Abstract
The high sensitivity of male reproductive cells to high temperatures may be due to an inadequate heat stress response. The results of a comprehensive expression analysis of HsfA2 and Hsp17-CII, two important members of the heat stress system, in the developing anthers of a heat-tolerant tomato genotype are reported here. A transcriptional analysis at different developmental anther/pollen stages was performed using semi-quantitative and real-time PCR. The messengers were localized using in situ RNA hybridization, and protein accumulation was monitored using immunoblot analysis. Based on the analysis of the gene and protein expression profiles, HsfA2 and Hsp17-CII are finely regulated during anther development and are further induced under both short and prolonged heat stress conditions. These data suggest that HsfA2 may be directly involved in the activation of protection mechanisms in the tomato anther during heat stress and, thereby, may contribute to tomato fruit set under adverse temperatures.
Keywords: Anther development, heat stress, HsfA2, Hsp17-CII, pollen, tomato
Introduction
Exposure to environmental stresses, such as high temperature, can severely reduce the productivity/yield of tomato plants grown under field conditions (Bar-Tzur et al., 1985). Decreased fruit set due to heat stress (hs) has been associated with several alterations in the morphology of tomato flower structures and with physiological imbalances in stress-protective metabolites, such as carbohydrates, polyamines, and proline (Pressman et al., 2002; Song et al., 2002; Sato et al., 2006). The major factor responsible for the failure of tomato fruit to set under suboptimal temperature conditions is considered to be the higher sensitivity of flower developmental processes to temperature changes, with the anthers being reported to be more vulnerable than the female organs (Peet et al., 1998; Sato et al., 2000). Alterations in tomato anthers, such as a failure of adequate dehiscence and of tapetum development, have been found when hs occurs during the early phases of pollen development, i.e. 7–15 d before anthesis (Sato et al., 2000, 2002). However, hs also affects late pollen development. Yang et al. (2009) recently demonstrated that TMS1, encoding a Hsp40 heat shock protein with DnaJ and PDI domains, is required for thermotolerance in growing pollen tubes in Arabidopsis. A mutation in this gene led to the reduction of seed production when plants were exposed to high temperatures.
The sensitivity of pollen development to hs has also been attributed to reduced thermotolerance due to the inability of the pollen to provide a strong hs response and, consequently, to produce large quantities of heat shock proteins (Hsps) as compared with vegetative tissues (Mascarenhas and Crone, 1996). However, both low and high molecular weight Hsps have been found to be expressed in the early and late stages of pollen development in various plant species (Becker et al., 2003; Honys and Twell, 2003; Volkov et al., 2005; Sheoran et al., 2007; Frank et al., 2009). One hypothesis is that the activation of hs gene expression during plant development is correlated to developmental programmes rather than to the response of the plant under stressful environmental conditions (Waters et al., 1996; Sun et al., 2002). According to this proposal, during pollen formation, Hsps may function as molecular chaperones for the folding/refolding of proteins involved in meiosis and tetrad formation (Bouchard, 1990; Atkinson et al., 1993; Reynolds, 1997). The developmental regulation of Hsp expression has been well characterized in animals, where they are induced by distinct heat stress transcription factors (Hsfs) activated by endogenous stimuli (Pirkkala et al., 2001). In contrast, few members of the large Hsf family in plants have been shown to be developmentally regulated (Gagliardi et al., 1995; Honys and Twell, 2004; Frank et al., 2009). Kotak et al. (2007) observed an elevated expression of HsfA9 during embryogenesis and seed maturation in Arabidopsis thaliana, and showed that its transcriptional control was strictly mediated by the ABI3 signal cascade. The function of other Hsf genes as potential transcriptional activators of Hsp expression in plant developmental programmes, however, remains poorly understood.
Researchers have expended the most effort on analysing the network of Hsfs activated in plant tissues, such as leaves, under hs conditions (von Koskull-Döring et al., 2007). In particular, one of the components of the tomato Hsf family, HsfA2, shows a number of striking characteristics that suggest it would be an ideal potential transcriptional activator during the hs response (Baniwal et al., 2004). Mishra et al. (2002) reported that HsfA2 was strongly accumulated in tomato cells under conditions of high temperature, following its induction by the master regulator, HsfA1, and that it became the dominant Hsf component in thermotolerant cells. Three major functional states of HsfA2 are distinguished: a soluble nuclear form, a soluble cytoplasmic form, and a stored form (Nover et al., 1989; Scharf et al., 1998; Heerklotz et al., 2001, Port et al., 2004). After the plant has sensed hs, HsfA2 is accumulated in the nucleus of tomato cells in the presence of HsfA1, and both Hsfs then form hetero-oligomeric complexes that markedly enhance the expression of hs genes (Chang-Schaminet et al., 2009). In contrast, during ongoing hs conditions, a considerable quantity of HsfA2 is present in large cytoplasmic aggregates (the hs granules). Ultimately, following the hs and during the recovery period, most of the HsfA2 is found in a soluble form in the cytoplasm. Port et al. (2004) demonstrated that Hsp17.4-CII acts as a specific repressor of HsfA2 activity, suggesting that this complex is responsible for the recruitment of HsfA2 into hs granules.
Analyses of HsfA2 and Hsp17.4-CII transcripts revealed a differential regulation during flower development under normal and hs conditions (F Giorno and S Grillo, unpublished data). Frank et al. (2009) subsequently demonstrated the expression of these hs genes by transcriptional profiling of tomato microspores.
The results reported here were obtained from an in-depth investigation on the expression of these genes and the accumulation of their corresponding proteins in the tomato anther during its development under experimental hs conditions. Unravelling the transcriptional mechanisms that regulate Hsf and, thereby, Hsp expression during tomato flower development will undoubtedly be a major step towards improving genetic traits in tomato plants, such as pollen thermotolerance and fruit set.
Materials and methods
Plant material, growth, and HS treatments
Tomato seeds from Solanum lycopersicum cv Saladette, a heat-tolerant line that has normal fruit set under high temperature conditions (Rudich et al., 1977), were germinated and grown at control temperatures (CT: 26 °C / 19° C day/night) under a long-day photoperiod (16/8 h, day/night), with low-intensity light supplied by high-pressure sodium lamps (600W; Philips, Eindhoven, The Netherlands). Following initiation of the flowering period, tomato buds containing anthers 2, 4, 6, and 8 mm long, respectively, corresponding to the flower morphological stages described by Brukhin et al. (2003), were harvested from the plants. Mature microspores and dry pollen were obtained from the 8 mm long anthers (200–300) and from flowers at anthesis, respectively.
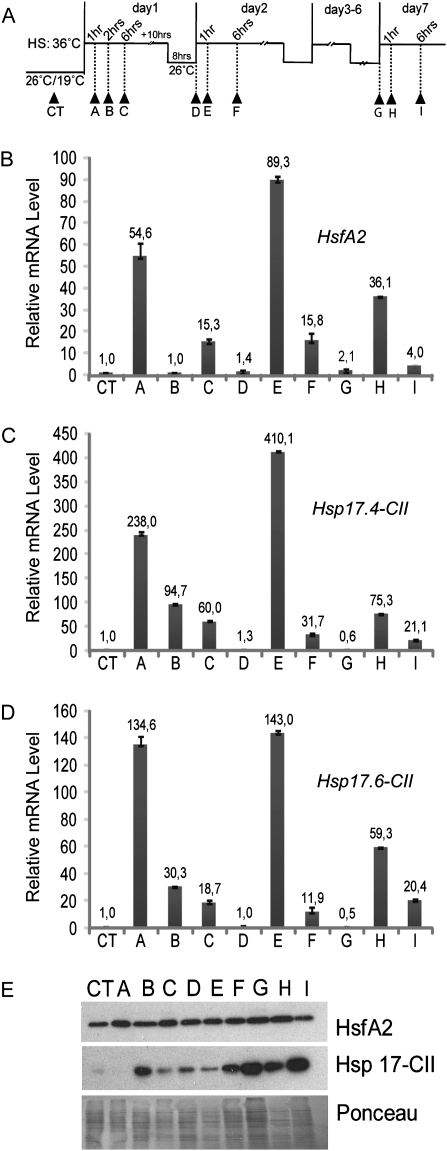
Heat stress (HS) was applied to plants in the form of high temperature (36 °C). Anthers at the 2 mm stage were collected after being subjected to short HS treatments at 36 °C for 1, 2, and 6 h, respectively (Fig. 4A, samples A, B, and C), after recovery at 26 °C and on the second day of the HS treatments (Fig. 4A, samples D, E, and F). Anthers at the 2 mm stage were harvested from the same treated plants (Fig. 4A, samples G, H and I) at day 7 after daily repeated cycles of mild HS and recovery (36 °C / 26 °C day/night). Recovery samples D and G (Fig. 4) were also harvested at 26 °C after 30 min of light acclimatization to avoid interference from the circadian rhythm.
Fig. 4.
Expression analyses of HsfA2 and Hsp17-CII in 2 mm anthers after HS treatments. (A) The pictogram shows the time course of HS treatments. Arrows indicate the time points when the 2 mm anthers were harvested (CT-I). (B–D) qRT-PCR of HsfA2 (B), Hsp17.4-CII (C), and Hsp17.6-CII (D). Expression data were normalized using LeEF1 and 18S rRNA as housekeeping genes. The mRNA levels of the target genes are relative to that of the sample CT (value 1). (E) Immunoblotting analyses showing protein levels of HsfA2 and Hsp17-CII in anthers at the same stages as in B, C, and D. Ponceau staining of total protein was used as control for equal loading.
From the same plants, flower buds containing anthers 4, 6, and 8 mm in length, mature microspores from 8 mm anthers, and dry pollen from the anthesis stage were collected after 2 h at 36 °C.
In this manuscript, the abbreviation HS refers to the heat stress treatment, while hs refers to heat stress as a stress condition.
RNA isolation and real-time PCR
Total RNA was isolated from anther cone tissues (n > 6) using a one-step RNA isolation method (TRIzol® Reagent; Invitrogen, Carlsbad, CA, USA). RNA quantity was measured spectrophotometrically, and its quality was checked by agarose gel electrophoresis. For reverse transcription, 1 μg of total RNA for each sample was incubated with RNase-free DNase (RQ1; Promega, Madison, WI, USA), and 400 ng of total RNA was used for reverse transcription according to the manufacturer's instructions (iScript™ cDNA Synthesis Kit; Bio-Rad Laboratories, Hercules, CA, USA). PCR analyses were carried out in 25 μl volumes containing 0.125 μl of cDNA synthesis reaction mixture, 400 nM of each primer, and 12.5 μl of iQ SYBR Green Supermix (Bio-Rad Laboratories). The PCRs were performed in a 96-well thermocycler (Bio-Rad iCycler; Bio-Rad Laboratories) using a controlled temperature program starting with 3 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 45 s at 60 °C. To verify the presence of a specific product, the melting temperature of the amplified products was determined. In addition, a fraction of each PCR mixture was analysed on a 2% agarose–ethidium bromide gel to verify the size of the amplified DNA fragment. The primers used for the real-time quantitative PCRs were designed using a computer program (BEACON DESIGNER 5.01; Premier Biosoft International, Palo Alto, CA, USA) to obtain primers that have close to identical melting temperatures and do not form secondary structures with each other under the given PCR conditions (Supplementary Table S1 available at JXB online).
The primers used to quantify Hsp17.4–CII and Hsp17.6-CII mRNAs were gene specific. Relative mRNA levels of the target genes were calculated following the Bio-Rad outlined methodology based on Vandesompele et al. (2002). The LeEF1 and 18S rRNA genes were used as references. For each sample, the mRNA quantity was calculated relative to the calibrator sample for the same gene. All reactions were performed on two independently collected series of RNA samples. The gene expression patterns were comparable in the two biological replicas. All figures show one series, with the error bars based on one technical repeat.
Semi-quantitative PCRs were performed using 5 μl of 25-fold diluted cDNA, buffer IV, 2.5 mM MgCl2, 0.5 U of Red Hot Taq DNA polymerase (all from ABgene Limited, Epson, Surrey, UK), 0.4 mM dNTPs (Fermentas, St. Leon-Rot, Germany), and 0.1 μM of primers. Amplification consisted of 30 cycles of 30 s at 95 °C (denaturation), 30 s at 58 °C (primer annealing), and 30 s at 72 °C (extension time).
In situ RNA hybridization
For in situ experiments, a 250 bp fragment was amplified from the pGEM5zf clone containing the total coding sequence of Hsp17.4-CII, previously amplified from tomato leaf cDNA. Primers 5′-TTCGTGATGCTAAGGCAATG-3′ (forward) and 5′-GCATTCTCCGGCAGACTAAA-3′ (reverse) were used for the amplification. The 250 bp fragment was then inserted into pGEM-T easy. The sense and antisense digoxigenin (DIG)-labelled ribo-probes were generated by in vitro transcription using T7 and SP6 RNA polymerases according to the manufacturer's protocol (Roche Diagnostics, Indianapolis, IN, USA). For the in situ experiments, 2 mm and 4 mm anthers were collected from tomato plants grown at CT and from those treated for 2 h at 36 °C. Anther fixation/paraplast embedding and in situ experiments were performed as described by Nitsch et al. (2009).
Protein isolation and immunoblotting
Proteins were extracted from anthers using TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. Following isolation, the protein concentration was measured using the bicinchoninic acid assay (BCA Assay Kit; Thermo Fisher Scientific, Waltham, MA, USA). One aliquot from each sample containing the same amount of total protein was precipitated with 4 volumes of acetone, and the proteins were dissolved in 1× SDS sample buffer. Aliquots of 25 μg of total proteins were separated on 14% SDS–polyacrylamide gels and transferred to a nitrocellulose membrane (pore size: 45 μm) (Protran; Schleicher and Schuell, Dassel, Germany). For immunodetection, the membranes were blocked with 5% (w/v) milk powder in phosphate-buffered saline (PBS) for 1 h and then incubated overnight at 4 °C with rabbit polyclonal antibodies against HsfA2 and Hsp17-CII, as described by Port et al. (2004). The blots were then rinsed and incubated with secondary antibody against rabbit IgG conjugated to horseradish peroxidase and were further processed for chemiluminescence detection using a kit as described by the manufacturer's protocol (Super Signal West Pico Chemiluminscent Substrate; Thermo Fisher Scientific).
Results
Expression of HsfA2 and Hsp17.4-CII in different tomato tissues and during anther development
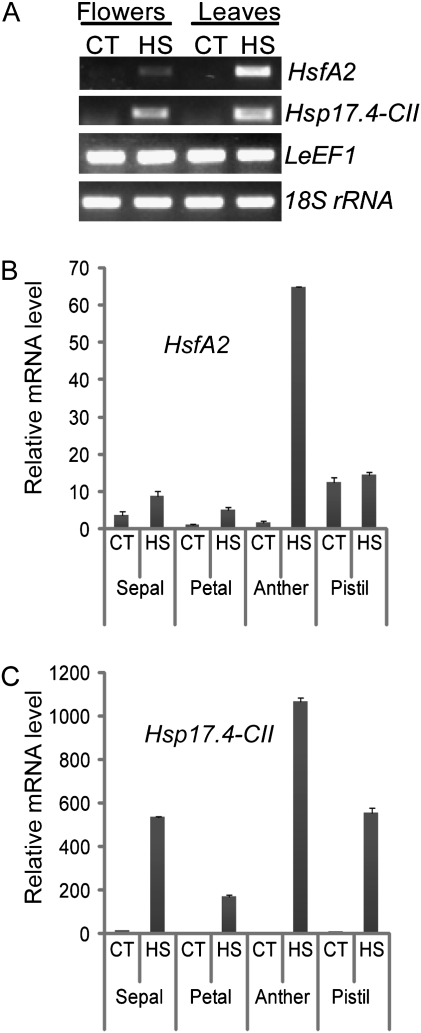
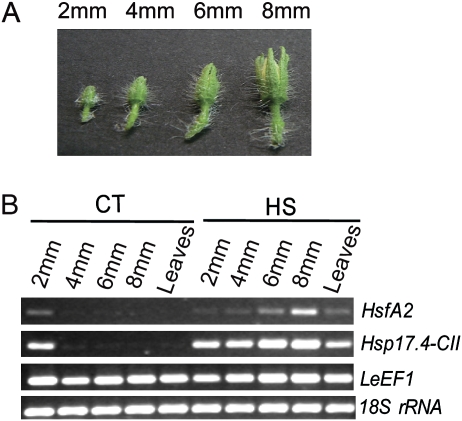
Previous results (F Giorno and S Grillo, unpublished data) suggested that HsfA2 and Hsp17.4-CII genes are induced in response to HS during tomato flower development, particularly at anthesis, when pollination occurs. The expression of these genes in tomato microspores has also been reported (Frank et al., 2009). The influence of HS on the expression of these genes in the flower and during anther development was therefore studied here in more detail. As shown in Fig. 1A, HsfA2 and Hsp17.4-CII were induced in the tissues of flowers at anthesis and in leaves harvested from cv Saladette treated with daily repeated cycles of mild HS and recovery for 3 weeks (HS: 36 °C / 26 °C, day/night). The real-time PCR (qRT-PCR) results showed that HsfA2 and Hsp17.4-CII were more highly induced in the anther than in the other flower tissues (Fig. 1B, C). The observation that both HsfA2 and Hsp17.4-CII were more highly expressed in the tomato male reproductive organs suggests that they may play a role in the protection system of sporophytic and/or sporogenic tissues in the anther. The timing of the activation of this protection system during anther development was investigated here by first determining the transcript levels of HsfA2 and Hsp17.4-CII in 2, 4, 6, and 8 mm long anthers using semi-quantitative PCR (Fig. 2). As shown in Fig. 2B, the transcripts of HsfA2 and Hsp17.4-CII were more detectable during development at the 2 mm stage under CT conditions, while both mRNAs were present at all stages of development after HS for 2 h at 36 °C.
Fig. 1.
Transcriptional changes of HsfA2 and Hsp 17.4-CII in tomato tissues under HS. Total flower tissues at anthesis and leaf tissues were harvested from cv Saladette treated with daily repeated cycles of mild HS and recovery for 3 weeks (HS: 36 °C / 26 °C, day/night) or maintained under control conditions (CT: 26 °C / 19 °C day/night). From these same treated and untreated plants, flowers harvested at anthesis were dissected into sepal, petal, anther, and pistil. (A) Semi-quantitative PCR shows increased mRNA levels of HsfA2 and Hsp17.4-CII in tomato flowers and leaves under prolonged HS conditions. (B) Relative transcript levels of HsfA2 were measured by qRT-PCR using LeEF1 and 18S rRNA as housekeeping genes to normalize the data. The highest induction of HsfA2 was identified in the anther tissues treated at high temperatures. (C) A similar transcript profile to that of (B) was also observed for Hsp17.4-CII, which was strongly induced in the male reproductive organs under HS.
Fig. 2.
Expression analyses of HsfA2 and Hsp17.4-CII during anther development under CT and HS conditions. (A) Tomato flower buds were harvested from cv Saladette and used to obtain different sized anthers (2, 4, 6, and 8 mm). (B) mRNAs of HsfA2 and Hsp17.4-CII, from anthers at different stages and leaves treated with HS (2 h at 36 °C) or kept at CT conditions (26 °C / 19 °C, day/night) were analysed by semi-quantitative PCR. LeEF1 and 18S rRNA transcript levels were monitored as controls. (This figure is available in colour at JXB online.)
HsfA2 and Hsp 17-CII are differentially modulated in young anther stages
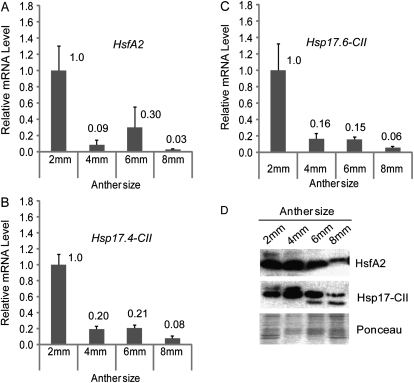
The observation that 2 mm anthers from CT plants had the highest expression of HsfA2 and Hsp17.4-CII led to a more detailed investigation of transcript levels and protein accumulation under CT conditions. As shown in Fig. 3A, HsfA2 expression in 2 mm anthers from CT plants was higher than that in older ones. Correspondingly, the amount of the protein was higher at the 2 mm and 4 mm anther stages, subsequently decreasing in 6 mm and 8 mm anthers (Fig. 3D). The Hsp17.4-CII gene was also expressed during anther development (Fig. 3B), particularly at the younger stages, when the accumulation of Hsp17-CII proteins was also observed (Fig. 3D). However, because tomato small Hsps, such as Hsp17.4-CII and Hsp17.6-CII, belong to the same subfamily and differ only by a few amino acid residues, it was impossible to discriminate them by the antibody used in this study (Port et al., 2004). This led to an analysis of the Hsp17.6-CII transcript profiles in order to distinguish the Hsp17.4-CII and Hsp17.6-CII mRNAs that are translated into the respective proteins. As observed for Hsp17.4-CII, the Hsp17.6-CII gene was also expressed during anther development under CT conditions, particularly in 2 mm anthers (Fig. 3C), indicating that the Hsp17.4-CII and Hsp17.6-CII proteins may be detected together with the Hsp17-CII antibody.
Fig. 3.
Expression analyses of HsfA2 and Hsp17-CII during anther development. (A–C) Relative mRNA levels of HsfA2 (A), Hsp17.4-CII (B), and Hsp17.6-CII (C) in different sized anthers under CT conditions (26 °C / 19 °C, day/night). Expression data were normalized using LeEF1 and 18S rRNA as housekeeping genes. The mRNA levels of the target genes in 4, 6, and 8 mm anthers are relative to that from 2 mm anthers (value 1). The experiment shown here is one of two biological replicates, which showed a comparable expression pattern. Each graph represents the data of two technical repeats. (D) Immunoblotting analyses of the proteins isolated from the same samples as in A, B, and C using specific antisera against HsfA2 and Hsp17-CII. Ponceau staining of total protein was used as control for equal loading.
An HS time course experiment was designed to analyse the influence of HS on HsfA2 and Hsp17-CII expression modulations in the youngest anther under stressful conditions. The results are shown in Fig. 4. Plants were grown in the growth chamber at CT. At the beginning of flowering, temperatures were raised to 36 °C during the day and 26 °C during the night. This treatment was repeated for 7 d to mimic the long-day hs condition that can occur in the field. Anthers at the 2 mm stage were collected at various intervals (samples CT-I, Fig. 4A). Controls (untreated samples) were harvested during the day at 26 °C, and samples D and G were harvested at the end of the recovery periods, after HS, at 26 °C and after 30 min of light. The results show that HsfA2 was rapidly induced in cv Saladette after 1 h of HS but that the mRNA levels transiently declined quickly after 2 h HS (Fig. 4B, samples A and B). Interestingly, the HsfA2 transcript was more strongly induced during the second HS treatments (sample E in Fig. 4B) and was still detectable in the recovery periods and under prolonged HS treatments (samples D, G, H, and I in Fig. 4B). However, its protein level remained comparable with that in the control sample (samples D, G, H, and I in Fig. 4E). The mRNA expression patterns of both Hsp17-CII were similar to that of HsfA2 (Fig. 4C, D), but Hsp17-CII protein accumulation was particularly strong only after prolonged HS regimes (samples F, G, H, and I in Fig. 4E).
HsfA2 and Hsp17-CII expressed in immature pollen
In situ RNA hybridization on anthers harvested at various developmental stages was performed to determine in which anther tissues the HsfA2 and Hsp17-CII transcripts were present.
A riboprobe corresponding to a fragment of the coding sequence of Hsp17.4-CII was synthesized. However, some sequence domains of Hsp17.4-CII and Hsp17.6-CII are known to be highly conserved, implying that both messengers may hybridize to this probe.
As expected, Hsp17-CII mRNA was detected in anthers from plants subjected to HS for 2 h at 36 °C, but not in the controls (Fig. 5). In contrast, HsfA2 transcripts could not be detected in any of these stages, most probably due to low mRNA abundance.
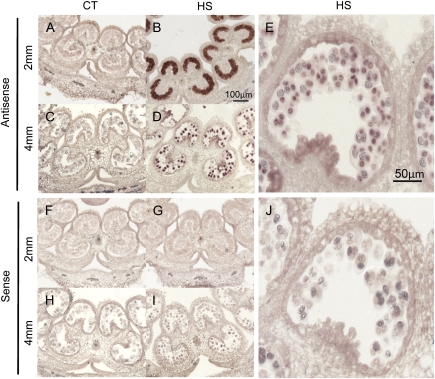
Fig. 5.
In situ RNA localization of Hsp17-CII at the 2 mm and 4 mm stages. Sections of 2 mm and 4 mm anthers from CT (26 °C / 19 °C, day/night) and HS samples (2 h at 36 °C) were hybridized with antisense (A–E) and sense (F–J) probes. Accumulation of Hsp17-CII transcripts was detected in HS samples, particularly in pollen mother cells at the 2 mm stage (B) and in tetrads at the anther stage of 4 mm (D, E). (This figure is available in colour at JXB online.)
In agreement with the results of the qRT-PCR, Hsp17-CII transcripts were strongly accumulated in anthers at young stages, particularly in the pollen mother cells (Fig. 5B). A signal was detected in 4 mm anthers in the microspores, bound together as tetrads in the pollen sac (Fig. 5D, E). Hsp17-CII transcripts were not detected in 6 mm and 8 mm anthers (data not shown), which is in contrast to the results of the semi-quantitative PCR analysis reported in Fig. 2B. One possible explanation for this discrepancy is that mature microspores at these stages are surrounded by an exine wall that obstructs the penetration of the probe.
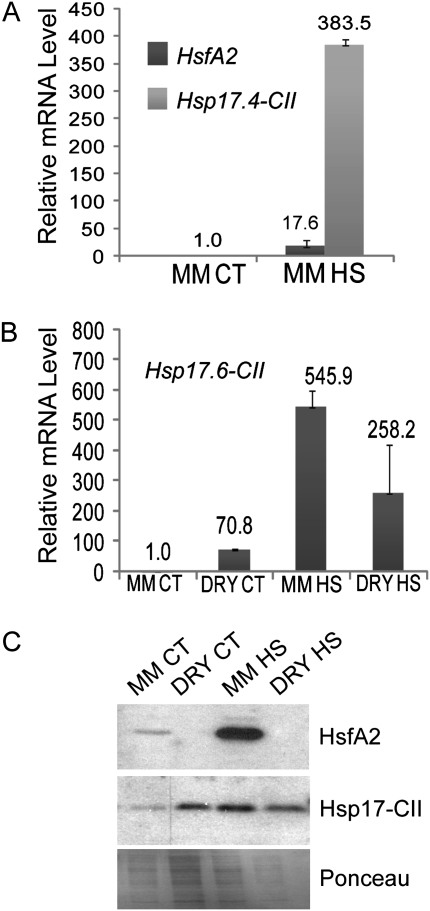
qRT-PCR and western blotting analyses were carried out on the developing pollen grains to confirm the presence of HsfA2 and Hsp17-CII. An increased level of HsfA2 and Hsp17.4 CII transcripts was observed in mature microspores isolated from 8 mm anthers harvested from plants subjected to the HS treatment for 2 h at 36 °C (Fig. 6A). The expression of these genes in dry pollen was also tested, as dry pollen grains develop from the microspores and are released at anther dehiscence. However, mRNAs of HsfA2 and Hsp17.4-CII were not detectable in the dry pollen from flowers at anthesis (data not shown), whereas the mRNA of Hsp17.6-CII was expressed at this stage in the CT and HS samples (Fig. 6B). The accumulation of HsfA2 and the two Hsp17-CII proteins was observed in mature microspores in the CT and HS samples, as shown in Fig. 6C. In contrast, HsfA2 protein was not detectable in the dry pollen, whereas Hsp17-CII persisted, most probably as a product of Hsp17.6-CII mRNA (Fig. 6B, C).
Fig. 6.
HsfA2, Hsp17.4-CII, and Hsp17.6-CII expression analyses in mature microspores (MM) and dry pollen (DRY) under CT conditions (26 °C / 19 °C, day/night) and HS (2 h at 36 °C). (A and B) mRNA levels of HsfA2 and Hsp17.4-CII (A) and Hsp17.6-CII mRNA (B) in MM and DRY pollen. The relative mRNA levels were measured using LeEF1 and 18S rRNA as housekeeping genes. (C) Protein levels of HsfA2 and Hsp17-CII in MM and DRY pollen. Ponceau staining of total proteins is given as the control for equal loading.
Discussion
High temperatures can potentially disrupt tomato fruit set by causing damage to developing pollen grains (Sato et al., 2002). The high sensitivity of the developing pollen grain to hs is partly attributable to its incomplete hs response due to a defect in the accumulation of Hsf and Hsp mRNAs (Frova et al., 1989; Gagliardi et al., 1995; Mascarehans and Crone, 1996). Hsp genes are also induced in a number of developmental pathways, such as seed maturation, embryogenesis, and/or fruit maturation, in various other plant species (Waters et al., 1996; Neta-Sharin et al., 2005; Volkov et al., 2005; Kotak et al., 2007). However, the overproduction of Hsps can also interfere with normal developmental processes, as shown by maize plants carrying a mutation in the Empty Pericarp2 gene. This gene is normally a negative regulator of the heat shock response, but the presence of the mutation results in abortion or retarded embryogenesis (Fu et al., 2002).
The results reported here on the temporal and spatial regulation of HsfA2 and Hsp17-CII suggest possible roles for the Hsf and Hsp genes in developmental processes.
HsfA2 was more highly expressed in anthers than in other organs and was strongly activated when exposed to hs; this is similar to its expression in leaves (Mishra et al., 2002). HsfA2 was expressed early in young anthers, at the stage when, according to Brukhin et al. (2003), pollen mother cells are developing. Frank et al. (2009) reported the expression of these genes at the post-meiotic stage, presumably in the tetrads, under non-hs conditions.
It has been suggested that during these developmental processes Hsps may function as molecular chaperones for the folding/refolding of proteins involved in meiosis and tetrad formation (Bouchard, 1990; Atkinson et al., 1993; Reynolds, 1997). HsfA2 is an activator of Hsp genes and is likely to be involved in the regulation of Hsp activation during tomato pollen/anther development (Nishizawa et al., 2006; Schramm et al., 2006; Oyawa et al., 2007). A similar function of transcriptional activator is well documented for Hsf members in animals and also for HsfA9 in Arabidopsis, where HsfA9 induces Hsp expression during seed maturation coinciding with dormancy and desiccation tolerance (Kotak et al., 2007). Honys and Twell (2004) meticulously analysed the transcriptome of the male gametophyte of Arabidopsis but were unable to identify transcripts of HsfA2. Given these data from earlier studies, the present results suggest that HsfA2 expression is differentially modulated between tomato and Arabidopsis. Consequently, the species-specific functional diversification previously documented for HsfA1 and HsfB1 may also be extended to HsfA2 in terms of its involvement in different developmental pathways (Bharti et al., 2004; von Koskull-Döring et al., 2007).
HsfA2 is known to be markedly activated in young anthers in response to hs regimes. Frank et al. (2009) reported that HsfA2 is strongly expressed in maturing tomato microspores experimentally treated with a short hs at 45 °C. However, in the present study, HsfA2 was activated even with a mild hs (36 °C), and its expression was maintained for several days when plants were subjected to this type of stress. This result indicates that HsfA2 may even be involved in the hs response to milder stresses in order to enhance the expression of Hsp genes and to protect the pollen grains during development. Scharf et al. (1998) and Schramm et al. (2006) examined the kinetic expression characteristics of HsfA2 and demonstrated that this protein is also present at relatively high levels during the recovery period following the hs. As such, the behaviour of HsfA2 resembles that of typical Hsp proteins in thermotolerant cells. In agreement with these data, in this study, the HsfA2 protein also remained at high levels in the anthers during the recovery periods. However, the decline in mRNA levels of Hsp17-CII observed during the recovery periods suggests that the HsfA2 protein was inactive. Port et al. (2004) observed that HsfA2 inactivation depends on its interaction with Hsp17.4-CII protein. The experimental data reported here suggest that a similar control mechanism may operate in anther/developing pollen grains.
The elucidated characteristics of HsfA2 make it an ideal and potent transcriptional activator in many environmental stress responses (Busch et al., 2005; Nishizawa et al., 2006; Zhang et al., 2009). In tomato, Mishra et al. (2002) reported that the essential component of the tolerance to high temperature is HsfA1 but that HsfA2 is sufficient to restore the thermotolerance in HsfA1-silenced protoplasts.
An Arabidopsis T-DNA insertion mutant lacking HsfA2 expression is more sensitive to prolonged hs due to its decreased expression of Hsps and genes such as ascorbate peroxidase 2 (Schramm et al., 2006; Charng et al., 2007). Meiri and Breiman (2009) recently demonstrated that HsfA2 contributes to the thermotolerance of Arabidopsis by mediating the translocation of the ROF1 and Hsp90.1 protein complex from the cytoplasm to the nucleus. Interference in this shuttle system severely affects the expression of Hsps during hs recovery periods in Arabidopsis mutant lines and therefore also hinders the acquisition of thermotolerance following exposure to prolonged hs conditions. Recent evidence that maturing microspores of an hs-tolerant line have a higher HsfA2 basal level than those of an hs-sensitive line also suggests that this gene may contribute to pollen thermotolerance (Frank et al., 2009).
However, it cannot be excluded that other Hsfs are active during microspore development in tomato and that HsfA2 is only one example of such transcription factors.
HsfA2 expression in dry pollen was not observed here. However, this absence of expression may be due to the fact that dry pollen is metabolically inactive as the cytoplasm is greatly dehydrated; therefore, no further protection should be required at this stage. In contrast, the Hsp17.6-CII transcript was detected in the dry pollen. It is therefore possible that this latter gene is activated at this stage by other transcriptional factors that are, in turn, regulated by the desiccation processes, as has been observed for other Hsps in Arabidopsis during seed maturation (Kotak et al., 2007).
In conclusion, the results from the study reported here show that HsfA2 and Hsp17-CII are activated early in the tomato diploid pollen mother cells before the microspores develop and that their expression is persistent under prolonged HS conditions until mature dry pollen is produced. The data suggest that HsfA2 may be directly involved in the activation of protection mechanisms in the tomato anther during hs and, thereby, may contribute to tomato fruit set under adverse temperatures.
Supplementary data
Supplementary data ara available at JXB online.
Table S1. Primer sequences used for semi- and real-time quantitative PCR.
Supplementary Material
Acknowledgments
We would like to thank Peter de Groot for technical assistance, Gerard van der Weerden for the excellent support with greenhouse facilities, Dr Elisabeth Pierson for help with the in situ images, and all of our colleagues in our lab for helpful suggestions and advice on this research. Dr Antonella Leone and Dr Immacolata Massarelli are thanked for helpful discussions during the early phase of this project. This work was supported by Accademia Nazionale dei Lincei and IWWR Institute, Radboud University.
Glossary
Abbreviations
- Hsp
heat shock protein
- Hsf
heat stress transcriptional factor
- hs
heat stress
References
- Atkinson BG, Raizada M, Bouchard RA, Frappier J, Walden D. The independent stage-specific expression of the 18-kDa heat shock protein genes during microsporogenesis in Zea mays L. Developmental Genetics. 1993;14:15–26. doi: 10.1002/dvg.1020140104. [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. Journal of Biosciences. 2004;29:471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- Bar-Tzur A, Rudich J, Bravdo B. High temperature effects on CO2 gas exchange in heat tolerant and sensitive tomatoes. Journal of the American Society for Horticultural Science. 1985;110:582–586. [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiology. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. The Plant Cell. 2004;16:1521–1535. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard R. Characterization of expressed meiotic prophase repeat transcript clones of Lilium: meiosis-specific expression, relatedness, and affinities to small heat-shock protein genes. Genome. 1990;33:68–79. doi: 10.1139/g90-012. [DOI] [PubMed] [Google Scholar]
- Brukhin V, Hernould M, Gonzalez N, Chevalier C, Mouras A. Flower development schedule in tomato Lycopersicon esculentum cv. sweet cherry. Sexual Plant Reproduction. 2003;15:311–320. [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal. 2005;41:1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf KD. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. Journal of Biological Chemistry. 2009;284:20848–20857. doi: 10.1074/jbc.M109.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiology. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. Journal of Experimental Botany. 2009;60:3891–3908. doi: 10.1093/jxb/erp234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frova C, Taramino G, Binelli G. Heat-shock proteins during pollen development in maize. Developmental Genetics. 1989;10:324–332. [Google Scholar]
- Fu S, Meeley R, Scanlon MJ. Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. The Plant Cell. 2002;14:3119–3132. doi: 10.1105/tpc.006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D, Breton C, Chaboud A, Vergne P, Dumas C. Expression of heat shock factor and heat shock protein 70 genes during maize pollen development. Plant Molecular Biology. 1995;29:841–856. doi: 10.1007/BF00041173. [DOI] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Molecular and Cellular Biology. 2001;21:1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiology. 2003;132:640–652. doi: 10.1104/pp.103.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. The Plant Cell. 2007;19:182–195. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP, Crone EC. Pollen and the heat shock response. Sexual Plant Reproduction. 1996;9:370–374. [Google Scholar]
- Meiri D, Breiman A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. The Plant Journal. 2009;59:387–399. doi: 10.1111/j.1365-313X.2009.03878.x. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes & Development. 2002;16:1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neta-Sharir I, Isaacson T, Lurie S, Weiss D. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. The Plant Cell. 2005;17:1829–1838. doi: 10.1105/tpc.105.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. The Plant Journal. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- Nitsch LM, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, Mariani C, Vriezen WH. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta. 2009;229:1335–1346. doi: 10.1007/s00425-009-0913-7. [DOI] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Molecular and Cellular Biology. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. Journal of Experimental Botany. 2007;58:3373–3383. doi: 10.1093/jxb/erm184. [DOI] [PubMed] [Google Scholar]
- Peet MM, Sato S, Gardner RG. Comparing heat stress effects on male-fertile and male-sterile tomatoes. Plant, Cell and Environment. 1998;21:225–231. [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB Journal. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Port M, Tripp J, Zielinski D, Weber C, Heerklotz D, Winkelhaus S, Bublak D, Scharf KD. Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiology. 2004;135:1457–1470. doi: 10.1104/pp.104.042820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressman E, Peet MM, Pharr DM. The effect of heat stress on tomato pollen characteristics is associated with changes in carbohydrate concentration in developing anthers. Annals of Botany. 2002;90:631–636. doi: 10.1093/aob/mcf240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T. Pollen embryogenesis. Plant Molecular Biology. 1997;33:1–10. doi: 10.1023/a:1005748614261. [DOI] [PubMed] [Google Scholar]
- Rudich J, Zamski E, Regev Y. Genotype variation for sensitivity to high temperature in the tomato: pollination and fruit set. Botanical Gazette. 1977;138:448–452. [Google Scholar]
- Sato S, Kamiyama M, Iwata T, Makita N, Furukawa H, Ikeda H. Moderate increase of mean daily temperature adversely affects fruit set of Lycopersicon esculentum by disrupting specific physiological processes in male reproductive development. Annals of Botany. 2006;97:731–738. doi: 10.1093/aob/mcl037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Peet MM, Thomas JF. Physiological factors limit fruit set of tomato (Lycopersicon esculentum Mill.) under chronic, mild heat stress. Plant, Cell and Environment. 2000;23:719–726. [Google Scholar]
- Sato S, Peet MM, Thomas JF. Determining critical pre- and post-anthesis periods and physiological processes in Lycopersicon esculentum Mill exposed to moderately elevated temperatures. Journal of Experimental Botany. 2002;53:1187–1195. doi: 10.1093/jexbot/53.371.1187. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Molecular and Cellular Biology. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Molecular Biology. 2006;60:759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- Sheoran IS, Ross AR, Olson DJ, Sawhney VK. Proteomic analysis of tomato (Lycopersicon esculentum) pollen. Journal of Experimental Botany. 2007;58:3525–3535. doi: 10.1093/jxb/erm199. [DOI] [PubMed] [Google Scholar]
- Song J, Nada K, Tachibana S. Suppression of S-adenosylmethionine decarboxylase activity is a major cause for high-temperature inhibition of pollen germination and tube growth in tomato (Lycopersicon esculentum Mill.) Plant and Cell Physiology. 2002;43:619–627. doi: 10.1093/pcp/pcf078. [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants. Biochimica and Biophysica Acta. 2002;1577:1–9. doi: 10.1016/s0167-4781(02)00417-7. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Schöffl F. Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Molecular Biology. 2005;57:487–502. doi: 10.1007/s11103-005-0339-y. [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends in Plant Science. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. Journal of Experimental Botany. 1996;47:325–338. [Google Scholar]
- Yang KZ, Xia C, Liu XL, et al. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. The Plant Journal. 2009;57:870–882. doi: 10.1111/j.1365-313X.2008.03732.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Y, Xing D, Gao C. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. Journal of Experimental Botany. 2009;60:2073–2091. doi: 10.1093/jxb/erp078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.