Abstract
Germination and early seedling development are coordinately regulated by glucose and phytohormones such as ABA, GA, and ethylene. However, the molecules that affect plant responses to glucose and phytohormones remain to be fully elucidated. Eukaryotic release factor 1 (eRF1) is responsible for the recognition of the stop codons in mRNAs during protein synthesis. Accumulating evidence indicates that eRF1 functions in other processes in addition to translation termination. The physiological role of eRF1-2, a member of the eRF1 family, in Arabidopsis was examined here. The eRF1-2 gene was found to be specifically induced by glucose. Arabidopsis plants overexpressing eRF1-2 were hypersensitive to glucose during germination and early seedling development. Such hypersensitivity to glucose was accompanied by a dramatic reduction of the expression of glucose-regulated genes, chlorophyll a/b binding protein and plastocyanin. The hypersensitive response was not due to the enhanced accumulation of ABA. In addition, the eRF1-2 overexpressing plants showed increased sensitivity to paclobutrazol, an inhibitor of GA biosynthesis, and exogenous GA restored their normal growth. By contrast, the loss-of-function erf1-2 mutant exhibited resistance to paclobutrazol, suggesting that eRF1-2 may exert a negative effect on the GA signalling pathway. Collectively, these data provide evidence in support of a novel role of eRF1-2 in affecting glucose and phytohormone responses in modulating plant growth and development.
Keywords: Arabidopsis, eRF1-2, germination, gibberellin, glucose response
Introduction
Seed germination and early seedling development are the processes that are regulated by sugars and phytohormones such as ABA, GA, and ethylene (Peng and Harberd, 2002; Koornneef et al., 2002; Wang et al., 2002; Rolland et al., 2006). Glucose at high concentrations has been shown to delay seed germination and early seedling development including cotyledon expansion and greening (Dekkers et al., 2004; Rolland et al., 2006). This phenomenon has been commonly used to screen for mutants with altered glucose responses. Many signal transducers in the glucose sensing and signalling pathways have been discovered (Rolland et al., 2006). Interestingly, analysis of several sugar-resistant mutants revealed that they are allelic to the genes implicated in plant hormone responses or phenocopied by plant hormone biosynthesis or signalling mutants. For example, the Arabidopsis mutants gin1, sis4, and isi4 were found to be allelic to aba2; gin5 was allelic to aba3; gin6, sis5, and isi3 were allelic to abi4; and gin4 and sis1 were allelic to ctr1 (Zhou et al., 1998; Arenas-Huertero et al., 2000; Gibson et al., 2001). Furthermore, gibberellins (GAs) have been demonstrated to stimulate seed germination. Arabidopsis GA-deficient or GA-insensitive mutants fail to germinate normally (Koornneef and Veen, 1980; Steber et al., 1998). Factors influencing GA biosynthesis also affect seed germination (Toyomasu et al., 1998; Yamaguchi et al., 1998; Yamauchi et al., 2004). Despite the fact that the processes of germination and early seedling development are known to be regulated by sugars as well as ABA, GA, and other phytohormones (Gazzarrini and McCourt, 2001; Finkelstein and Gibson, 2002; Leon and Sheen, 2003), factors that affect sugar and phytohormone responses remain to be fully elucidated.
In eukaryotes, protein biosynthesis is terminated by the heterodimetric complex which is composed of two releasing factors, eukaryotic release factor 1 (eRF1) and eukaryotic release factor 3 (eRF3) (Frolova et al., 1994; Stansfield et al., 1995; Zhouravleva et al., 1995). Human eRF1 forms a crystal structure mimicking tRNA, with three domains resembling the anticodon loop, aminoacyl acceptor stem, and the T-stem of a tRNA molecule (Song et al., 2000). The structural similarity between eRF1 and a tRNA could account for the function of eRF1 in the process of translation termination, i.e. specific recognition of nonsense codons followed by the hydrolysis of a peptide–tRNA bond to release the completed polypeptide from the ribosome (Frolova et al., 1994).
The eRF1 homologues identified from various eukaryotes share a high degree of sequence and functional similarity, indicating conservation of translation termination throughout eukaryotes (Urbero et al., 1997; Karamyshev et al., 1999; Chapman and Brown, 2004). Although the biochemical activity of eRF1 proteins has been well established, several lines of evidence indicate that eRF1 may function in other processes in addition to translation termination. Mutation in the SUP45 (eRF1) gene affected the sensitivity of Saccharomyces cerevisiae to the microtubule poisoning drug benomyl and chromosome segregation at anaphase (Borchsenius et al., 2000). Repression of eRF1 caused the accumulation of unbudded yeast cells carrying 2C or even more DNA content, whereas repression of eRF3 caused different morphological changes, including enlarged cells with large buds, disappearance of the actin cytoskeleton, and defective mitosis, suggesting that the phenotypic changes caused by the down-regulation of eRF1 were not just the consequences of a disturbance of translation termination (Valouev et al., 2002). In Arabidopsis, cosuppression of eRF1-1 led to the broomhead phenotype with reduced internode elongation and altered cell division in fascicular cambial regions (Petsch et al., 2005). Collectively, these observations indicate that there are additional functional roles of eRF1 in the growth and development in eukaryotes.
By comparison with numerous studies on eRFs in other eukaryotes, there is only limited work on the functional characterization of a translation release factor in plants. Arabidopsis carries an eRF1 gene family with three members that fall into two subclasses: eRF1-1 and eRF1-2/eRF1-3. The eRF1-1 protein shares 86% and 85% sequence identity with eRF1-2 and eRF1-3, respectively, while eRF1-2 and eRF1-3 share up to 94% identity (Chapman and Brown, 2004). Although the involvement of eRF1-1 in cell elongation and radicle cell division has been documented (Petsch et al., 2005), the additional functions of this protein family in plant growth and development remain largely unknown.
In this study, the physiological role of eRF1-2, a member of the eRF1 gene family, in Arabidopsis was examined through genetic, physiological, and molecular analyses. The eRF1-2 gene encoded a protein localized in both the cytoplasm and the nucleus. It was expressed ubiquitously throughout plant development, but accumulated highly in young tissues, vascular tissues, root tips, and guard cells. Its transcript level was specifically stimulated by glucose. Transgenic Arabidopsis plants overexpressing eRF1-2 showed hypersensitivity to glucose with delayed germination and reduced expression of glucose-regulated genes. Such hypersensitivity appeared not to be due to an increased level of ABA accumulation. Overexpression and knock-out of eRF1-2 reversely regulated the plant responses to paclobutrazol (PAC), an inhibitor of GA biosynthesis. These findings suggest a novel role of eRF1-2 in affecting plant responses to glucose and GAs.
Materials and methods
Plant materials and growth conditions
Arabidopsis plants were grown either on MS plates containing Murashige and Skoog salts supplemented with 1% sucrose, or in soil in a growth chamber with a 16 h light period at 24 °C. The Arabidopsis thaliana SALK T-DNA insertion line of erf1-2 (SALK_150931) was obtained from the Arabidopsis Biological Resource Center (Columbus, Ohio). Screening of the homozygous T-DNA insertion line was performed by PCR with gene-specific primers SALK_150931LP: 5′TGGCTAACATCGTTATCTCCG-3′; SALK_150931RP: 5′ACAGTCTGCACTTCGTTTTGC-3′ and T-DNA border primer Lba1 (5′-TGGTTCACGTAGTGGGCCATCG-3′).
RNA extraction, reverse transcription, RT-PCR, quantitative RT-PCR
Total RNA from various tissues of Arabidopsis plants was extracted using the TRIzol reagent according to the manufacturer's instruction (Invitrogen). One microgram of total RNA was primed with oligo(dT) and reverse-transcribed into cDNA using Superscript III (Invitrogen). Gene-specific primers used for RT-PCR are as follows: ACTIN 8 (At1g49240), 5′-ATGAAGATTAAGGTCGTGGC-3′ (forward) and 5′-TCCGAGTTTGAAGAGGCTAC-3′ (reverse); ERF1-2 (At1g12920), 5′-ACAGTCTGCACTTCGTTTTGC-3′ (forward) and 5′TTAATCAGAATCTTCGTAAACTTC-3′ (reverse); and green fluorescent protein (AB434768), 5′-ATGGGCACAAATTTTCTGTCAG-3′ (forward) and 5′-AGGACCATGTGGTCTCTCTCTTTTCGT-3′ (reverse).
Quantitative RT-PCR was performed using the SYBR Green Mastermix (Applied Biosystems) with primers as listed in Table 1 and in Supplementary Table S1 at JXB online. Expression of all genes was assayed in triplicate. The PCR program was 50 °C for 10 min and 95 °C for 2 min followed by 40 cycles of denaturation for 15 s at 95 °C and annealing/extension at 60 °C for 1 min.
Table 1.
Primers used for qRT-PCR analysis of glucose-induced and GA biosynthetic and signalling genes
| Gene | Sequences | AGI number |
| Actin 8 F | TGCAGACCGTATGAGCAAAG | At1g49240 |
| Actin 8 R | CTGGAAAGTGCTGAGGGAAG | |
| ERF1-2 F | GTCTGATACAAGCAACTTCCA | At1g12920 |
| ERF1-2 R | TTAATCAGAATCTTCGTAAACTTC | |
| CAB1F | CACTGGTAAGGGACCGATAGAG | At1g29930 |
| CAB1R | ACACTCACGAAGCAAAGACTGA | |
| PCF | CCGTCAGCTCAAAACCTAAGAC | At1g76100 |
| PCR | GACACCGAAATCCTTCAAAGAG | |
| CPSF | GATCGATGCCGGAGATAAAA | At4g02780 |
| CPSR | GATACGCATCTCCCCAAGAA | |
| KSF | CAATCGCAGCAAAGAAGTGA | At1g79460 |
| KSR | TCTTTGCATTCCCTTGGAAC | |
| KO1F | GCTAGGGACCATCCACAAGA | At5g25900 |
| KO1R | TCCACATCTTTCCCAAAAGC | |
| AtGA3ox1-F | ACGTTGGTGACCTCTTCCAC | At1g15550 |
| AtGA3ox1-R | CCCCAAAGGAATGCTACAGA | |
| AtGA3ox2-F | GGCGTAGCTCGTATTGCTTC | At1g80340 |
| AtGA3ox2-R | GGAGAGCCAATAACGGTGAA | |
| AtGAIF | ACTCGTTGGAAGGTGTACCG | AT1G14920 |
| AtGAIR | CTCAACTCGGTCAGGTCCAT | |
| AtGID1AF | AAGAAAGCGGGTCAAGAGGT | AT3G05120 |
| AtGID1AR | ACAAACGCCGAAATCTCATC | |
| AtSLY1F | ACGTCGACGCAAAGACCTTA | AT4G24210 |
| AtSLY1R | GCAGCCGATGTTAGTCCAGT |
Immunoblot analysis
Total proteins from rosette leaves of 4-week-old Arabidopsis plants were extracted using CelLytic P Plant Cell Lysis/Extraction Reagent (Sigma). Proteins (10 μg) were resolved in a 10% SDS-PAGE and analysed by Western blot with anti-GFP antibody (sc-8334, Santa Cruz Biotech). Signals were detected using the ECL Plus Western blotting detection system (Amersham) and visualized by a STORM 860 PhosphorImager (Amersham).
Sugar treatment
Ten-day-old Arabidopsis seedlings of the wild type (WT) grown on MS plates were immersed in solutions of 6% glucose, 6% mannitol or water, respectively, and gently shaken at room temperature in the dark. Seedlings were collected at different time points, immediately frozen in liquid nitrogen, and stored at –80 °C until use.
Glucose, ABA, and paclobutrazol response assay
Arabidopsis seeds harvested at the same time were dried at room temperature for over 2 weeks. Seeds were surface-sterilized with 30% bleach for 10 min and washed five times with sterile water. The glucose response assay was carried out following the method as described previously by Arenas-Huertero et al. (2000).
For the ABA response assay, the surface-sterilized seeds were placed on MS plates with 0, 0.3, 0.6, and 1 μM ABA, respectively. Seeds were stratificated at 4 °C for 48 h and germinated under a 16/8 h light/dark photoperiod at 24 °C. Germination, scored by radicle emergence from the seed coat, was recorded.
For the PAC response assay, the surface-sterilized seeds were placed on MS plates with 0, 1, 2, 3, 4, 5, and 10 μM PAC, respectively.
Plasmid construction and plant transformation
For endogenous gene expression analysis, a 483 bp promoter sequence of the eRF1-2 gene, which is the DNA sequence between eRF1-2 and the upstream gene, was amplified from the Arabidopsis genome using the primers of 5′-AACTGCAGGAGGCAAACCAATAAACGGC-3′ (forward) and 5′-AACCATGGTTTTAGCTTCTCTCGAGGTC-3′ (reverse) with added PstI and NcoI sites, respectively (underlined). The amplified PCR product was fused to the GUS gene in pSG565 vector (Gan and Amasino, 1995). The entire cassette was then subcloned into the binary vector pCAMBIA1300 (CAMBIA) between the BamHI and PstI sites to make the ProeRF1-2:GUS construct.
For overexpressing the eRF1-2 gene, the full-length eRF1-2 cDNA was amplified using primers eRF1-2F 5′-AACTCGAGATGGCAGAAGAAGCGGATAC-3′ (forward, XhoI underlined) and eRF1-2R 5′-AACCATGGGATCAGAATCTTCGTAAACTTCAG-3′ (reverse, NcoI underlined). The amplified product was fused into a GFP coding sequence in pAVA395 via XhoI and NcoI digestion (von Arnim et al., 1998), and then subcloned into pCAMBIA1300S between the KpnI and BamHI sites (Xiong and Yang, 2003) to yield the 35S:eRF1-2:GFP construct.
The constructs and empty vectors were electroporated into Agrobacterium tumefaciens strain GV3101 and introduced into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS media containing 50 mg ml−1 hygromycin.
Confocal microscopy analysis and GUS histochemical staining
GFP fluorescence in 3-d-old stable transgenic Arabidopsis plants overexpressing the eRF1-2-GFP fusion protein was observed with a Leica TCS SP5 confocal microscope as described previously (Lu et al., 2006). Histochemical GUS activity was examined as described by Jefferson et al. (1987). Three transgenic lines with more than five plants each were used.
ABA extraction and determination
ABA extraction was performed as described by Artsaenko et al. (1995). In brief, 50 mg of 10-d-old Arabidopsis seedlings were homogenized in 1 ml of 80% acetone, shaken continuously at 4 °C for 24 h, and centrifuged. The supernatant was diluted 10-fold in TRIS-buffered saline. The ABA content was determined with a Phytodeteck ABA kit following the manufacturer's instruction (Agdia).
Results
Structure and expression patterns of Arabidopsis eRF1-2
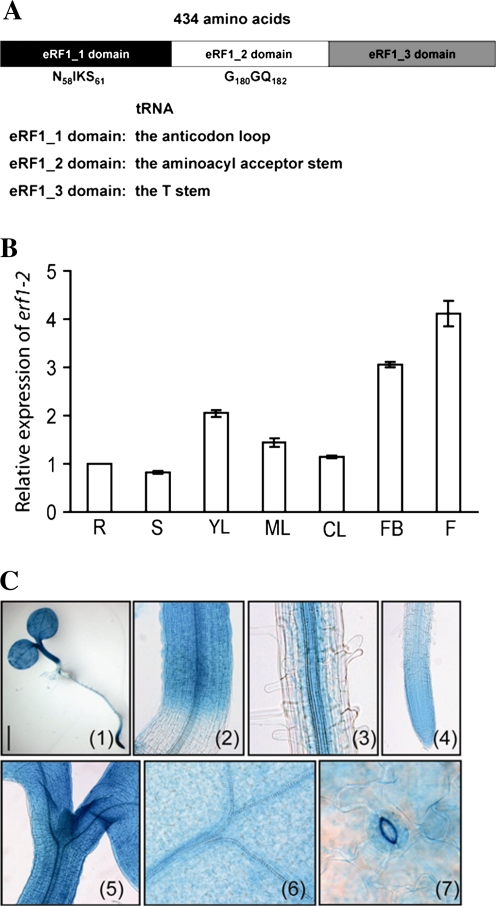
The predicted eRF1-2 protein of Arabidopsis contains 434 amino acid residues with a deduced molecular mass of 48.9 kDa. Arabidopsis eRF1-2 shares 73% amino acid sequence identity with human eRF1 and carries three conserved domains, which correspond to the anticodon loop, the aminoacyl acceptor stem, and the T stem of tRNA, respectively (Fig. 1A). The NIKS and GGQ motifs, which are shown to be responsible for interaction with the ribosome and hydrolysis of peptidyl-tRNA (Frolova et al., 1999, 2002), are located between amino acids 58 and 61, and amino acids 180 and 182, respectively (Fig. 1A).
Fig. 1.
Structure and expression patterns of Arabidopsis eRF1-2. (A) Structure analysis of the eRF1-2 protein. The essential NIKS and GGQ motifs in the anticodon loop and the aminoacyl acceptor stem of eRF1-2 are indicated. (B) qRT-PCR analysis of the expression pattern of eRF1-2. R, roots; S, stems; YL, 28-d-old rosette leaves; ML, 40-d-old rosette leaves; CL, cauline leaves; FB, flower buds; F, open flowers. (C) Localization of GUS activity in 3-d-old transgenic Arabidopsis seedlings expressing ProeRF1-2:GUS. GUS staining of 3-d-old seedling (1), hypocotyl (2), root elongation zone (3), root tip (4), shoot meristem tissue (5), cotyledon (6), and guard cell (7). Bar=1 mm. (This figure is available in colour at JXB online.)
To investigate the physiological role of eRF1-2 in plants, its expression pattern was examined. Quantitative RT-PCR analysis revealed that eRF1-2 was expressed ubiquitously in various tissues of the plants under normal growth conditions (Fig. 1B). Higher levels of transcript were found in young leaves, flower buds, and flowers (Fig. 1B). To examine the expression pattern of eRF1-2 further, the promoter sequence of eRF1-2 was fused to the β-D-glucuronidase (GUS) gene and transformed into Arabidopsis plants. Thirteen independent stable transgenic lines were generated and analysed. High GUS activity was observed in very young leaves and flowers, whereas relatively low expression was found in mature or senescent tissues (see Supplementary Fig. S1 at JXB online). The overall GUS expression pattern in these transgenic plants recapitulated the eRF1-2 transcript levels in plants. A detailed examination of the GUS activity staining of 3-d-old transgenic seedlings revealed that eRF1-2 was expressed highly in root tips, vascular bundles of roots and hypocotyls, root and shoot meristem, veins of cotyledons, and guard cells (Fig. 1C). The vascular and guard cell-preferred expression of the gene indicates its involvement in the vascular and guard cell-specific processes (Wenzel et al., 2008).
eRF1-2 was localized in the cytoplasm and nucleus, and specifically induced by glucose
The eRF1-2 protein contains no obvious predicted localization signal sequence. To experimentally determine the subcellular localization of eRF1-2 in plants, the eRF1-2:GFP chimeric gene under the control of the CaMV 35S promoter was introduced into Arabidopsis plants. Fifteen independent transformants were obtained and two homozygous lines (OV1-4 and OV13-11) of T3 plants were selected. RT-PCR analysis showed that eRF1-2 was highly expressed in these two overexpressing lines, which were further verified by PCR using GFP specific primers (Fig. 2A). Immunoblot analysis with the GFP antibody revealed that, the eRF1-2 transgenic lines contained a specific band close to the expected molecular weight of the fusion protein at approximately 85 kDa and an extra band at higher molecular weight (Fig. 2B), confirming that eRF1-2 was expressed in the transgenic Arabidopsis.
Fig. 2.
Analysis of eRF1-2:GFP transgenic lines, subcellular localization of eRF1-2, and qRT-PCR analysis of the induction of eRF1-2 expression by sugar. (A) RT-PCR analysis revealed that eRF1-2 and GFP were highly expressed in the homozygous OV1-4 and OV13-11 overexpressing lines in comparison with WT plants (Col). (B) Immunoblot analysis of the eRF1-2 overexpressing plants. Ten microgram of proteins extracted from leaf tissues were separated by SDS-PAGE and immunoblotted with a GFP antibody. Arrow points to the fusion protein at approximately 85 kDa. Molecular sizes are indicated on the left. (C) Subcellular localization of eRF1-2 protein. GFP signal in the root elongation zone of a 3-d-old transgenic seedling expressing 35S:eRF1-2:GFP (1) and 35S:GFP (2). GFP signal in the guard cells of a 3-d-old transgenic seedling expressing 35S:eRF1-2:GFP (3) and 35S:GFP (4). (D) Expression of eRF1-2 was specifically induced by glucose. Arabidopsis seedlings (10-d-old) of WT plants were treated with 6% glucose, 6% mannitol, and water for 0, 2, 4, 6, 8, and 24 h. (This figure is available in colour at JXB online.)
These two transgenic lines along with vector-only control lines were examined by a confocal laser-scanning microscope. In the cells of the root elongation region, a green fluorescent signal was observed in the cytoplasm between the cell membrane and the central vacuole as well as in the nucleus (Fig. 2C, 1). Similarly, an intense GFP signal could be detected in the nucleus and cytoplasm of the guard cells in cotyledon tissue (Fig. 2C, 3). These results clearly suggest that eRF1-2 was expressed in both the cytoplasm and the nucleus. A stronger GFP signal was observed in every subcellular localization in the control transgenic Arabidopsis expressing CaMV 35S:GFP (Fig. 2C, 2, 4).
Protein synthesis is tightly regulated and requires cellular resources (Kallmeyer et al., 2006). To see whether eRF1-2, as a component of the protein synthesis machinery, is regulated by sugar, its expression in response to glucose and mannitol treatment in WT Arabidopsis was examined. Quantitative RT-PCR analysis showed that the eRF1-2 transcript abundance gradually increased with time and reached a level of over 2-fold at 24 h after treatment with 6% glucose, a glucose concentration commonly used to study glucose response (Zhou et al., 1998; Arenas-Huertero et al., 2000). No increased eRF1-2 mRNA levels could be observed following exposure of the plant to 6% mannitol or water (Fig. 2D). The results indicate that the expression of eRF1-2 was specifically induced by glucose.
Transgenic Arabidopsis plants overexpressing eRF1-2 conferred hypersensitivity to glucose
Germination and early seedling development are known to be modulated by sugar signals (Rolland et al., 2006). To test whether eRF1-2 affects the Arabidopsis plant responses to glucose, germination and early seedling growth of the OV1-4 and OV13-11 overexpressing lines, an erf1-2 mutant, and wild-type plants were examined by placing the seeds on MS plates containing various concentrations of glucose. The erf1-2 mutant had a T-DNA insertion site at 897 bp of the open reading frame and did not contain a detectable amount of endogenous eRF1-2 transcript (see Supplementary Fig. S2A, B at JXB online). Southern blot analysis of the transgenic plant showed that this line carried a single copy of T-DNA (see Supplementary Fig. S2C at JXB online).
As shown in Fig. 3A, there was no difference in seed germination and early seedling development when the plants were grown in the medium containing no glucose. However, the germination rate of the overexpressing lines was reduced in the presence of 5% and 6% glucose compared with the wild-type plants (Fig. 3A). At 6% glucose, the eRF1-2 overexpressing lines had 25% seed germination, whereas wild-type seeds germinated 65% (Fig. 3B). The primary root length of these transgenic plants was shorter than that of the wild type when the medium contained more than 3% glucose (Fig. 3C). These results show that transgenic plants overexpressing eRF1-2 resulted in hypersensitivity to glucose. No significant difference between wild type and the erf1-2 mutant in response to glucose treatment was observed; indicating that knock-out of the single gene was not enough to affect the glucose response, possibly because of the functional redundancy among the three eRF1 family members. Double knockout lines of eRF1-2 and eRF1-3 were generated. Examination of their response to glucose treatment showed that they exhibited the same response as the eRF1-2 single mutant (data not shown). Extensive efforts were made to generate triple knockout lines but without success. This may be due to the essential role of eRF1 proteins in protein synthesis termination. Similarly, homozygous eRF1-1 cosuppression lines could not be obtained (Petsch et al., 2005).
Fig. 3.
Transgenic Arabidopsis plants overexpressing eRF1-2 were hypersensitive to glucose during germination and early seedling development. (A) Effect of glucose on the growth phenotype of plants with altered levels of eRF1-2 expression. Six-day-old seedlings grown on MS plates containing 0%, 5%, and 6% glucose, respectively. Approximately 30 seeds from each line were used in the assay. The experiments were repeated three times. (B, C) Germination rate and primary root length of Arabidopsis plants after 6 d of incubation at 24 °C. The data represent an average of 30 plants +SD. Asterisks indicate significantly different means (P <0.05). (D, E) qRT-PCR analysis of expression of CAB1 and PC in 6-d-old Arabidopsis seedlings. The open bar and black bar stand for the 0% and the 6% glucose treatment, respectively. (This figure is available in colour at JXB online.)
To examine the involvement of eRF1-2 in the glucose response further, the expression of several glucose-regulated genes was examined in the eRF1-2 overexpressing plants and the erf1-2 mutant by qRT-PCR. Among them, chlorophyll a/b-binding protein (CAB1) and plastocyanin (PC) showed differential expression. In the absence of glucose, CAB1 and PC were expressed similarly among the plants with different level of eRF1-2 expression (Fig. 3D, E). In the presence of 6% glucose, however, the transcript levels of these two genes were dramatically reduced in the overexpressing lines compared with the wild type and the erf1-2 mutant (Fig. 3D, E). CAB1 and PC are nuclear-encoded photosynthesis genes and regulated by glucose via the HXK-dependent glucose signalling pathway (Sheen et al., 1999). These results suggest that eRF1-2 might moderate the glucose response by affecting glucose signalling components during germination and early seedling development.
Overexpression of eRF1-2 led to increased sensitivity to ABA
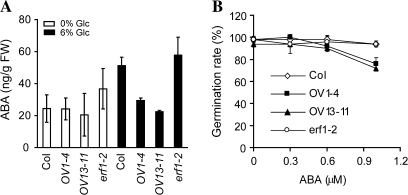
Glucose can increase cellular ABA concentration by either increasing ABA synthesis or inhibiting degradation during germination (Arenas-Huertero et al., 2000; Cheng et al., 2002; Price et al., 2003). To test whether the hypersensitivity of the eRF1-2 overexpressing plants to glucose was attributable to the excessive accumulation of ABA in the cells, the ABA levels were examined in the 6-d-old transgenic seedlings grown on MS plates containing no glucose and 6% glucose, respectively. No significant difference in the ABA levels could be seen among WT, the two overexpressing lines, and the erf1-2 mutant in the absence of glucose (Fig. 4A). In the presence of 6% glucose, the ABA levels in the WT plants were increased as reported previously (Cheng et al., 2002). Interestingly, while the erf1-2 mutant showed a slightly enhanced level of ABA accumulation following glucose treatment, the eRF1-2 overexpressing lines contained less ABA than the WT (Fig. 4A), indicating that the hypersensitivity of the eRF1-2 overexpressing lines to glucose was not caused by excessive accumulation of cellular ABA in these plants.
Fig. 4.
ABA responses of Arabidopsis plants with altered expression of eRF1-2. (A) ABA levels in 10-d-old Arabidopsis seedlings grown on MS and MS with 6% glucose. Fifty milligrams of seedlings were used in ABA extraction. (B) Germination rate of a 6-d-old seedling grown on MS media containing 0, 0.3, 0.6, and 1 μM ABA. Emergence of the radicle is ascribed to germination. The data represent the average of two independent experiments. Bars indicate standard deviation.
To examine whether the hypersensitivity to glucose was due to altered sensitivity to ABA, the response of these Arabidopsis plants to ABA treatment were tested. Seeds were grown on MS plates containing various concentrations of ABA. The rate of germination, as indicated by radicle emergence from the seed coats, was measured. On the ABA-free medium, the eRF1-2 overexpressing lines and the erf1-2 mutant exhibited similar germination rates as the WT plants (Fig. 4B). With increased levels of ABA, the eRF1-2 overexpressing lines showed hypersensitivity to ABA. Whereas the germination rate of the erf1-2 mutant like the WT was not affected by 1 μM ABA, the germination rate of eRF1-2 overexpressing lines was reduced to less than 80% at 1 μM ABA (Fig. 4B).
Expression of ABA biosynthetic genes, such as ABA1/ZEP1 (At5g67030), ABA2 (At1g52340), ABA3 (At1g16540), AAO3 (At2g27150), and NCED3 (At3g14440) as well as a catabolic gene CYP707A2 (Okamoto et al., 2006) was examined in the WT, eRF1-2 overexpressing lines, and the erf1-2 mutant treated without or with 6% glucose. Interestingly, no significant difference in their expression levels was observed in spite of reduced levels of ABA in the eRF1-2 overexpressing lines (see Supplementary Fig. S3 at JXB online).
Transgenic Arabidopsis overexpressing eRF1-2 showed increased sensitivity to PAC and the erf1-2 mutant exhibited resistance to PAC
GA has been known to control seed germination and early seedling development. To explore whether eRF1-2 also affected plant responses to GA, the responses of these Arabidopsis plants to PAC, an inhibitor of GA biosynthesis, were examined. All the plants germinated equally well on medium containing no PAC (Fig. 5A, left image). When sown on MS plates supplemented with PAC, the overexpressing lines showed an increased sensitivity to PAC, while the erf1-2 mutant exhibited a clearly enhanced resistance to PAC (Fig. 5A, middle image). The two transgenic overexpressing lines OV1-4 and OV13-11 were unable to germinate in the presence of 3 μM or more PAC (Fig. 5B). At a concentration of 3 μM PAC, the green cotyledon rates for the erf1-2 and wild-type plants were 75% and 40%, respectively (Fig. 5B). When 3 μM PAC and 1 μM GA3 were applied together, the green cotyledon percentage of the eRF1-2 overexpressing plants and the wild type reached a similar level as that of the erf1-2 mutant (Fig. 5C), indicating that inhibition of germination in the overexpressing lines and the wild type by PAC could be overcome by exogenous application of GA. Resistance to PAC could result from ABA-deficiency or GA-hypersensitivity in some cases (Jacobsen and Olszewski, 1993; Léon-Kloosterziel et al., 1996). The possibility of ABA-deficiency was ruled out because no reduction in the ABA level of the erf1-2 mutant was observed.
Fig. 5.
Responses of eRF1-2 transgenic Arabidopsis to paclobutrazol (PAC). (A) Six-day-old seedlings grown on MS plates containing 0 and 3 μM PAC, as well as 3 μM PAC with 1 μM GA3. (B) Percentage of seedlings with green cotyledons at 6 d after treatment. (C) Percentage of seedlings with green cotyledon grown for 6 d on MS plates containing 3 μM PAC and 1 μM GA3. (D) Expression of GA biosynthetic and signalling genes in 10-d-old seedlings grown on the MS plates. CPS, ent-copalyl diphosphate synthase; KO, ent-kaurene oxidase; KS, ent-kaurene synthase; AtGA3ox, GA3-oxidase; AtGAI (GA INSENSITIVE); AtGID1C, (GA INSENSITIVE DWARF1C); and AtSLY1 (SLEEPY1). The data represent the average of two biological repeats. (This figure is available in colour at JXB online.)
To investigate the involvement of eRF1-2 in the GA response, expression of GA biosynthetic and signalling component genes in 6-d-old seedlings grown on MS plates was examined by qRT-PCR using gene-specific primers (Table 1). Among these genes, AtGA3ox2 had a higher expression in the erf1-2 mutant with over 4-fold higher than that in wild-type plants. Interestingly, AtGA3ox2 was expressed with a 2-fold increase in the eRF1-2 overexpressing plants than in the wild type (Fig. 5D), perhaps due to feedback regulation under GA deficiency condition or other unknown mechanisms. The expression of the other GA biosynthetic and GA signalling genes did not significantly differ from that in the wild-type seedlings. These results collectively suggest that eRF1-2 affects the GA response probably via regulating the expression of AtGA3ox2 during germination.
Discussion
Arabidopsis eRF1-2 has been demonstrated to possess protein translation termination activity (Chapman and Brown, 2004). The ribosome is assembled in the nucleus and functions in the cytoplasm, and eRF1 along with eRF3 bind the ribosome (Mitkevich et al., 2006). The subcellular localization of eRF1-2 at both the nucleus and cytoplasm in Arabidopsis plants is consistent with its role in protein translation termination.
In addition to the important role as essential components of the protein synthesis machinery, eRF1 proteins have been shown to exert additional functional roles in growth and development in eukaryotes (Valouev et al., 2002). Several lines of evidence provided here showed that eRF1-2 affected plant responses to glucose in Arabidopsis. First, the eRF1-2 overexpressing lines showed delayed germination and arrested early seedling development in response to 5% or more glucose. The inhibitory effect of sugar on germination and early seedling development is a characteristics of many sugar responsive genes. It has been commonly used to identify the sugar responsive components, such as GIN1/ABA2, GIN2/HXK1, GIN4/SIS1/CTR1, GIN5/ABA3/LOS5, and GIN6/SIS5/ABI4 (Zhou et al., 1998; Arenas-Huertero et al., 2000; Gibson et al., 2001; Moore et al., 2003). Second, it is well known that an increased level of glucose represses photosynthetic gene expression (Rolland and Sheen, 2005). CAB1 and PC are nuclear-encoded photosynthetic genes that are regulated by glucose via the HXK-dependent pathway (Zhou et al., 1998). CAB1 and PC mRNA levels were significantly reduced in the eRF1-2 overexpressing plants compared with wild-type plants. Collectively, the enhanced sensitivity of the eRF1-2 overexpressing transgenic plants to glucose and reduced expression of the glucose-repressed genes indicate that eRF1-2 can affect the glucose response.
ABA plays an important role in plant vegetative development regulated by sugars. A number of sugar-resistant mutants were found to be allelic to the genes involved in either ABA biosynthesis or signalling (Arenas-Huertero et al., 2000). The ABA levels of the eRF1-2 overexpressing lines were found to be lower than that of WT when treated with 6% glucose. The low level could be due to enhanced activity of ABA catabolic enzymes despite the fact that one such gene CYP707A2 showed no differential expression among plants with altered eRF1-2 expression. The low ABA levels in the eRF1-2 overexpressing lines rule out the possibility that hypersensitivity of the overexpressing plants to glucose was caused by the enhanced accumulation of ABA in these transgenic plants. Examination of the ABA response during germination revealed that the eRF1-2 overexpressing plants were hypersensitive to ABA, establishing a possible involvement of ABA in the hypersensitivity of the eRF1-2 overexpressing plants to glucose.
eRF1-2 also appears to affect the plant responses to GA during plant growth and development. There is a possibility that eRF1-2 affects the GA response through regulating the expression of AtGA3ox2. GAs are derived from geranylgeranyl diphosphate via a series reactions catalysed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS), GA 20-oxidase (AtGA20ox), and GA 3-oxidase (AtGA3ox) in higher plants (Olszewski et al., 2002). AtGA3ox catalyses the conversion of precursors to bioactive GAs, which is the rate-limiting step in GA biosynthesis (Seo et al., 2006). Of four Arabidopsis GA3ox family members, AtGA3ox1 and AtGA3ox2 have been demonstrated to be responsible for GA synthesis during vegetative growth (Mitchum et al., 2006). In this study, higher expression of AtGA3ox2 was observed in the erf1-2 mutant. Although the AtGA3ox2 transcript level was also higher in the eRF1-2 overexpressing line than wild-type plants, such a higher level could be due to the feedback regulation of the gene expression by GA-deficiency as shown in other cases (Dai et al., 2007).
Gibberellins regulate plant growth and development, including seed germination, internode elongation, and flowering (Yamaguchi, 2008). Previously, SPINDLY (SPY) has been identified as a negative regulator of GA signalling. While the spy mutants are able to germinate in the presence of PAC (Jacobsen and Olszewski, 1993), overexpression of SPY in Arabidopsis produces enhanced sensitivity to PAC (Swain et al., 2001). Similarly, the erf1-2 mutant exhibited enhanced resistance to PAC during seed germination and early seedling development, and overexpression of eRF1-2 caused an enhanced sensitivity to PAC, suggesting that eRF1-2 might exert a negative role in the GA responsive pathway. Although PAC-resistant germination is a characteristic typical of ABA-deficient mutants (Léon-Kloosterziel et al., 1996), the altered GA sensitivity observed in the erf1-2 mutant appears not to be associated with a deficiency in ABA biosynthesis. The increased resistance of the erf1-2 mutant might be due to increased levels of GA as indicated by higher expression of GA3ox2, or hypersensitivity to GA. In addition, as demonstrated in the spy mutants and overexpressing lines (Jacobsen and Olszewski, 1993; Swain et al., 2001), alteration of eRF1-2 expression also appeared to change the plant flowering time. Transgenic plants overexpressing the eRF1-2 were observed to have a slightly delayed flowering time while the erf1-2 mutant flowered earlier (data not shown), which is consistent with the involvement of eRF1-2 in the GA response.
Recent studies have revealed an interesting link between sugar and phytohormone in regulating plant growth and development (Gazzarrini and McCourt, 2001; Finkelstein et al., 2002; Leon and Sheen, 2003). There is a possibility that high sugar may affect mRNA stability or protein translation of some ABA or GA catabolic proteins. Overexpression/knockout of eRF1-2 may affect these processes. Future experiments will test out these hypotheses.
As an essential component of the protein synthesis machinery, it is intriguing how eRF1-2 mediates the glucose and GA responses. One possibility is that eRF1-2 may confer its function by regulating mRNA metabolism. In eukaryotes, release factors eRF1 and eRF3 are involved in the nonsense-mediated mRNA decay (Czaplinski et al., 1998). The nonsense-mediated mRNA decay has diverse roles in the regulation of gene expression (Green et al., 2003; Singh and Lykke-Andersen, 2003), which might specifically regulate genes involved in glucose and GA metabolism. The other possibility is that eRF1-2 may exert its role through interacting with various proteins of distinct biological functions. eRF1 is known to be associated with a number of proteins in participating different cellular processes, and the eRF1 binding partners determine eRF1 function (Valouev et al., 2004). For example, when eRF1 binds to eRF3, it acts as a translation termination factor. When it interacts with the myosin light chain (Mlc1p) in yeast, it affects cytokinesis (Valouev et al., 2004). In Drosophila, eRF1 was shown to interact with DnaJ-1 and Sap47 in a two-hybrid-based protein-protein-interaction network (Giot et al., 2003). The other eRF1 interacting proteins include protein phosphatase 2A, UPF1, Itt1p, and Mtt1 in yeast and human (Andjelkovic et al., 1996; Czaplinski et al., 2000; Urakov et al., 2001). Of these eRF1-interacting proteins, the loss-of-function mutant upf1 (lba1) causes hypersensitivity to glucose and early flowering in Arabidopsis (Mita et al., 1997; Yoine et al., 2006). The UPF1 has been suggested to modulate sugar signalling via altering the expression of an unknown target gene that affects the expression of a subset of sugar-induced gene (Yoine et al., 2006).
In conclusion, our findings showed that eRF1-2 can affect glucose and phytohomone responses in addition to its activity in protein synthesis termination. The fact that the erf1-2 mutant exhibited a clearly enhanced resistance to PAC but was not enough to interfere with the glucose response, possibly due to functional redundancy among the three eRF1 family proteins, suggests that eRF1-2 may exert a stronger functional role in affecting the GA response. Further investigation of the molecular mode of action of eRF1-2 will facilitate a better understanding of its precise role in regulating plant growth and development.
Supplementary data
The following supplementary materials are available at JXB online:
Supplementary Fig. S1. Expression pattern of GUS driven by the AteRF1-2 promoter in Arabidopsis.
Supplementary Fig. S2. Analysis of the erf1-2 mutant.
Supplementary Fig. S3. Expression of ABA biosynthetic and catabolic genes in wild-type and AteRF1-2 transgenic plants.
Supplementary Table S1. Primers used for qRT-PCR analysis of ABA biosynthetic and catabolic genes.
Supplementary Material
Acknowledgments
We are grateful to Dr Yinong Yang for providing the pCAMBIA1300S vector and Dr Susheng Gan for the pSG565 vector. We also thank Randy Clark for technical assistance with photography, and Drs Lu Shan, Xin Zhou, and Youxi Yuan for helpful discussions. This work was supported by USDA National Research Initiative Competitive Grant 2007-35318-17794.
References
- Andjelkovic N, Zolnierowicz S, Van HC, Goris J, Hemmings BA. The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. The EMBO Journal. 1996;15:7156–7167. [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes and Development. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Artsaenko O, Peisker M, Zur NU, Fiedler U, Weiler EW, Muntz K, Conrad U. Expression of a single-chain Fv antibody against abscisic acid creates a wilty phenotype in transgenic tobacco. The Plant Journal. 1995;8:745–750. doi: 10.1046/j.1365-313x.1995.08050745.x. [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Tchourikova AA, Inge-Vechtomov SG. Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Current Genetics. 2000;37:285–291. doi: 10.1007/s002940050529. [DOI] [PubMed] [Google Scholar]
- Chapman B, Brown C. Translation termination in Arabidopsis thaliana: characterisation of three versions of release factor 1. Gene. 2004;341:219–225. doi: 10.1016/j.gene.2004.06.053. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signalling and abscisic acid biosynthesis and functions. The Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Majlesi N, Banerjee T, Peltz SW. Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA. 2000;6:730–743. doi: 10.1017/s1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes and Development. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiology. 2007;144:121–133. doi: 10.1104/pp.107.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC. Glucose delays seed germination in Arabidopsis thaliana. Planta. 2004;218:579–588. doi: 10.1007/s00425-003-1154-9. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signalling in seeds and seedlings. The Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI. ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Current Opinion in Plant Biology. 2002;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, Justesen L, Kisselev L. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signalling pathways. Current Opinion in Plant Biology. 2001;4:387–391. doi: 10.1016/s1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochemical and Biophysical Research Communications. 2001;280:196–203. doi: 10.1006/bbrc.2000.4062. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, Rio DC, Brenner SE. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19:i118–i121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. The Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev AL, Ito K, Nakamura Y. Polypeptide release factor eRF1 from Tetrahymena thermophila: cDNA cloning, purification and complex formation with yeast eRF3. FEBS Letters. 1999;457:483–488. doi: 10.1016/s0014-5793(99)01089-3. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theoretical and Applied Genetics. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Current Opinion in Plant Biology. 2002;5:33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- Leon P, Sheen J. Sugar and hormone connections. Trends in Plant Science. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. The Plant Cell. 2006;18:3594–3605. doi: 10.1105/tpc.106.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K. Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. The Plant Journal. 1997;11:841–851. doi: 10.1046/j.1365-313x.1997.11040841.x. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. The Plant Journal. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Research. 2006;34:3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiology. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. Gibberellin signalling: biosynthesis, catabolism, and response pathways. The Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP. The role of GA-mediated signalling in the control of seed germination. Current Opinion in Plant Biology. 2002;5:376–381. doi: 10.1016/s1369-5266(02)00279-0. [DOI] [PubMed] [Google Scholar]
- Petsch KA, Mylne J, Botella JR. Cosuppression of eukaryotic release factor 1-1 in Arabidopsis affects cell elongation and radial cell division. Plant Physiology. 2005;139:115–126. doi: 10.1104/pp.105.062695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Li TC, Kang SG, Na JK, Jang JC. Mechanisms of glucose signalling during germination of. Arabidopsis. Plant Physiology. 2003;132:1424–1438. doi: 10.1104/pp.103.020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signalling in plants: conserved and novel mechanisms. Annual Review of Plant Physiology. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Rolland F, Sheen J. Sugar sensing and signalling networks in plants. Biochemical Society Transactions. 2005;33:269–271. doi: 10.1042/BST0330269. [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. The Plant Journal. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC. Sugars as signalling molecules. Current Opinion in Plant Biology. 1999;2:410–418. doi: 10.1016/s1369-5266(99)00014-x. [DOI] [PubMed] [Google Scholar]
- Singh G, Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends in Biochemical Sciences. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1: mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. The EMBO Journal. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Olszewski NE. Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiology. 2001;126:1174–1185. doi: 10.1104/pp.126.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiology. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakov VN, Valouev IA, Lewitin EI, Paushkin SV, Kosorukov VS, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Itt1p, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Molecular Biology. 2001;2:9. doi: 10.1186/1471-2199-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbero B, Eurwilaichitr L, Stansfield I, Tassan JP, Le Goff X, Kress M, Tuite MF. Expression of the release factor eRF1 (Sup45p) gene of higher eukaryotes in yeast and mammalian tissues. Biochimie. 1997;79:27–36. doi: 10.1016/s0300-9084(97)87622-5. [DOI] [PubMed] [Google Scholar]
- Valouev IA, Kushnirov VV, Ter-Avanesyan MD. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motility and the Cytoskeleton. 2002;52:161–173. doi: 10.1002/cm.10040. [DOI] [PubMed] [Google Scholar]
- Valouev IA, Urakov VN, Kochneva-Pervukhova NV, Smirnov VN, Ter-Avanesyan MD. Translation termination factors function outside of translation: yeast eRF1 interacts with myosin light chain, Mlc1p, to effect cytokinesis. Molecular Microbiology. 2004;53:687–696. doi: 10.1111/j.1365-2958.2004.04157.x. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. Ethylene biosynthesis and signalling networks. The Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel CL, Hester Q, Mattsson J. Identification of genes expressed in vascular tissues using NPA-induced vascular overgrowth in Arabidopsis. Plant and Cell Physiology. 2008;49:457–468. doi: 10.1093/pcp/pcn023. [DOI] [PubMed] [Google Scholar]
- Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. The Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annual Review of Plant Biology. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. The Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. The Plant Cell. 2004;16:367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in. Arabidopsis. The Plant Journal. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proceedings of the National Academy of Sciences, USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. The EMBO Journal. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.