Abstract
The micropylar endosperm is a major regulator of seed germination in endospermic species, to which the close Brassicaceae relatives Arabidopsis thaliana and Lepidium sativum (cress) belong. Cress seeds are about 20 times larger than the seeds of Arabidopsis. This advantage was used to construct a tissue-specific subtractive cDNA library of transcripts that are up-regulated late in the germination process specifically in the micropylar endosperm of cress seeds. The library showed that a number of transcripts known to be up-regulated late during germination are up-regulated in the micropylar endosperm cap. Detailed germination kinetics of SALK lines carrying insertions in genes present in our library showed that the identified transcripts do indeed play roles during germination. Three peroxidases were present in the library. These peroxidases were identified as orthologues of Arabidopsis AtAPX01, AtPrx16, and AtPrxIIE. The corresponding SALK lines displayed significant germination phenotypes. Their transcripts were quantified in specific cress seed tissues during germination in the presence and absence of ABA and they were found to be regulated in a tissue-specific manner. Peroxidase activity, and particularly its regulation by ABA, also differed between radicles and micropylar endosperm caps. Possible implications of this tissue-specificity are discussed.
Keywords: Arabidopsis thaliana, cress, Lepidium sativum, micropylar endosperm cap, peroxidases, seed germination, subtractive cDNA library
Introduction
Most mature angiosperm seeds consist of an embryo surrounded by the triploid living endosperm and the dead maternal testa. Seeds show great diversity of morphological and physiological features which have evolved to control germination and dormancy in response to different environments. One of the obvious morphological differences in mature angiosperm seeds is their ‘embryo to seed’ size ratio. This ratio expresses the extent to which the endosperm is obliterated during seed development by incorporating its nutrients into the growing embryo (reviewed by Finch-Savage and Leubner-Metzger, 2006). In mature seeds, the endosperm varies in thickness between filling almost the whole seed and covering a tiny embryo (many Ranunculaceae seeds), via medium-type seeds like Nicotiana tabacum to seeds where the endosperm was completely obliterated and the nutrients absorbed by thick storage cotyledons, as is the case in many legumes.
Mature seeds of the model species Arabidopsis thaliana (Arabidopsis) and its close relative Lepidium sativum (cress) both have a thin endosperm (one to two cell layers) that covers the embryo. Nutrients are stored predominantly in the large storage cotyledons (Müller et al., 2006). While the thin endosperm is unlikely to play a major role as nutritional storage for the embryo, its role in controlling germination timing is coming into focus (Müller et al., 2006; Penfield et al., 2006; Bethke et al., 2007). Arabidopsis and cress germinate in two sequential steps: testa rupture occurs along a preformed breaking line and leaves the radicle covered with only the micropylar endosperm (endosperm cap) (Liu et al., 2005; Müller et al., 2006). Only after the endosperm cap has sufficiently weakened to allow the growth potential of the radicle to overcome the resistance of the micropylar endosperm cap can endosperm rupture take place. Endosperm rupture marks the endpoint of the germination process (Bewley, 1997a).
Endosperm weakening, as well as the growth potential of the radicle, are known to be regulated by plant hormones: abscisic acid (ABA) inhibits endosperm weakening and rupture, while gibberellins (GA) act as its antagonists in a complex network integrating environmental signals such as light, temperature, water availability, and nutrient status (reviewed by Kucera et al., 2005). Ethylene and brassinosteroids also counteract ABA, but their effects on endosperm weakening are not known. Seeds of GA-insensitive Arabidopsis mutants, as well as dormant wild-type seeds, are only able to germinate when the endosperm layer is artificially opened, which further proves the ability of this thin tissue layer to inhibit radicle protrusion (Bethke et al., 2007; Bentsink and Koornneef, 2008). Circumstantial evidence supports the view that the endosperm cap of Arabidopsis weakens during the germination process, and that this weakening process does not take place in GA-insensitive mutants. In cress seeds, which are about 20 times larger than the seeds of Arabidopsis, endosperm weakening can be measured by the puncture force method (Müller et al., 2006). Using this technique, it was shown that the cress endosperm cap weakens during the germination process, allowing the radicle to protrude when the endosperm tissue resistance has decreased to a low enough threshold value. ABA specifically inhibits this weakening process and GA promotes it.
Seeds can enter a state of dormancy, in which they fail to complete germination under favourable conditions, even though they are viable (Finch-Savage and Leubner-Metzger, 2006). Dormancy ensures the proper temporal distribution of seed germination in natural environments and is critical for a species’ fitness. Dormant seeds must be exposed to species-specific environmental cues for dormancy to be terminated. Dormancy release also occurs during seed after-ripening, i.e. air-dry storage of seeds at room temperature. A loss of sensitivity to the germination-inhibiting hormone ABA is characteristic of after-ripening (Cadman et al., 2006; Holdsworth et al., 2008). Testa rupture and endosperm rupture are both promoted in after-ripened seeds, and after-ripened seed batches germinate more homogeneously than fresh ones (Leubner-Metzger, 2005; Müller et al., 2006).
Seeds are known to contain stored RNAs deposited during seed maturation (first described by Dure and Waters, 1965). These RNAs are used upon imbibition to restart full metabolism quickly. 12 000 different stored RNAs coding for proteins from a variety of functional categories were found in dry Arabidopsis seeds (Nakabayashi et al., 2005). While these stored RNAs are probably crucial for the first steps of germination, de novo transcription is quickly activated. After only 6 h of imbibition, the Arabidopsis seed transcriptome differs significantly from that of dry seeds (Nakabayashi et al., 2005).
So far the only transcriptome analysis of the whole Arabidopsis endosperm, sampled after radicle protrusion, revealed large differences between transcript abundance and regulation between endosperm and embryo (Penfield et al., 2006). A similar difference is to be expected during the germination process, i.e. prior to radicle protrusion. Known changes on the transcript and protein level in the endosperm of germinating seeds include changes in transcript abundance for cell-wall-modifying proteins such as expansins (Chen and Bradford, 2000), β-1,4-mannanase (Bewley, 1997b; Nonogaki et al., 2000) and β-1,3-glucanase (Leubner-Metzger et al., 1995). Isoforms of the latter two accumulate specifically in the endosperm cap which covers the radicle tip, but not in the non-micropylar endosperm. These cell-wall-modifying proteins probably play a role in cell wall loosening during endosperm weakening. It has also been shown that reactive oxygen species (ROS) play a role in this process by targeted attacks on cell wall polysaccharides in a hormonally and developmentally controlled manner (Müller et al., 2009). Polysaccharide targets differ between radicle and endosperm cap and, in general, in vivo formation of apoplastic ROS is inhibited by ABA and promoted by GA.
Thus, it is of interest to investigate transcript changes in the micropylar endosperm separately from those in other seed tissues, in order to further our understanding of the regulatory role of the micropylar endosperm in the germination process. As this is not easily possible with Arabidopsis seeds due to their small size, its close relative, cress, was used here. Cress seeds are anatomically very similar to Arabidopsis seeds, but are big enough for dissection (Müller et al., 2006) in order to create a subtractive cDNA-library of transcripts whose abundance increases during the germination process and thereby during endosperm weakening. This library was then used to identify genes whose mutation leads to changes in germination behaviour. It was found that peroxidases were overrepresented among the transcripts up-regulated during late germination in the micropylar endosperm of cress and among the genes leading to a germination phenotype when transcript levels were reduced. Therefore a closer look was taken at the transcript abundance of three specific peroxidases present in our cDNA library and at tissue-specific peroxidase activity during germination.
Materials and methods
Germination conditions and tissue preparation
Cress (Lepidium sativum, ‘Gartenkresse, einfache’, Juliwa, Germany) and tobacco (Nicotiana tabacum W38) seed germination analyses were performed in Petri dishes on two layers of filter paper with 6 ml 10% (w/v) Murashige–Skoog salts (MS) in continuous light (101.2 μmol m−2 s−1) at 18 °C and 24 °C, respectively. Where mentioned, 10 μM cis-S(+)-ABA was added to the medium. For protein or RNA preparation of different tissues, seeds were dissected with a scalpel and the separate parts collected on ice or frozen in liquid nitrogen until further use.
For Arabidopsis thaliana 10% MS salts solidified with 1% agar in continuous light at 18 °C or 24 °C was used as the germination medium. Where mentioned, 1 μM cis-S(+)-ABA was added. Stratification was performed where mentioned at 4 °C in darkness for 1 d. For after-ripening, seeds were stored at 25 °C for 2 months over saturated Ca(NO3)2 solution (relative humidity 51–54%).
For the preliminary germination screens of the SALK lines, one plate with 50 seeds was used per treatment and testa and endosperm rupture counted every 6 h (every 24 h for ABA-treated seeds). For the detailed germination kinetics, three times 50 seeds were used and germination was counted every 4 h.
RNA extraction and creation of the subtractive cDNA library
Total RNA from endosperm caps dissected from seeds after 8 h and 18 h imbibition was prepared as described in Cadman et al. (2006).
A Suppression Subtraction Hybridization library was constructed using the PCR Select™ cDNA Subtraction kit (Clontech, Palo Alto, CA, USA). cDNA from endosperm caps dissected after 18 h imbibition of the whole seed was used as tester against cDNA from endosperm caps dissected after 8 h imbibition as a driver. The cDNA fragments obtained were cloned in pUC19 and pCR®4-TOPO, multiplied in E. coli Top10, and sequenced (GATC, Konstanz, Germany). The 56 cDNAs sequences obtained were deposited as ESTs in GenBank/EMBL data libraries and their NCBI accession numbers are listed in Table 1.
Table 1.
Homozygous Arabidopsis SALK lines used in the germination screen
| AGI locus | Protein | SALK number | Insert location | Germination fresh seeds |
Germination after-ripened seeds |
||||||
| 18 °C | 24 °C | 24 °C S | 24 °C ABA S | 18 °C | 24 °C | 24 °C S | 24 °C ABA S | ||||
| AT1G01300 | Aspartyl protease | SALK_021485.37.45.x | exon | − | − | − | − | − | 0 | − | − |
| AT1G07890 | APX01 | SALK_000249.38.45.x | exon | + | + | + | + | nd | nd | nd | nd |
| AT1G64970 | G-TMTa | SALK_072105.54.50.x | promoter | − | − | 0 | 0 | − | − | 0 | − |
| AT1G66200 | GSR2 | SALK_102291.50.50.x | exon | nd | 0 | nd | 0 | nd | 0 | nd | 0 |
| AT2G18980 | Prx16 | SALK_028328.55.00.x | exon | + | + | + | + | nd | nd | nd | nd |
| AT2G34040 | API5 | SALK_008073.51.20.x | exon | nd | nd | nd | nd | − | 0 | 0 | 0 |
| AT2G36530 | LOS2 | SALK_100086.52.65.x | promoter | − | 0 | 0 | 0 | − | 0 | − | 0 |
| AT3G12620 | PP2C | SALK_016641.56.00.x | exon | − | 0 | 0 | − | − | 0 | − | − |
| AT3G13920 | EIF4A1 | SALK_107633.50.85.x | promoter | 0 | 0 | 0 | − | 0 | 0 | + | 0 |
| AT3G21720 | Isocitrate lyase | SALK_004127.35.10.x | promoter | 0 | 0 | 0 | 0 | nd | nd | nd | nd |
| AT3G52960 | PrxIIE | SALK_064512.50.75.x | exon | − | − | = | − | nd | nd | nd | nd |
| AT4G02890 | UBQ14 | SALK_107827.16.05.x | exon | − | − | − | − | − | 0 | 0 | − |
| AT4G05050 | UBQ11 | SALK_069877.55.25.x | promoter | − | − | − | 0 | − | − | − | − |
| AT5G05750 | DNAJ | SALK_032572.56.00.x | promoter | 0 | 0 | 0 | 0 | nd | nd | nd | nd |
(–) Lines for which endosperm rupture is reached later than for WT, (0) is equal to WT, (+) endosperm rupture earlier than WT; nd, not determined. S, cold stratified, ABA, 1 μM ABA, incubation was in continuous light. AGI locus identifier in bold means that lines were selected for further investigation.
Propagation failed, therefore this line is not included in the detailed germination analysis.
Quantitative PCR
Total RNA was prepared from four samples each of 1000 endosperm caps and 150 radicles after 8 h and 18 h imbibition in medium without further additions and at 8, 18, 30, and 72 h imbibition in medium containing 10 μM ABA, respectively. 5 μg RNA was reverse transcribed using a mixture of random hexamers and oligo(dT)-primers with the Superscript III kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The cDNA was diluted to a final volume of 50 μl. 1 μl was used for quantitative PCR with the ABsolute QPCR SYBR Green ROX Mix (ABgene, Epsom, Surrey, UK) using the ABI PRISM 7300 Sequence Detection System (Applied Biosystems, Scoresby, Victoria, Australia). Amplification of a single product was confirmed with a melting curve according to the manufacturer's instructions. The efficiency (E) for every single well was determined using the LinRegPCR program and the relative expression for each well was calculated as E–Ct. Expression data were normalized to the geometric mean of actin-7, previously shown to be constitutive in cress seeds during germination (data not shown): E–Ctsample/E–Ctstamdard (Pfaffl, 2001).
The means are averages from 3–4 biological replicates. Primers used for reference gene actin were Act7-F (GGTCGTACAACCGGTATTGT) and Act7-R (GAAGAGCATACCCCTCGTA). Primers for the transcripts to be quantified were qPAt3g52969-F (CAGCTGTTAAGGAGGAGAC), qPAt3g52969-R (CTGACTCAACTCTCTCCTAC), qPAt1g07890-F (CGAAGGTGCCTGGACTTCA), qPAt1g07890-R (AGACCAGCTGAATGAGACCT), qPAt2g18980-F (CAGGAGCACATACAATAGGA) and qPAt2g18980-R (TAAGAGTCGGATCGATACGT).
Protein extraction and peroxidase activity assay
All protein extracts were produced from after-ripened seeds. Proteins were extracted from radicles and endosperm caps of cress seeds or from embryos and covering tissues of tobacco by homogenizing the tissue in 200 mM TRIS, 0.25 mM EDTA, 5 mM DTT, pH 8. The extraction buffer used for tobacco seeds contained, in addition, 1 M NaCl. The extract was centrifuged at 12 000 rpm at 4 °C. The clear supernatant was used for the enzyme assays. Protein content was determined with the Bio-Rad protein assay, using bovine γ-globulin as standard. Peroxidase activities were measured as oxidation of guaiacol (0.4 mM in 50 mM K-phosphate buffer, pH 7) in the presence of 2 mM H2O2. The reaction was followed spectrophotometrically as the increase of absorption at 450 nm for 30 min. A standard curve was produced using defined amounts of horseradish peroxidase (Sigma-Aldrich, Steinheim, Germany).
Histochemical detection of peroxidase activity
Seeds were cut in half and incubated in 0.2% w/v TMB (3,5,3′,5′-tetramethylbenzidine-HCl, Sigma, Germany) and 1 mM H2O2 in 20 mM phosphate buffer, pH 6.5. When staining was visible, seeds were washed in the buffer and photographed with a Leica DCF480 digital camera (Bensheim, Germany) attached to a stereomicroscope (Leica Mz 12.5).
SALK line genotyping
Appropriate SALK lines with a T-DNA insertion in the exon or putative promoter region were chosen according to the best Arabidopsis NCBI Blast hit for each cress cDNA identified in the subtractive library. Supplementary Table S1 (available at JXB online) shows the accession numbers of lines and the insertion details. Plants were propagated and genotyped. For genotyping of each line, leaf DNA was extracted according to Edwards et al. (1991). 1 μl of DNA per PCR reaction was used as a template. Primers to confirm T-DNA insertion were designed as suggested by the T-DNA Express homepage (http://signal.salk.edu/cgi-bin/tdnaexpress) using the LBA1 Primer (5′-TGGTTCACGTAGTGGGCCAT-3’) and PCRs were run with 60 °C annealing temperature. Only seeds of homozygous plants were used for germination kinetics.
To test if the genes that were analysed in the SALK lines were expressed during germination, RNA was extracted from 24 h imbibed seeds of WT Columbia and reverse transcribed into cDNA as described above. PCRs were run with primer pairs specific to each of the transcripts.
Statistical analysis
One-way ANOVA followed by Tukey's multiple comparison test was calculated for peroxidase assay data and germination data using GraphPad Prism software 4.03 (GraphPad Software Inc., La Jolla, USA).
Results
Generation and analysis of an endosperm cap Suppression Subtraction Hybridization (SHH) cDNA library
Under the conditions used in our study, populations of cress seeds completed testa rupture at 8–10 h after imbibition. Shortly after, at around 12 h, endosperm weakening set in, and the population reached 70–80% endosperm rupture at 18 h (CON in Fig. 2A; Müller et al., 2009). It was decided to sample seeds for dissection at 8–10 h (after testa rupture, before the onset of endosperm weakening) and at 18 h. At the latter point, only intact endosperm caps, which would by this time be strongly weakened, were sampled. RNA was extracted from these endosperm caps and a subtractive cDNA library was constructed with the SSH-technique of transcripts specifically up-regulated in the endosperm cap between 8 h and 18 h. Our choice of samples led to an enrichment of transcripts expressed during endosperm weakening and late germination. Several thousand clones were obtained, of which 144 were sequenced, yielding 118 unique sequences that were subjected to a BLAST search in the NCBI database. Out of these 118 sequences 62 were encoding rRNAs, 3 ribosomal proteins and 2 translation factors, indicating an increase in translation rate during late germination. Of the 56 non-rRNA sequences, 10 cDNA sequences code for proteins with unknown function. The 56 cDNA sequences are available in the category EST on the NCBI website (www.ncbi.nlm.nih.gov).
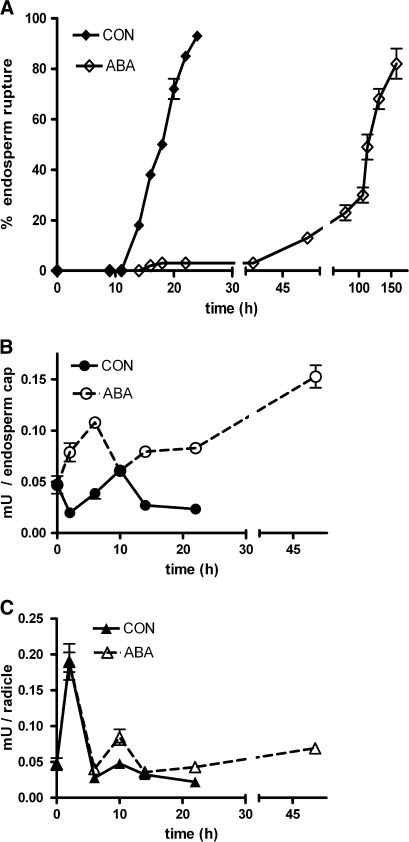
Fig. 2.
Tissue-specific peroxidase activity during cress seed germination. (A) Germination kinetics of cress seeds in continuous light at 18 °C in medium without (CON, control) or with 10 μM ABA added; the percentage of endosperm rupture was scored over time. (B, C) Peroxidase activity of protein extracts from micropylar endosperm caps (B) and radicles (C) dissected at different points during the germination process after imbibition in medium without (CON) or with 10 μM ABA added. Means ±SE of at least three biological replicates are presented.
For 90% of the remaining 46 sequences, the closest homologues were Arabidopsis cDNA sequences with an average of around 88% nucleotide sequence similarity (see Supplementary Table S1 at JXB online). This confirms the validity of our approach, as we would expect most hits to be Arabidopsis transcripts considering the close phylogenetic relationship between cress and Arabidopsis and the abundance of Arabidopsis data in the NCBI database. The NCBI accession numbers of the cDNAs obtained, as well as the identity of the most significant Arabidopsis BLAST hit and the corresponding AGI locus identifier are listed in Supplementary Table S1 at JXB online.
The list of transcripts identified by their close homology to Arabidopsis transcripts contained several transcripts which are known to be up-regulated during seed germination in other endospermic species, although in most cases not specifically in the micropylar endosperm cap. Examples include isocitrate lyase (Chibani et al., 2006) and kat2 (Footitt et al., 2006). Isocitrate lyase is a key enzyme of the glyoxylate cycle which plays a role in storage lipid mobilization (Eckardt, 2005), which, in turn, is important for seedling establishment following germination (Footitt et al., 2006). An orthologue of kat2 (At2g33150) was identified: a peroxisomal 3-ketoacyl-CoA thiolase which Footit et al. (2006) found to be involved in lipid mobilization via beta-oxidation and to influence germination potential and subsequent seedling establishment. Both Arabidopsis and cress are known to store lipids. Significant amounts of storage lipids are present in the endosperm layer and are required for the maximum elongation of hypocotyls in darkness (Penfield et al., 2004). Lipid mobilization in the endosperm starts prior to its rupture. The early onset is evident in Arabidopsis (Pritchard et al., 2002; Penfield et al., 2004), castor bean (Marriott and Northcote, 1975), and tobacco (Manz et al., 2005).
The fact that we could confirm the up-regulation of these transcripts validates our subtractive library for transcripts up-regulated during seed germination. It also validates the cross-species approach using the non-sequenced species cress, for which the genome has not yet been sequenced, but which has a close phylogenetic relationship to Arabidopsis (Franzke et al., 2009), to yield new candidate genes involved in germination.
A screen was made of the germination behaviour of 15 homozygous transgenic Arabidopsis-lines from the SALK collection with inserts in an exon or putative promoter region of the genes identified in our subtractive cDNA library (Table 1). Three lines with insertion in the peroxidase genes that were identified in the cDNA library were chosen, as the strong representation of peroxidases was noticed, plus 12 lines for which homozyguous seeds plants were available in the SALK collection. Germination was screened by comparing percentages of endosperm rupture at two points after imbibition of fresh and after-ripened mutant (SALK line) seeds under four conditions to two wild-type (WT) seed batches from the same harvest. The conditions were 24 °C with and without stratification, 18 °C without stratification, and 24 °C in the presence of 1 μM ABA with stratification (Table 1). One Petri dish with 50 seeds was used for each treatment in the screen.
Lines which strongly differed in the timing of endosperm rupture from the WT under at least half of these conditions were considered to have a germination phenotype. More than 50% of the screened lines showed a germination phenotype (Table 1), which further validates the usefulness of a tissue-specific subtractive cDNA library to identify genes with roles in germination.
For lines which showed a germination phenotype in the screen, detailed germination kinetics of triplicates of 50 seeds per treatment were recorded (Table 2; see Supplementary Fig. S1 at JXB online). The expression of the transcript affected in the SALK lines was confirmed in WT seeds at 24 h after imbibition, i.e. before radicle protrusion (data not shown).
Table 2.
Percentages of testa and endosperm rupture at selected times after imbibition for fresh and after-ripened seeds of Arabidopsis WT and SALK lines that showed a germination phenotype in the screen
| Mutated gene | Line | Fresh (48 h) |
AR (41 h) |
||
| Testa rupture (%) | Endosperm rupture (%) | Testa rupture (%) | Endosperm rupture (%) | ||
| WT Columbia | 52±4 | 45±8 | 83±7 | 76±6 | |
| Aspartyl protease | SALK_021485.37.45.x | 24±2 | 18±3 | 58±1 | 49±1 |
| PP2C | SALK_016641.56.00.x | 22±5 | 13±3 | 26±6 | 22±5 |
| UBI11 | SALK_069877.55.25.x | 19±4 | 15±4 | 47±6 | 39±4 |
| UBI14 | SALK_107827.16.05.x | 25±2 | 17±3 | 49±6 | 44±5 |
| AtPrxIIE | SALK_064512.50.75.x | 19±2 | 13±2 | nd | nd |
| AtPrx16 | SALK_028328.55.00.x | 79±4 | 71±3 | nd | nd |
| AtAPX01 | SALK_000249.38.45.x | 100±0 | 91±3 | nd | nd |
Endosperm rupture of imbibed seeds was recorded over time at 24 °C in continuous light. Means of three replicates of 50 seeds ±SE are given. AR, after-ripened; nd, not determined. All differences in endosperm rupture to the WT are statistically significant at P <0.05 in the after-ripened and at P <0.01 in the fresh state.
Germination kinetics of fresh and after-ripened seeds were recorded and one point in time after imbibition was selected to compare the percentages of endosperm rupture that had been reached by the different lines (Table 2). As expected, after-ripened seeds germinated faster and had reached around 80% endosperm rupture for WT at 41 h, while the SALK lines listed in Table 2 had reached a maximum of around 50% endosperm rupture. Fresh seeds took longer to complete germination, with the WT reaching around 50% endosperm rupture at 48 h. All lines showing a delay in endosperm rupture in the after-ripened state also showed a delay in the fresh state. The germination phenotypes were thus independent of the progression of after-ripening. It was also noted that testa and endosperm rupture were equally affected, thus the time span between testa and endosperm rupture remained roughly constant.
Only two lines completed germination earlier compared to the WT. These were the SALK lines for At1g07890 (AtApx01, ascorbate peroxidase) and At2g18980 (AtPrx16, putative class III peroxidase). Here and in the following text, the nomenclature for peroxidases used in the PeroxiBase database (Bakalovic et al., 2006) is followed. Figure 1 shows the germination kinetics in the fresh state for these two lines as well as for the third peroxidase SALK line, which carries an insertion in AtPrxIIE (atypical 2-cysteine peroxiredoxin (type II, type V).
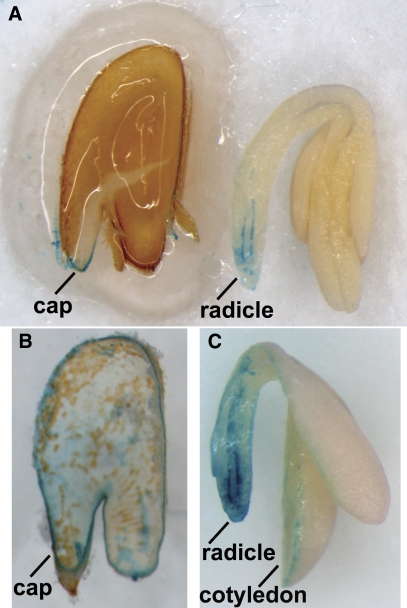
Fig. 1.
Peroxidase activity histostain of longitudinal sections of cress seeds after 16 h of imbibition. (A) Longitudinal section of cress embryo and seed-covering layers (endosperm, testa). After 2 min in the staining solution, the micropylar endosperm and the radicle display staining on the cut surface. (B) Longitudinal section of a cress endosperm separated from the testa after 15 min of staining. Cell outlines can be observed over the whole endosperm surface, and the cut surface is stained completely. (C) Longitudinal section of a cress embryo. After 8 min staining, the colour spread over the whole radicle, but not the cotyledons. ‘cap’=micropylar endosperm cap.
Thus, all three peroxidase SALK lines that were obtained from the library showed a germination phenotype. It was therefore decided to quantify transcript levels of the three individual peroxidases in different tissues of germinating cress seeds. In addition, total peroxidase activity was investigated in the micropylar endosperm cap as well as in the radicle of cress seeds.
Peroxidase activity develops specifically in the radicle and endosperm cap of germinating cress seeds and is increased by ABA
It was decided both to localize and to quantify peroxidase activity in different tissues of cress seeds. Peroxidase activity could be localized by TMB staining in longitudinal sections of germinating cress seeds (Fig. 1). Peroxidase activity was first detected in the endosperm cap, but not the non-micropylar endosperm and the radicle tip (Fig. 1A). When seeds were left longer in the staining solution, staining proceeded to the whole radicle and the complete endosperm (Fig. 1B, C), indicating activity in these parts, but not in the cotyledons. Staining mostly outlined the cells, indicating peroxidase activity in the cell wall rather than the cytoplasm, although accessibility might be an issue. Such apoplastic peroxidases can either be soluble (extractable with low-salt buffer) or more tightly bound to the cell wall (extractable only with high-salt buffer) (Fecht et al., 2001). In longitudinal sections of Arabidopsis seeds, peroxidase activity could be observed in the whole endosperm, the radicle tip, and at the edges of the cotyledons (data not shown). No staining was visible without the addition of H2O2 for both Arabidopsis and cress, confirming the specificity of the stain for peroxidase activity.
To quantify the peroxidase activity in the two most active cress seed tissues, that is in the radicle and the endosperm cap, the ability of protein extracts (sampled at different points in time during germination) to oxidize guaiacol in the presence of H2O2, was measured (Fig. 2). Since ABA is an inhibitor of cress seed germination which specifically targets endosperm weakening and rupture (Fig. 2A; Müller et al., 2006), peroxidase activity in seeds imbibed in medium with 10 μM ABA was also analysed. The peroxidase activity time-course presented in Fig. 2 was obtained with samples of soluble proteins extracted with a TRIS buffer, thus peroxidases bound tightly to the cell wall will not be present in the extracts. Peroxidase activity in micropylar endosperm caps (Fig. 2B) and radicles (Fig. 2C) of cress seeds was assayed in dry seeds and at six points during the germination process. No clear difference in activity between the two tissues was observed in the control treatment apart from one very high, but reproducible value in the radicle at 2 h. This peak was not affected by ABA in the germination medium. ABA led to tissue-specific activity induction: peroxidase activity was strongly enhanced by the addition of ABA in endosperm caps (this effect is statistically significant at all points except 10 h, P < 0.01). Radicles were less affected by ABA treatment, the only significant (P < 0.05) effect was observed at 22 h.
The endosperm-specific induction of peroxidase activity by ABA was also observed in tobacco seeds as measured in a guaiacol-assay: activity was below the detection limit in protein extracts of embryos isolated from germinating seeds, while the covering layers, consisting mostly of the thick endosperm, possessed a measurable activity which doubled when seeds were incubated in ABA-containing medium. Activities in the covering layers were CON, 0.275±0.012 mU seed−1; ABA, 0.559±0.034 mU seed−1. Seeds were imbibed for 120 h, at which time all seeds had testa rupture, and c. 10% of the CON seeds and none of the ABA seeds had endosperm rupture. Only tissues from seeds with unruptured endosperm were used in the assay. No activity could be detected in whole dry seeds. Activity in whole seeds increased upon imbibition from 0.155±0.025 (0 h) to 0.183±0.016 at 48 h CON, and to 0.225±0.008 at 48 h ABA. Taken together, it is possible that the peroxidase activity at 48 h localizes to the endosperm.
Tissue-specific peroxidase gene expression
Quantitative PCR was used to quantify the expression of the three peroxidases found in our subtractive library to be up-regulated during cress endosperm weakening (Fig. 3). The three cDNAs showed the highest similarities to AtAPx01, AtPrx16, and AtPrxIIE. In addition to expression in the endosperm cap, transcript levels in the non-micropylar endosperm, the radicle, and the cotyledons were investigated since peroxidase activity histostains, as well as activity assays, showed different activity in all tissues. Expression was investigated at the two points used for the subtractive library, i.e. at 8 h and 18 h after imbibition for all four tissues. In addition, micropylar endosperm caps and radicles were sampled from seeds imbibed in medium containing ABA at four different time points. Transcript levels in the dry micropylar end were also investigated. In the dry state, radicles and endosperm caps cannot be separated. RNA was thus extracted from radicles with the surrounding micropylar endosperm cap and the attached testa. Primers were designed based on the partial cress cDNAs obtained from the library.
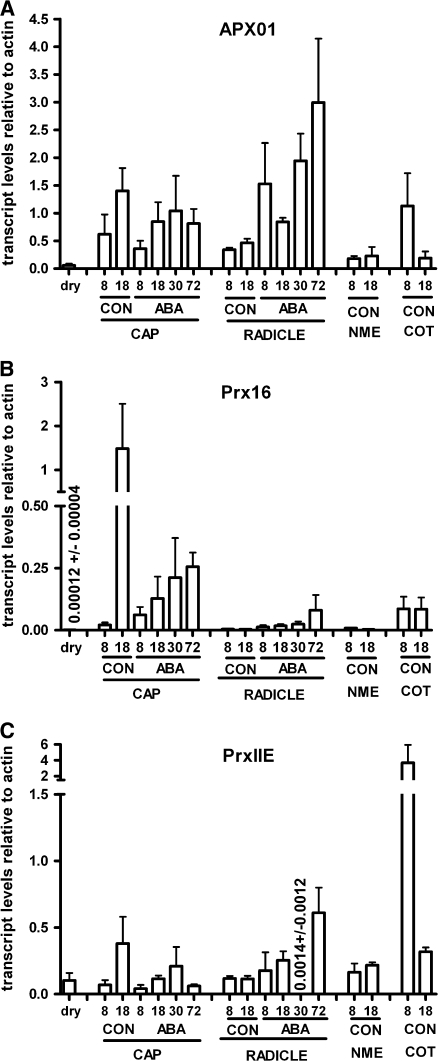
Fig. 3.
Transcript levels of the three peroxidases identified in the subtractive cDNA library in different cress seed tissues and in dry micropylar seed ends (radical + cap) at different points during germination (in hours) in the absence (CON, control) and presence of 10 μM ABA. Transcript abundance was determined by qPCR and normalized against actin abundance. Note the different scales on the y-axes. Means of three biological replicates ±SE are shown. Cap, micropylar endosperm cap; Nme, non-micropylar endosperm; Cot, cotyledons.
Transcripts of APX01 and PrxIIE were both already present in dry radicles and endosperm caps. Cress seeds thus store mRNAs of these two peroxidases during seed maturation and desiccation, which points to a function in the very early stages of germination before de novo mRNA synthesis is activated.
For all three transcripts a significant increase of transcript levels from 8 h to 18 h was apparent in the micropylar endosperm cap in accordance with the results from the subtractive library (Fig. 3). ABA inhibited the increase of transcript abundance in the micropylar endosperm cap that took place in its absence.
No corresponding increase was observed in the radicle or the non-micropylar endosperm in any of the transcripts. The increase was thus specific to the micropylar endosperm cap. APX01 showed a higher expression than Prx16 and PrxIIE. In contrast to the histostains, a rather high expression in cotyledons was evident for APX01 and PRXIIE. Interestingly, the abundance of both transcripts decreases in the cotyledons during germination from 8 h to 18 h imbibition. A low level of peroxidases would be in accordance with our histostains, which showed only a weak signal in the cotyledons at 16 h after imbibition. The accumulation of APX01 transcripts was strongly induced in the radicle by ABA.
Prx16 showed a strong differential expression between the micropylar endosperm cap and all other seed parts that were tested: at all sampling points, transcript levels were significantly higher in the endosperm caps than in the radicles, in accordance with the observations of tissue-specific peroxidase-activity in tobacco seeds.
In PrxIIE, endosperm cap transcript levels did not change significantly in the presence of ABA, remaining at the level also found at 8 h CON. Expression in the radicle, on the other hand, increased over time in the presence of ABA, interrupted by a strong, but highly reproducible, decrease in expression in the radicle at 30 h in the presence of ABA, followed by a strong increase at 72 h ABA.
Discussion
A tissue-specific subtractive cDNA library for the micropylar endosperm of germinating cress seeds
A subtractive cDNA library was used to identify transcripts which are up-regulated in the micropylar endosperm cap late during the germination of cress seeds.
Our library included cDNAs of genes known to play roles during germination as well as novel candidates. Two genes that have been characterized in the context of seed germination and dormancy are transparent testa 4 and 5 (tt4 and tt5), which encode for chalcone synthase and chalcone flavonone isomerase, respectively, both of which play a role in flavonoid synthesis (Debeaujon et al., 2000). Seeds carrying mutations in these genes have light-coloured seed coats which are more permeable than WT testas. The mutants have been described as being less dormant, and to complete germination earlier than the corresponding WT.
The only transcript identified whose expression has been investigated in the context of endosperm weakening and rupture is β-1,3-glucanase. β-1,3-glucanase is expressed in the micropylar endosperm of germinating tobacco seeds, and antisense lines show a delayed endosperm rupture (Leubner-Metzger, 2003; Leubner-Metzger and Meins, 2001). A role in endosperm weakening has been hypothesized in tobacco and other Solanaceae seeds in which the transcript was found to be expressed in the endosperm cap (Petruzzelli et al., 2003).
Other transcripts have not yet been investigated in the context of seed germination, but the corresponding mutants show a phenotype in our germination screen. An orthologue of At3g12620, which encodes for a member of the protein phosphatase 2C (PP2C) family, was identified. The insertion mutant completed germination later than the WT. PP2C enzymes are a large family with over 70 members in Arabidopsis, many of which play roles in various plant signal transduction processes. For example, the PP2Cs ABI1 and ABI2 are negative regulators of ABA signalling (Rodriguez, 1998), and the family has been hypothesized to play a role in stress signalling by negatively regulating stress-induced MAP kinase pathways (Schweighofer et al., 2004), which in turn is interconnected with ABA signalling.
Nearly half of the subtractive cDNA library consisted of clones encoding rRNA or ribosomal proteins, indicating an increase in translation during germination. De novo synthesis of ribosomal components appears to be required for normal and rapid germination and subsequent seedling growth (reviewed by Holdsworth et al., 2007). In accordance with this hypothesis, the transcriptome of non-germinating COMATOSE (cts-1) mutant seeds resembled that of germinating wild-type seeds, while the proteome of cts-1 resembles dormant seeds, which suggests that this mutant cannot translate the accumulated germination-related RNAs (Footitt et al., 2006).
An insertion in the orthologue of the aspartyl protease At1g01300 also led to a delay in endosperm rupture compared to the WT. In Arabidopsis seeds, vacuolation of protein storage vacuoles is first evident in the endosperm cap prior to endosperm rupture and takes place only much later in the non-micropylar endosperm and the embryo (Bethke et al., 2007). The vacuolation is promoted by GA and inhibited by ABA in accordance with these substances’ influence on endosperm weakening and rupture. In other species such as Datura and tomato, protein body degradation also occurs only when the seeds are induced to germinate (Mella et al., 1995; Nonogaki et al., 1998). In Arabidopsis and cress seeds with their single cell layer of endosperm, the mobilization of protein bodies in the endosperm cap is likely to serve a non-nutritional function in the control of germination in addition to delivering amino acids for translation.
The three ubiquitins that were identified in our subtractive library as being up-regulated during late germination in the cress endosperm cap, UBI10, UBI11, and UBI14, belong to the same subtype of ubiquitin genes (Chih-Wen Sun, 1997). It is noteworthy that so many up-regulated ubiquitins were found, considering that these transcripts are often used for normalization of quantitative PCR data on the assumption that they are constantly expressed. The same goes for elongation factor 1-α (At5g60390). Ubiquitins mediate proteolytic pathways: they can be covalently linked to proteins, which are thereby flagged for proteolysis by proteasome complexes (reviewed by Smalle and Vierstra, 2004).
Mutants with an insertion in UBI11 and UBI14, respectively, germinated more slowly than the corresponding WT. This might be due to a reduced function of proteolysis in hormonal pathways known be involved in germination, particularly the GA pathway. GA signal transduction, which promotes germination, involves the degradation of negative regulators by the proteasome, for example, the DELLA proteins RGA in Arabidopsis or Slender1 in barley (Dill et al., 2001; Fu et al., 2002). Intact DELLA proteins repress germination by inhibiting testa rupture and by stimulating ABA accumulation which, in turn, inhibits endosperm rupture (Piskurewicz et al., 2009). It is possible that the absence of one of the ubiquitins in our mutants leads to reduced DELLA proteolysis, which in the endosperm cap would lead to reduced endosperm weakening and a delay in endosperm rupture. In addition, protein turnover is important during seed germination, and will be impaired if ubiquitins are non-functional. Ubiquitins are also involved in lupin (Lupinus albus) seed germination (Ferreira et al., 1995).
Seed peroxidases are regulated by ABA in a tissue-specific manner
Tissue-specific peroxidase activity was investigated in germinating cress and tobacco seeds. Histostaining showed peroxidase activity to be higher in the micropylar endosperm cap than in the non-micropylar endosperm, and higher in the radicle than in the cotyledons. Peroxidase activity was also found specifically in the micropylar endosperm of germinating tomato seeds (Morohashi, 2002). This spatial pattern might therefore be more general.
A peak in peroxidase activity was observed in the radicle at 2 h after imbibition. Imbibition and the return to full metabolism is a strong stress for the seed. (Bewley, 1997a; Bailly, 2004; Kranner and Birtic, 2005). Stress commonly leads to an increase in ROS production, leading to a more oxidative environment in the cell, oxidative damage to macromolecules (Möller et al., 2007) and, ultimately, to cell death (Kranner et al., 2006). Seeds activate their antioxidant system upon rehydration (Kranner and Birtic, 2005), as has been demonstrated for wheat and pine seeds (De Gara et al., 1997; Tommasi et al., 2001). The importance of the antioxidant machinery for successful rehydration has also been documented in the resurrection plant Myrothamnus flabellifolia (Kranner et al., 2002).
Of the peroxidases identified in our subtractive cDNA library, APX, which localizes to the cytosol, as well as PrxIIE which is predicted by its protein sequence to localize to the mitochondria or peroxisome (SherLoc prediction, Höglund et al., 2006) are considered to be part of the antioxidant machinery, as they scavenge H2O2. A rapid rise in ascorbate content and APX activity during the first stages of germination has been demonstrated in wheat seeds. (Cakmak et al., 1993; De Gara et al., 1997).
Interestingly, the insertion mutants atapx01 as well as atprx16 completed germination earlier than the corresponding WT. Davletova et al. (2005) showed that a different apx1 mutant is characterized by a higher incident of protein oxidation, as the concentration of H2O2 in the cytoplasm increases. Protein oxidation has been associated with loss of dormancy in sun flowers (Oracz et al., 2007) and with germination of Arabidopsis seeds (Job et al., 2005). ROS have also been shown to elicit growth responses in different plant parts (Pasternak et al., 2005), which fits with the earlier testa and endosperm rupture of our atapx01 and atprx16 SALK lines.
Furthermore, H2O2 promotes the oxidative catabolism of ABA and, potentially, other inhibitors, as Wang et al. (1998) showed in dormant barley seeds. H2O2 also breaks dormancy and promotes germination of a range of other species including barley (Fontaine et al., 1994) and wild almond (Zeinalabedini et al., 2009). Dormant seeds generally have higher concentrations of endogenous ABA than non-dormant seeds and are more sensitive to its effects, so that a degradation of ABA would have a greater effect in dormant seeds (reviewed by Finch-Savage and Leubner-Metzger, 2006). The seeds used in our study showed only a very shallow dormancy in the fresh state, but germination was promoted in the case of the atapx01 and atprx16 mutants. It should be kept in mind that germination-promoting substances such as GA might also be oxidized by H2O2 and subsequently degraded.
The putative class III peroxidase AtPrx16 was predicted by its amino acid sequence to be targeted to the apoplast (SherLoc prediction; Höglund et al., 2006), as is characteristic for class III peroxidases (Cosio and Dunand, 2009). Schopfer et al. (2001) found that peroxidases are released by radish seed coats during late germination and early seedling development. Peroxidase release is promoted by GA, and prevented by ABA. Apoplastic peroxidases have been postulated to have two opposing functions in cell wall remodelling: They can cross-link cell wall macromolecules, leading to a stiffer cell wall, but are also involved in the production of apoplastic hydroxyl radicals (Chen and Schopfer, 1999), which can cleave cell wall polysaccharides, leading to cell wall loosening. Cell wall loosening by hydroxyl radicals has been shown to play a role in cress seed germination and seedling growth (Müller et al., 2009).
Besides acting as antioxidants, peroxidases can also be involved in ROS production. This is particularly true of class III peroxidases, which can, under certain physiological conditions, produce superoxide (Minibayeva et al., 2009) as well as hydroxyl radicals (Chen and Schopfer, 1999).
A reduced ROS production caused by a mutated class III peroxidase as well as an increase in ROS caused by the lack of detoxyfiying peroxidase activity will influence ROS signalling. ROS signals play a role in a variety of hormonal signalling pathways, for example, ethylene and ABA signalling include ROS produced by NADPH-oxidases (Kwak et al., 2006). Shifting the ROS-balance in either direction might also influence redox signal transduction pathways (reviewed by Shao et al., 2008).
In our samples, overall peroxidase activity was increased by ABA in the micropylar endosperm cap, but to a much lesser extent in the radicle (Fig. 2). As Arabidopsis contains 115 known peroxidases listed in the PeroxiBase (Bakalovic et al., 2006), the changes in activity cannot be attributed to any particular peroxidase. Peroxidase activity in the radicle peaked at 2 h after imbibition both in the presence and in the absence of ABA. This early after imbibition, it is possible that the peroxidases have been translated from stored mRNAs. It could be shown for two out of three peroxidases found in our cDNA library that their mRNA is already present in the dry state (Fig. 3). The insensitivity of the peroxidase activity peak to ABA brings to mind the insensitivity of the testa rupture to ABA. The ABA-insensitivity points to peroxidases being regulated by ABA at the transcriptional rather than the translational or post-translational level: only when de novo transcription takes place does ABA have a stimulating effect. This hypothesis is in accordance with our qPCR data, where ABA seems to stimulate transcription of the three peroxidases tested. Transcripts of all three peroxidases tested accumulated specifically in the endosperm cap between 8 h and 18 h, and their expression was influenced by ABA in a tissue-specific manner (Fig. 3).
In conclusion, the close relatedness of the two Brassicaceae species cress and Arabidopsis is not only evident morphologically in that they have retained a thin endosperm layer in their mature seeds but also molecularly in the high similarities of their cDNA sequences. It was found that the high molecular relatedness was also true for the three specific peroxidases that were investigated in more detail. So far, germination phenotypes of large gene families are rare and our cross-species experiments, tissue-specific SSH library in cress combined with Arabidopsis mutants, appears to be a successful approach to this. Our results demonstrate that peroxidases have roles in germination and, as distinct phenotypes were obtained, particular peroxidases may serve distinct functions. This opens the field for future experiments on the specific roles of specific peroxidase genes. Our seed-centred approach with both species showed that a subtractive cress cDNA library specific for the micropylar endosperm provides genes of cross-species importance. A considerable portion of the Arabidopsis SALK lines that were screened, based on the cress endosperm cap cDNA library, turned out to have germination phenotypes. Gene orthologues thus seem to have retained conserved roles in seed germination of endospermic Brassicaceae.
Supplementary data
The supplementary data are available at JXB online. Table S1 lists genes identified in the subtractive cDNA library, and Fig. S1 presents germination kinetics of the three peroxidase-SALK lines.
Supplementary Material
Acknowledgments
Our work is funded by grants of the Deutsche Forschungsgemeinschaft (grant no. DFG LE720/6) to GL-M, which is gratefully acknowledged.
References
- Bailly C. Active oxygen species and antioxidants in seed biology. Seed Science Research. 2004;14:93–107. [Google Scholar]
- Bakalovic N, Passardi F, Ioannidis V, Cosio C, Penel C, Falquet L, Dunand C. PeroxiBase: a class III plant peroxidase database. Phytochemistry. 2006;67:534–539. doi: 10.1016/j.phytochem.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M. The Arabidopsis Book. American Society of Plant Biologists; 2008. Seed dormancy and germination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung YY, Still DW, Jones RL. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiology. 2007;143:1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. The Plant Cell. 1997a;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends in Plant Science. 1997b;2:464–469. [Google Scholar]
- Cadman C, Toorop P, Hilhorst H, Finch-Savage W. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Cakmak I, Strbac D, Marschner H. Activities of hydrogen peroxide-scavenging enzymes in germinating wheaseeds. Journal of Experimental Botanty. 1993;44:127–132. [Google Scholar]
- Chen F, Bradford KJ. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiology. 2000;124:1265–1274. doi: 10.1104/pp.124.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Schopfer P. Hydroxyl-radical production in physiological reactions. A novel function of peroxidase. European Journal of Biochemistry. 1999;260:726–735. doi: 10.1046/j.1432-1327.1999.00199.x. [DOI] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiology. 2006;142:1493–1510. doi: 10.1104/pp.106.087452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih-Wen Sun JC Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. The Plant Journal. 1997;11:1017–1027. doi: 10.1046/j.1365-313x.1997.11051017.x. [DOI] [PubMed] [Google Scholar]
- Cosio C, Dunand C. Specific functions of individual class III peroxidase genes. Journal of Experimental Botany. 2009;60:391–408. doi: 10.1093/jxb/ern318. [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. The Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gara L, Pinto MC, Arrigoni O. Ascorbate synthesis and ascorbate peroxidase activity during the early stage of wheat germination. Physiologia Plantarum. 1997;100:894–900. [Google Scholar]
- Debeaujon I, Léon-Kloosterziel K, Koornneef M. Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiology. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun T-P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proceedings of the National Academy of Sciences, USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L, Waters L. Long-lived messenger RNA: evidence from cotton seed germination. Science. 1965;147:410–412. doi: 10.1126/science.147.3656.410. [DOI] [PubMed] [Google Scholar]
- Eckardt NA. Peroxisomal citrate synthase provides exit route from fatty acid metabolism in oilseeds. The Plant Cell. 2005;17:1863–1865. [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecht M, Maier P, Horst WJ. Peroxidase activity in the leaf apoplast is a sensitive marker for Mn toxicity and Mn tolerance in Vigna unguiculata (L.) Walp. Developments in Plant and Soil Sciences. 2001;92:264–265. [Google Scholar]
- Ferreira RMB, Ramos PCR, Franco E, Ricardo CPP, Teixeira ARN. Changes in ubiquitin and ubiquitin–protein conjugates during seed formation and germination. Journal of Experimental Botany. 1995;46:211–219. [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Fontaine O, Huault C, Pavis N, Billard JP. Dormancy breakage of Hordeum vulgare seeds: effect of hydrogen peroxide and stratification on glutathione level and glutathione reductase activity. Plant Physiology and Biochemistry. 1994;32:677–683. [Google Scholar]
- Footitt S, Marquez J, Schmuths H, Baker A, Theodoulou FL, Holdsworth M. Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. Journal of Experimental Botany. 2006;57:2805–2814. doi: 10.1093/jxb/erl045. [DOI] [PubMed] [Google Scholar]
- Franzke A, German D, Al-Shehbaz IA, Mummenhoff K. Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon. 2009;58:425–437. [Google Scholar]
- Fu X, Richards DE, Ait-alia T, Hynes LW, Ougham H, Peng J, Harberd NP. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. The Plant Cell. 2002;14:3191–3200. doi: 10.1105/tpc.006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglund A, Blum T, Brady S, Dönnes P, Miguel J, Rocheford M, Kohlbacher O, Shatkay H. Significantly improved prediction of subcellular localization by integrating text and protein sequence data. Pacific Symposium on Biocomputing. 2006 Maui, Hawaii, USA. [PubMed] [Google Scholar]
- Holdsworth M, Bentsink L, Soppe W. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- Holdsworth M, Finch-Savage W, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends in Plant Science. 2007;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiology. 2005;138:790–802. doi: 10.1104/pp.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranner I, Beckett R, Wornik S, Zorn M, Pfeifhofer H. Revival of a resurrection plant correlates with its antioxidant status. The Plant Journal. 2002;31:13–24. doi: 10.1046/j.1365-313x.2002.01329.x. [DOI] [PubMed] [Google Scholar]
- Kranner I, Birtíc S. A modulating role for antioxidants in desiccation tolerance. Integrative and Comparative Biology. 2005;45:734–740. doi: 10.1093/icb/45.5.734. [DOI] [PubMed] [Google Scholar]
- Kranner I, Birtíc S, Anderson K, Pritchard H. Glutathione half-cell reduction potential: A universal stress marker and modulator of programmed cell death? Free Radicals in Biology and Medicine. 2006;40:2155–2165. doi: 10.1016/j.freeradbiomed.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Science Research. 2005;15:281–307. [Google Scholar]
- Kwak JM, Nguyen V, Schroeder JL. The role of reactive oxygen species in hormonal responses. Plant Physiology. 2006;141:323–329. doi: 10.1104/pp.106.079004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G. Functions and regulation of β-1,3-glucanase during seed germination, dormancy release and after-ripening. Seed Science Research. 2003;13:17–34. [Google Scholar]
- Leubner-Metzger G. β-1,3-Glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. The Plant Journal. 2005;41:133–145. doi: 10.1111/j.1365-313X.2004.02284.x. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G, Fründt C, Vögeli-Lange R, Meins F., Jr Class I β-1,3-glucanase in the endosperm of tobacco during germination. Plant Physiology. 1995;109:751–759. doi: 10.1104/pp.109.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G, Meins F., Jr Antisense-transformation reveals novel roles for class I β-1,3-glucanase in tobacco seed after-ripening and photodormancy. Journal of Experimental Botany. 2001;52:1753–1759. doi: 10.1093/jexbot/52.362.1753. [DOI] [PubMed] [Google Scholar]
- Liu P-P, Koizuka N, Homrichhausen T, Hewitt J, Martin R, Nonogaki H. Large-scale screening of Arabidopsis enhancer-traps lines for seed germination-associated genes. The Plant Journal. 2005;41:936–944. doi: 10.1111/j.1365-313X.2005.02347.x. [DOI] [PubMed] [Google Scholar]
- Manz B, Müller K, Kucera B, Volke F, Leubner-Metzger G. Water uptake and distribution in germinating tobacco seeds investigated in vivo by nuclear magnetic resonance imaging. Plant Physiology. 2005;138:1538–1551. doi: 10.1104/pp.105.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott K, Northcote D. The induction of enzyme activity in the endosperm of germinating castor-bean seeds. Biochemical Journal. 1975;152:65–70. doi: 10.1042/bj1520065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mella RA, Maldonado S, Sanchez RA. Phytochrome-induced structural changes and protein degradation prior to radicle protrusion in Datura ferox seeds. Canadian Journal of Botany. 1995;73:1371–1378. [Google Scholar]
- Minibayeva F, Kolesnikov O, Chasov A, Beckett RP, Lüthje S, Vylegzhanina N, Buck F, Böttger M. Wound-induced apoplastic peroxidase activities: their roles in the production and detoxification of reactive oxygen species. Plant, Cell and Environment. 2009;32:497–508. doi: 10.1111/j.1365-3040.2009.01944.x. [DOI] [PubMed] [Google Scholar]
- Möller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Morohashi Y. Peroxidase activity develops in the micropylar endosperm of tomato seeds prior to radicle protrusion. Journal of Experimental Botany. 2002;53:1643–1650. doi: 10.1093/jxb/erf012. [DOI] [PubMed] [Google Scholar]
- Müller K, Linkies A, Vreeburg RAM, Fry SC, Krieger-Liszkay A, Leubner-Metzger G. In vivo cell wall loosening by hydroxyl radicals during cress (Lepidium sativum L.) seed germination and elongation growth. Plant Physiology. 2009;150:1855–1865. doi: 10.1104/pp.109.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:864–877. doi: 10.1093/pcp/pcj059. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. The Plant Journal. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- Nonogaki H, Gee Oh, Bradford KJ. A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiology. 2000;130:1235–1245. doi: 10.1104/pp.123.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Nomaguchi M, Okumoto N, Kaneko Y, Matsushima H, Morohashi Y. Temporal and spatial pattern of the biochemical activation of the endosperm during and following imbibition of tomato seeds. Physiologia Plantarum. 1998;102:236–242. [Google Scholar]
- Oracz K, Bouteau HE, Farrant JM, Cooper K, Belghazi M, Job C, Job D, Corbineau F, Bailly C. ROS production and protein oxidation as novel mechanisms for seed dormancy alleviation. The Plant Journal. 2007;50:452–465. doi: 10.1111/j.1365-313X.2007.03063.x. [DOI] [PubMed] [Google Scholar]
- Pasternak T, Potters G, Caubergs R, Jansen MAK. Complementary interactions between oxidative stress and auxins control plant growth responses at plant, organ, and cellular level. Journal of Experimental Botany. 2005;56:1991–2001. doi: 10.1093/jxb/eri196. [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday A, Graham S, Graham I. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination in the endosperm. The Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE. The Plant Cell. 2004;16:2705–2718. doi: 10.1105/tpc.104.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzelli L, Müller K, Hermann K, Leubner-Metzger G. Distinct expression patterns of β-1,3-glucanases and chitinases during the germination of Solanaceous seeds. Seed Science Research. 2003;13:139–153. [Google Scholar]
- Piskurewicz U, Turečková V, Lacombe E, Lopez-Molina L. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO Journal. 2009;28:2259–2271. doi: 10.1038/emboj.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2001–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard S, Charlton W, Baker A, Graham I. Germination and storage reserve mobilization are regulated independently in Arabidopsis. The Plant Journal. 2002;31:639–647. doi: 10.1046/j.1365-313x.2002.01376.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez PL. Protein phosphatase 2C (PP2C) function in higher plants. Plant Molecular Biology. 1998;38:919–927. doi: 10.1023/a:1006054607850. [DOI] [PubMed] [Google Scholar]
- Schopfer P, Plachy C, Frahry G. Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, hydroxyl radicals) and peroxidase in germinating radish (Raphanus sativus L.) seeds controlled by light, gibberellin and abscisic acid. Plant Physiology. 2001;125:1591–1602. doi: 10.1104/pp.125.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Shao HB, Chu Ly, Lu ZH, Kang CM. Primary antioxidant free radical scavenging and redox signaling pathways in higher plant cells. International Journal of Biological Sciences. 2008;4:8–14. doi: 10.7150/ijbs.4.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annual Reviews of Plant Biology. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Tommasi F, Paciolla C, de Pinto MC, de Gara L. A comparative study of glutathione and ascorbate metabolism during germination of Pinus picea seeds. Journal of Experimental Botany. 2001;52:1647–1654. [PubMed] [Google Scholar]
- Zeinalabedini M, Majourhat K, Hernandez JA, Dicenta F, Martinez-Gomez P. Breaking seed dormancy in long-term stored seeds from Iranian wild almond species. Seed Science and Technology. 2009;37:267–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.