C (19). RNase A was from two sources. For degrading cellular RNAs in crude cell lysates, RNase A was obtained from Sigma. For functional reconstitution of salt-stripped PICs, Qiagen RNase A was used. Single-stranded DNA-binding protein was from Stratagene, BSA was from New England Biolabs, and digitonin, aprotinin, and Nycodenz were from Sigma.
Preparation of Uninfected Cell Extract.
Sup T1 T cells (8 × 107) were lysed in 2 ml of buffer K (20 mM Hepes, pH 7.5/5 mM MgCl2/1 mM DTT/40 μg/ml aprotinin) containing 150 mM KCl/0.025% digitonin. After centrifugation to remove cell debris, the supernatant was concentrated to 0.2 ml using a Centricon-3 Concentrator as recommended by the manufacturer (Amicon). Glycerol and 4-(2-aminoethyl)benzenesulfonyl fluoride were added to the retentate to the final concentrations of 10% (wt/vol) and 0.5 mM, respectively, and the extract was frozen in liquid N2 and stored at −80°C. This mixture contained approximately 7 mg/ml of total protein as determined by the Bio-Rad protein assay.
Isolation, Salt Stripping, Partial Purification, and Reconstitution of HIV-1 PICs.
HIV-1 infection was initiated by cocultivating uninfected Sup T1 T cells with chronically infected Molt IIIB cells essentially as described previously (4). Five hours postinfection, 2.4 × 108 cells were lysed in 6 ml of buffer K/150 mM KCl/0.025% digitonin, and RNase A was added to the final concentration of 0.1 mg/ml. After incubation at room temperature for 30 min, 2 ml were passed through a 12-ml Sepharose CL-4B spin column equilibrated in buffer K/150 mM KCl/0.025% digitonin. The spin column eluate was purified further on a 10-ml, 10–50% Nycodenz gradient prepared in buffer K/150 mM KCl. The gradient was centrifuged at 274,000 × g for 16 hr at 4°C in a swinging-bucket rotor and then was separated into 1-ml fractions by removing material from the top with a serological pipette.
For salt stripping, the concentration of KCl in the remaining cytoplasmic extract was adjusted to 1.2 M. After incubating on ice for 30 min, the extract was spun through 12-ml Sepharose CL-4B spin columns equilibrated in buffer K/1.2 M KCl/0.025% digitonin. Salt-stripped PICs were purified further by Nycodenz gradient centrifugation as described above.
For reconstitution, uninfected cell extract or purified proteins were added to 200 μl of the gradient-purified, salt-stripped PICs in the presence of 0.04% BSA. The mixtures were incubated on ice for 1 h before integration and MM-PCR assays.
Structure and Function Analyses of HIV-1 PICs.
In vitro integration assays were performed essentially as described previously (4). Integration reactions were deproteinized, electrophoresed through agarose, and analyzed by Southern blotting also as described previously (4). Integration activity was quantified by using a PhosphorImager (Molecular Dynamics) as the percentage of cDNA substrate converted into the integration product.
MM-PCR was performed essentially as described previously (18). Briefly, 200 μl of the assembled Mu A-Mu DNA donor complex was mixed with an equal volume of gradient-purified PICs or deproteinized DNA. Mu transposition proceeded for 30 min at 30°C, and the products were deproteinized and recovered by precipitation with ethanol. Transposition products were detected by autoradiography after two rounds of PCR and denaturing PAGE.
RESULTS
High Salt Disrupts the Structure and Function of HIV-1 PICs.
In MM-PCR, Mu transpososomes are used to cleave the protein–DNA complex to be footprinted. The advantage of this footprinting technique over others is that the DNA cleavage reagent itself provides a substrate for subsequent PCR amplification (18). Using MM-PCR footprinting, we recently described a distinctive nucleoprotein structure, termed the intasome, at the ends of HIV-1 cDNA in wild-type PICs. This native HIV-1 protein–DNA structure is similar to its Mo-MLV counterpart (18), in that 200–250 bp of each viral terminus is protected from nuclease digestion relative to internal DNA regions and strong transpositional enhancements are detected near each viral end (H.C., S.-Q. Wei, and A.E., unpublished work).
To investigate the functional relevance of the HIV-1 intasome, cytoplasmic extracts containing HIV-1 PICs were treated with increasing amounts of KCl, ranging from 0.3 to 1.2 M. Proteins that might be dislodged from PICs by salt stripping were separated from the bulk complexes by passing the mixtures through spin columns equilibrated with the corresponding salt concentrations. Whereas large nucleoprotein complexes are expected to pass through these columns, free proteins would be caught up in the matrices. Integration activity was tested after adjusting the concentration of KCl to 150 mM in the spin column eluates. Whereas PICs treated with 0.9 M KCl retained partial activity (data not shown), treatment with 1.2 M KCl completely abolished integration activity (see below).
Both native and functionally inactivated HIV-1 PICs were purified further by Nycodenz gradient centrifugation. After centrifugation, the gradients were fractionated and individual fractions were analyzed for HIV-1 DNA content by Southern blotting. Both native and salt-stripped PICs sedimented to similar positions in the gradients (fractions 6 and 7), indicating that high-salt treatment did not disrupt the global structure of HIV-1 PICs (data not shown; Fig. 1, lanes 1 and 2). Both types of gradient-purified PICs then were analyzed by MM-PCR footprinting. The transpositional enhancements and regions of nuclease protection indicative of the native HIV-1 intasome were abolished by high-salt treatment (Fig. 2A and B, lanes 1–3).
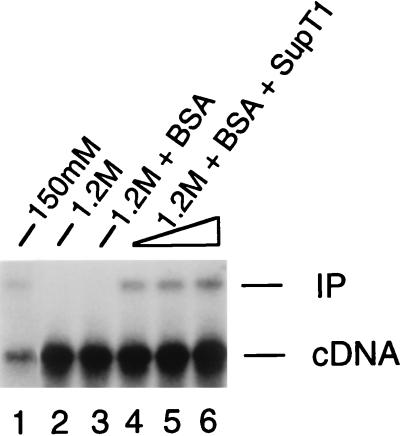
Figure 1.
Functional reconstitution of salt-stripped HIV-1 PICs with uninfected cell extract. Integration reactions were deproteinized and analyzed by Southern blotting. Lane 1, integration activity of untreated, gradient-purified PICs. Lane 2, HIV-1 PICs treated with 1.2 M KCl and then purified by spin column chromatography and gradient centrifugation before the integration assay. Lane 3, BSA included in the reconstitution buffer. Lanes 4–6 included 5, 10, and 15 μl, of crude cell extract, corresponding to approximately 35, 70, and 105 μg of total protein, respectively. cDNA, 9.7-kb HIV-1 linear DNA substrate; IP, 15.1-kb integration product. Approximately 3-fold more material was loaded in lanes 2–6, accounting for the increased level of cDNA.
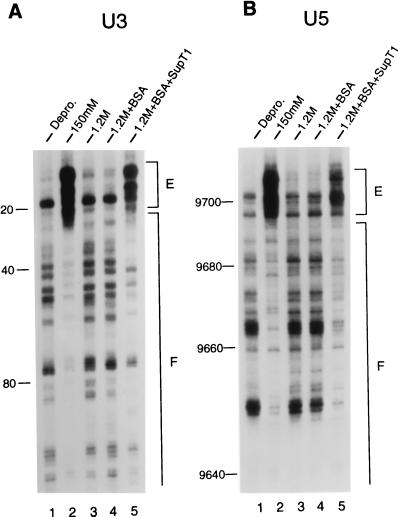
Figure 2.
Structural reconstitution of the HIV-1 intasome with uninfected cell extract. Mu transposition reactions were deproteinized and analyzed by denaturing sequencing gels after two rounds of PCR. The U3 end of HIV-1 DNA was analyzed in A, and the U5 end was analyzed in B. Lanes 1, the pattern of Mu transposition into deproteinized cDNA. Lanes 2, the native structure of the HIV-1 intasome. Note the regions of strong transpositional enhancements (E) and DNA footprints (F) relative to the deproteinized control. Lanes 3, salt-stripped PICs. Note the similarity in patterns between the salt-stripped and deproteinized PICs. Lanes 4, reconstitution buffer control. Lanes 5, salt-stripped PICs reconstituted with 5 μl of uninfected cell extract. The reaction products were run alongside DNA-sequencing ladders to determine the distance from the ends of HIV-1 (nucleotide position 1 in U3 and position 9,718 in U5). The U3 and U5 footprints extended approximately 250 and 200 bp, respectively, in the native structure.
An Extract from Uninfected Cells Restores HIV-1 PIC Structure and Function.
Crude cell extracts previously have been shown to restore integration activity to salt-stripped Mo-MLV (14, 16) and HIV-1 PICs (15). Therefore, we tested whether an extract from uninfected cells might restore integration activity to our salt-treated preparations. Adding BSA alone, a protein included in the reconstitution buffer, had no effect on integration activity (Fig. 1, lane 3). As expected, approximately 50% of the starting integration activity was recovered by including a cytoplasmic extract from uninfected cells (Fig. 1, compare lanes 4–6 with lane 1). This not only showed that host cell factors are important for HIV-1 PIC function in vitro, but also indicated that some functional integrase was retained after salt-treatment and gradient purification, as none of this viral protein was added back.
We next used MM-PCR footprinting to determine the protein–DNA structure of the functionally reconstituted HIV-1 PICs. As expected, incubating BSA alone with the salt-stripped PICs had no discernible effect on the viral end structure (Fig. 2 A and B, compare lanes 4 with lanes 3). In contrast, the crude cell extract, in part, reconstituted the structure of the HIV-1 intasome. The strong transpositional enhancements associated with the native structure were partially restored, as was the protein–DNA footprint near each end of HIV-1 (Fig. 2 A and B, compare lanes 5 with lanes 2 and 3).
BAF Restores HIV-1 PIC Activity and Intasome Structure.
Two host factors have been purified from crude cell extracts based on their abilities to restore intermolecular integration activity to salt-stripped retroviral PICs. BAF protein both blocked the autointegration of salt-stripped Mo-MLV PICs and efficiently restored their intermolecular integration activity (17). HMG I(Y) restored intermolecular integration activity to salt-stripped HIV-1 (15) and Mo-MLV (16) PICs, yet had no effect on blocking the autointegration of Mo-MLV (16).
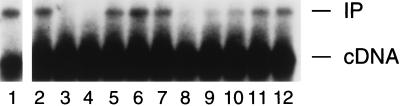
To further investigate how these two host factors might differentially contribute to retroviral integration, we first tested their abilities to reconstitute the activity of salt-stripped HIV-1 PICs. As shown in Fig. 3, as little as 6.25 ng of recombinant human BAF was able to partially restore integration activity (lane 3); maximum reconstitution was detected in this experiment with 12.5 ng of protein (lane 4). In repeated experiments, 5–20 nM BAF (corresponding to 12.5–50 ng of protein) was sufficient to restore the same level of integration activity as the crude cell extract, which ranged from 20 to 50% of the activity that was present before salt stripping (Fig. 3, lanes 3–7; data not shown). In contrast, dramatically different quantities of human HMG I(Y) (almost 1,000-fold more as compared with BAF) were required for functional reconstitution (Fig. 3, lanes 8–12). In repeated experiments, 5–10 μM HMG I(Y) (corresponding to 8–16 μg of protein) reconstituted about half of the maximum activity that was restored by either the crude cell extract or BAF alone (Fig. 3, compare lane 11 with lane 4). Incubation with either excess BAF or HMG I(Y) inhibited functional reconstitution (Fig. 3, lane 12; data not shown).
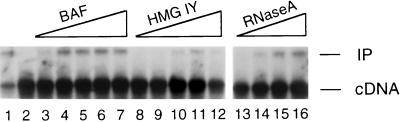
Figure 3.
Functional reconstitution of salt-stripped HIV-1 PICs by using purified proteins. Integration reactions were deproteinized and analyzed by Southern blotting. Lane 1, activity of PICs partially purified under native (150 mM KCl) conditions. Lane 2, activity of salt-stripped PICs. Lanes 3–7 included 6.25, 12.5, 25, 50, and 100 ng of BAF per reaction. Lanes 8–12 included 500, 2,000, 8,000, 16,000, and 32,000 ng of HMG I(Y) per reaction. Lanes 13–16 contained 500, 2,000, 8,000, and 16,000 ng of RNase A per reaction. Neither HMG I(Y) nor RNase A was tested at lower protein concentrations. Other labeling is the same as in Fig. 1.
In addition to HMG I(Y) and BAF, other DNA-binding proteins from diverse sources have been shown to stimulate the intermolecular integration activity of HIV-1 (15) and Mo-MLV (16) PICs. In an attempt to distinguish nonspecific stimulation from specific reconstitution, we also tested the abilities of RNase A from bovine pancreas and single-stranded DNA-binding protein from E. coli to reconstitute the activity of salt-stripped HIV-1 PICs. Whereas single-stranded DNA-binding protein had no complementation activity (data not shown), RNase A efficiently stimulated the intermolecular integration activity of our salt-stripped preparations (Fig. 3, lanes 13–16). In repeated experiments, 3–8 μM RNase A (corresponding to 8–20 μg of protein) reconstituted levels of activity similar to those of BAF (compare lane 16 with lane 4). However, the amount of RNase A protein required for functional reconstitution was similar to that of HMG I(Y). Increasing the concentration of RNase A in the reconstitution reaction also inhibited integration activity (data not shown).
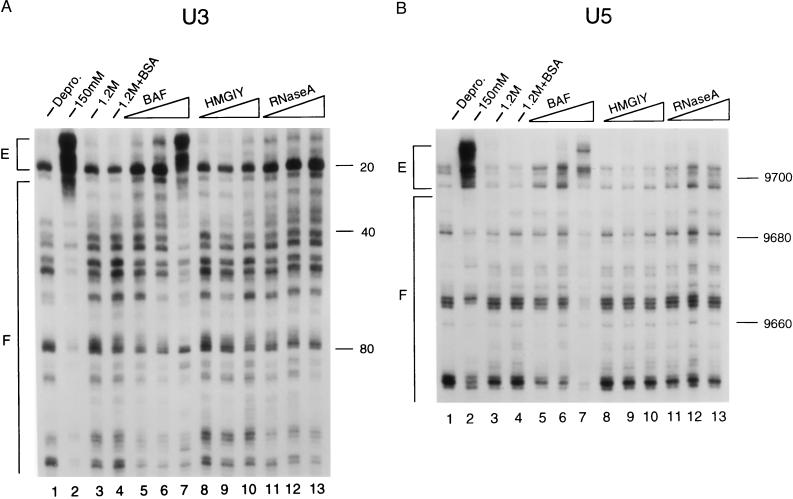
Salt-stripped PICs reconstituted with different amounts of each host factor next were analyzed by MM-PCR footprinting. Whereas increasing levels of BAF gradually restored the transpositional enhancements and footprints associated with the native HIV-1 structure (Fig. 4 A and B, compare lanes 5–7 with lanes 2), neither HMG I(Y) (lanes 8–10) nor RNase A (lanes 11–13) showed any hints of intasome reconstitution. Although increasing concentrations of each cofactor inhibited integration activity, excess levels of BAF yielded transpositional enhancements even stronger than those observed for the untreated native samples and DNA footprints that extended into the internal regions of the viral DNA (data not shown). Evidence for either the transpositional enhancements or DNA footprints associated with the native HIV-1 intasome were not observed for mixtures containing either HMG I(Y) or RNase A, even under conditions of protein excess (data not shown).
Figure 4.
Structural reconstitution of the HIV-1 intasome with purified BAF. Salt-stripped PICs were reconstituted with the indicated proteins and analyzed by MM-PCR footprinting. A and B present the results for the U3 and U5 ends, respectively. Lanes 1–4, as indicated. Lanes 5–7 included 12.5, 25, and 50 ng of BAF per reaction. The integration activities of these reconstitution mixtures were undetectable, and about 8% and 35% of the activity of the untreated native sample was detected, respectively (data not shown). Lanes 8–10 included 2,000, 8,000, and 16,000 ng of HMG I(Y) per reaction. These mixtures displayed undetectable, and about 7% and 11% of the activity of the untreated sample, respectively (data not shown). Lanes 11–13 contained 2,000, 8,000, and 16,000 ng of RNase A per reaction. The mixture containing 2 μg of RNase A displayed about 20% of the activity of the untreated sample; the mixture with 16 μg displayed about 35% activity. Other labeling is the same as in Fig. 2.
DISCUSSION
In this report, we used salt-stripping and functional reconstitution assays together with MM-PCR footprinting to probe the roles of various host cell factors in the structure and function of HIV-1 PICs isolated from infected cells. As shown previously by others (14, 15), high-salt-treated PICs purified away from their dislodged proteins required only an uninfected host cell extract, and not viral integrase, to restore intermolecular integration activity (Fig. 1). Moreover, including purified recombinant HIV-1 integrase in the reconstitution reactions did not stimulate integration activity beyond the level restored by the host cell extract alone (data not shown). This demonstrates that some functional endogenous HIV-1 integrase was retained with the PICs after salt stripping and purification. However, unlike the native intasome, this bound integrase was not detected by MM-PCR footprinting (Fig. 2).
Intact integrase also was required to form the native HIV-1 intasome, as MM-PCR footprints of PICs made from cells infected with an integrase mutant virus resembled those of deproteinized cDNA (H.C., S.-Q. Wei, and A.E., unpublished work). Which host factor collaborates with integrase to form active PICs in HIV-1-infected cells? Between two proteins previously purified by their ability to restore intermolecular DNA recombination to salt-stripped PICs, BAF efficiently restored integration activity (Fig. 3) and also restored the native HIV-1 structure detected by MM-PCR footprinting (Fig. 4). HMG I(Y), in contrast, behaved more like RNase A, a protein that most likely does not participate in the integration process. Although both HMG I(Y) and RNase A restored function to salt-stripped HIV-1 PICs (Fig. 3), the relatively large quantity of each of these proteins required for reconstitution suggests that they may not accomplish this function in vivo. We note that the amounts of HMG I(Y) and BAF proteins required for functional reconstitution in our hands is consistent with the levels reported by others (15–17), and that HMG I(Y) did not reconstitute the structure of the Mo-MLV intasome as detected by MM-PCR footprinting (S.-Q. Wei and R. Craigie, personal communication).
In an attempt to further distinguish the roles of the different host factors, integration activity was analyzed after a second round of Nycodenz gradient purification. PICs reconstituted with either BAF, HMG I(Y), or RNase A displayed the same level of activity after this purification step as they did before the step (data not shown). Thus, the different factors appear to reconstitute equally stable complexes.
Because HMG I(Y) was detected in HIV-1 PICs isolated from infected cells (15), we tested whether mixing HMG I(Y) and BAF together might stimulate the level of functional reconstitution achieved by either protein alone. In this experiment, 50 ng of BAF and 25 μg of HMG I(Y) restored about one-third and one-fifth, respectively, of the level of activity that was present before salt-stripping (Fig. 5, lanes 1, 2, and 5). Regardless of their order of addition, reactions containing both proteins displayed less activity as compared with reactions containing either factor alone (Fig. 5, compare lanes 8–10 with lanes 2 and 5). Thus, BAF and HMG I(Y) do not appear to cooperate in HIV-1 integration under these conditions.
Figure 5.
BAF and HMG I(Y) inhibit each other’s activity. Reconstituted salt-stripped PICs were tested for integration activity. Lane 1, activity of the preparation before salt stripping (approximately 25% of substrate converted to product). Lane 2 contained 50 ng of BAF (about 8% of substrate converted to product). Lanes 3–5 contained 1, 5, and 25 μg of HMG I(Y), respectively. About 5% integration activity was recovered in lane 5. In lanes 6–8, 50 ng of BAF and 1, 5, and 25 μg of HMG I(Y), respectively, were added simultaneously to the reconstitution reactions. The approximate levels of activity in lanes 6–8 were 9, 5, and 3%, respectively. In lane 9, 50 ng of BAF was preincubated with the stripped PICs for 30 min, and then 25 μg of HMG I(Y) was added (about 2% activity recovered); lane 10, the order of addition in lane 9 was reversed (about 4% activity recovered). In lane 11, 50 ng of BAF was preincubated, followed by 5 μg of HMG I(Y) (about 8% activity recovered); lane 12, the order of addition in lane 11 was reversed (about 8% activity recovered). Other labeling is the same as in Fig. 1.
How might host factors function in HIV-1 integration? Since PICs disrupted with high salt contained functional integrase yet displayed the same MM-PCR footprinting pattern as naked DNA, BAF may localize integrase to the ends of the viral cDNA. Although BAF is a DNA-binding protein (17), it is unknown where on HIV-1 cDNA it may bind or whether BAF and integrase interact with each other. It is puzzling that PICs functionally reconstituted with either HMG I(Y) or RNase A did not display the native pattern of viral-end footprinting and enhancement. It therefore is possible that BAF plays an indirect role in forming the protein–DNA intasome detected by MM-PCR footprinting. Some integrase may remain bound to the cDNA ends after salt stripping, but this is not detected by MM-PCR because the bulk of the cDNA is protein-free and therefore is an efficient target for Mu insertion (S.-Q. Wei and R. Craigie, personal communication).
BAF apparently protects Mo-MLV from autointegration by compacting the viral cDNA (17). Although salt-treated HIV-1 PICs do not appear to autointegrate as efficiently as salt-stripped Mo-MLV complexes, intramolecular HIV-1 integration has been detected in vivo (20) and can be induced in vitro by treating PICs with nucleoside triphosphates (21). Thus, BAF may play identical roles in Mo-MLV and HIV-1 integration, both protecting cDNA from autointegration and promoting intermolecular recombination once a suitable DNA target has been located in the nucleus. In this sense, BAF may restore the native intasome to salt-stripped PICs more efficiently than other DNA-binding proteins because its main function is suppression of autointegration, and retroviral integration and Mu transposition are mechanistically very similar to each other (22). Since BAF appears to play specific roles in both Mo-MLV and HIV-1 integration, it is tempting to speculate that it may play a role in the replication of a large variety of retroviruses. Deciphering the precise modes of host-factor function in retroviral integration may provide novel strategies for blocking retroviral replication.
Acknowledgments
We thank M. Mizuuchi for purified Mu A protein, R. Zheng and R. Craigie for purified recombinant human BAF, S.-Q. Wei, R. Zheng, and R. Craigie for sharing results before publication, R. Reeves for plasmid pET7C, and D. Harris for carefully reading the manuscript. This work was supported by National Institutes of Health Grant AI39394, by funds from the G. Harold and Leila Y. Mathers Foundation, and by a gift from the Friends 10.
ABBREVIATIONS
- PIC
preintegration complex
- HIV-1
HIV type 1
- BAF
barrier-to-autointegration factor
- Mo-MLV
Moloney murine leukemia virus
- MM-PCR
Mu-mediated PCR
References
- 1.Brown P O, Bowerman B, Varmus H E, Bishop J M. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y M H, Coffin J M. J Virol. 1990;64:5958–5965. doi: 10.1128/jvi.64.12.5958-5965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellison V H, Abrams H, Roe T, Lifson J, Brown P O. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farnet C M, Haseltine W A. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara T, Mizuuchi K. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 6.Brown P O, Bowerman B, Varmus H E, Bishop J M. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown P O. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 161–203. [Google Scholar]
- 8.Craigie R, Fujiwara T, Bushman F. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 9.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 10.Bushman F D, Craigie R. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vora A C, McCord M, Fitzgerald M L, Inman R I, Grandgenett D P. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodzari G, Im G-J, Brackmann K, Grandgenett D P. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnet C M, Haseltine W A. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M S, Craigie R. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnet C M, Bushman F D. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Farnet C M, Anderson W F, Bushman F D. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee M S, Craigie R. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei S-Q, Mizuuchi K, Craigie R. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissen M S, Langan T A, Reeves R. J Biol Chem. 1991;266:19945–19952. [PubMed] [Google Scholar]
- 20.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnet C M, Haseltine W A. J Virol. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuuchi K. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]