Abstract
Biliary bile salt composition of 677 vertebrate species (103 fish, 130 reptiles, 271 birds, 173 mammals) was determined. Bile salts were of three types: C27 bile alcohols, C27 bile acids, or C24 bile acids, with default hydroxylation at C-3 and C-7. C27 bile alcohols dominated in early evolving fish and amphibians; C27 bile acids, in reptiles and early evolving birds. C24 bile acids were present in all vertebrate classes, often with C27 alcohols or with C27 acids, indicating two evolutionary pathways from C27 bile alcohols to C24 bile acids: a) a ‘direct’ pathway and b) an ‘indirect’ pathway with C27 bile acids as intermediates. Hydroxylation at C-12 occurred in all orders and at C-16 in snakes and birds. Minor hydroxylation sites were C-1, C-2, C-5, C-6, and C-15. Side chain hydroxylation in C27 bile salts occurred at C-22, C-24, C-25, and C-26, and in C24 bile acids, at C-23 (snakes, birds, and pinnipeds). Unexpected was the presence of C27 bile alcohols in four early evolving mammals. Bile salt composition showed significant variation between orders but not between families, genera, or species. Bile salt composition is a biochemical trait providing clues to evolutionary relationships, complementing anatomical and genetic analyses.
Keywords: bile acids, cholesterol, enzymes, metabolism, molecular evolution, phylogeny
The purposes of this review are to summarize the bile salt composition of vertebrates and to relate differences in their bile salt structures to current concepts of vertebrate evolution.
Bile alcohols and bile acids are the amphipathic multifunctional end products of cholesterol metabolism in vertebrates (1). After their synthesis in the hepatocyte, they are converted to strong acids by “conjugation” at the terminal carbon of the side chain. Bile alcohols are conjugated by esterification with sulfate, whereas bile acids are conjugated by N-acylamidation with taurine or a taurine derivative or, less commonly, with glycine. Such conjugation results in the formation of molecules that are impermeant to the epithelium of the biliary tract and small intestine, a factor contributing to their high micellar concentrations in bile and small intestinal content. Bile alcohol sulfates and conjugated bile acids are collectively termed “bile salts”. Bile salts are reabsorbed from the distal intestine after secretion into the proximal intestine and returned to the liver, which removes them and resecretes them into bile. The end result is the accumulation of a bile salt pool that circulates between the intestine and the liver, this molecular flux being termed the enterohepatic circulation. Bile salts are not completely absorbed by the distal small intestine, with a fraction lost via fecal excretion; bile acid excretion is equivalent to bile acid biosynthesis from cholesterol in the steady state (1).
Bile acid biosynthesis, at least in mammals, results from at least two complex biochemical pathways (2, 3). Bile acid and presumably bile alcohol biosynthesis is under negative feedback control modulated at least in part by the nuclear hormone receptor farnesoid X receptor (FXR) (4–7) and the peptide fibroblast growth factor 19 (8). The ligand binding pocket of FXR varies in relation to bile salt structure (9).
Biliary bile alcohols and bile acids show remarkable structural diversity across animal species. No other class of small molecules in vertebrates exhibits such a variety of chemical structures. The diversity of structure was recognized at the end of the 19th century and identification of new bile alcohols and bile acids continues to this day. Structural variation occurs in the 19-carbon (C19) cyclopentanophenanthrene (steroid) nucleus in several ways: A/B ring juncture stereochemistry, sites of hydroxy or oxo groups, and orientation of hydroxyl groups (i.e., whether α or β). Structural variation in the side chain includes the length of the side chain, the presence and orientation of hydroxy groups, the presence of unsaturation, the stereochemistry of the C-25 carbon atom, and the site of the carboxyl group.
Because of the great structural diversity of bile alcohols and bile acids, attempts are being made by us and others to develop simplifying principles. Most but not all bile salts can be assigned to three great classes according to the length of the side chain and its terminal polar group. These classes are 27-carbon (C27) bile alcohols, C27 bile acids, and 24-carbon (C24) bile acids. It is useful to further simplify bile salt structure by introducing the idea of the “default” structure of nuclear hydroxylation in each of these three classes. The default structure has hydroxy groups at C-3 (from the 3-hydroxy group of cholesterol) and at C-7, as hydroxylation at C-7 by a cytochrome P450 enzyme (CYP7A1 or CYP7B1) is considered rate limiting in bile acid biosynthesis (2, 3). The default structure also has a functional group (primary alcohol or carboxyl group) at the terminal carbon atom of the side chain. Additional substituents may then be added to the default structure on the nucleus or the side chain or both.
A further complexity in bile acid metabolism is modification of bile salt structure by intestinal bacteria (1, 10). Bile acids whose hydroxy groups have been modified by bacteria are named secondary bile acids to distinguish them from primary bile acids that are formed from cholesterol in the hepatocyte. Secondary bile acids may be absorbed from the intestinal tract and thereby become part of the circulating bile acid pool. The major bacterial modification for C27 and C24 bile acids is dehydroxylation at C-7, which is preceded by deconjugation (10). The resulting unconjugated 7-deoxy bile acids are absorbed from the distal intestine, return to the liver, and are reamidated during hepatocyte transport and then resecreted into bile. Rehydroxylation at C-7, which results in formation of the original primary bile acid, occurs in some species (1). Rehydroxylation may also occur at a position other than C-7.
In addition to dehydroxylation at C-7, bacterial enzymes may also dehydrogenate (oxidize) hydroxy groups to form hydroxy-oxo bile acids or may epimerize hydroxy groups (α-hydroxy to β-hydroxy, or β-hydroxy to α-hydroxy). Such secondary bile acids may be further modified during hepatocyte transport. For example, 3β-hydroxy bile acids (so called “iso-bile acids”) are efficiently epimerized to 3α-hydroxy bile acids (11, 12). The oxo group of hydroxy-oxo bile acids may undergo reduction of the oxo group to form a hydroxyl group. Bacterial desaturation of the side chain of C24 bile acids to form Δ22,23-bile acids also occurs in some rodents. The process of bacterial modification and further hepatocyte modification of secondary bile acids has been termed “damage and repair” (1).
Bile alcohol sulfates are likely to undergo hydrolysis of their ester linkage by bacterial sulfatases, at least in some species. To what extent this occurs is not known. For 5α-cyprinol (3α,7α,12α,26,27-pentahydroxy-5α-cholestane), the main bile alcohol of cypriniform fish, the liberated bile alcohol is unlikely to be absorbed from the intestine because of the large number of hydroxy groups as well as the limited solubility of the unsulfated bile alcohol (13). As a result, in these fish, biliary bile alcohols are entirely primary, and the same is likely to be true for other species in which bile alcohol sulfates are present. Whether C27 bile alcohols undergo 7-dehydroxylation is not known.
Biliary bile acids, therefore, consist of a mixture of primary and secondary bile acids in conjugated form, whereas biliary bile alcohols are likely to be solely primary and present as ester sulfates. Each individual bile salt has an input, either de novo synthesis for primary bile acids and alcohols or intestinal absorption for newly formed secondary bile acids. Each bile salt is then conjugated and secreted into bile. Subsequent absorption from the distal intestine leads to the accumulation of a pool whose size depends on the rate of input and the efficiency of intestinal conservation. The size of the relative pools determines biliary biliary bile salt composition.
The synthesis of primary bile acids and bile alcohols is not thought to be influenced by diet. The formation and subsequent absorption of secondary bile acids is influenced in large part by the intestinal flora, which is itself shaped by intestinal anatomy (e.g., presence or absence of a cecum, length of the intestinal tract), intestinal peristalsis, diet, and genetic factors. The presence of a cecum permits an anaerobic flora to develop, and 7-dehydroxylation is mediated only by anaerobic bacteria. Information on the comparative anatomy of the large intestine has been ably summarized by Stevens (14)
FURTHER CONSIDERATIONS OF CHEMICAL STRUCTURE
Side chain functional group
The functional group at the end of the side chain determines the class of a bile salt. The mitochondrial enzyme cholesterol 27-hydroxylase (CYP27A1) mediates both hydroxylation at C-27 (forming C27 bile alcohols) and oxidation to a carboxyl group (forming C27 bile acids) (15). C24 bile acids are formed by a β-oxidation process in peroxisomes (2, 3, 16–19). A C24 bile alcohol (5α-petromyzonol; 3α,7α,12α,24-tetrahydroxy-5α-cholane) occurs uniquely in the lamprey (20).
Side chain length
The side chain of cholesterol is C8 as is that of C27 bile alcohols and C27 bile acids. Bile acids with 30, 29, 28, and 26 carbon atoms have been identified in amphibian bile (21, 22). C24 bile acids are the dominant bile acids in later evolving species including ray-finned fish, snakes, many birds, and most mammals (23). C25 bile acids (homo-cholanoic acids) have not been identified in biliary bile acids, probably because such compounds, if formed, undergo β-oxidation to form C23 (C24 nor) bile acids, at least in rodents (24). C23 (C24 nor) bile acids are present in small proportions in species having 23-hydroxy C24 bile acids (snakes, birds, and pinnipeds), as is shown in this paper.
A/B ring juncture
The cyclopentanophenanthrene nucleus is fully saturated in all primary bile salts. The junctures of the rings are denoted in en face views of the steroid nucleus by showing the configuration of the juncture hydrogen atom. When the hydrogen atom at C-5 is in the β configuration, the A/B ring juncture is cis; when it is in the α configuration, the A/B juncture is trans. The configuration of the A/B ring juncture determines the orientation of the hydroxy group at C-3. In 5β- (A/B cis) bile acids, the 3α-hydroxy group is equatorial; in 5α- (A/B trans) bile acids, the 3α-hydroxy group is axial. The B/C and C/D ring junctures are trans in all known bile acids. Enantiomers of bile acids have recently been synthesized (25).
Bile alcohols in the earliest evolving vertebrate species (the jawless fish or Agnatha, represented by extant hagfish and lampreys) have an A/B trans (5α) ring juncture, which leads to an overall flat (planar) orientation of the four rings of the steroid nucleus (1). In Chondrichthyes (sharks, skates, rays, and chimaerae), sometimes called “cartilaginous fish”, bile alcohols are A/B cis (5β), indicating that the ability to form 5β bile alcohols evolved quite early for vertebrates. In bile acids, the presence of an A/B trans (5α) ring juncture is denoted by the prefix “allo” in trivial nomenclature. Bile salts that have an A/B cis ring juncture have a ‘bent’ orientation of the A-ring relative to the other three rings of the steroid nucleus (1). To date, all C27 bile acids that have been characterized are A/B cis. A single C28 bile acid that is A/B trans (5α) has been isolated from toad bile (26). There remains the possibility that allo C27 bile acids exist and have not as yet been identified. There is one report of the chemical synthesis of allo C27 bile acids (27).
C24 allo (5α) bile acids do occur under three circumstances. First, they may be dominant primary bile acids, as occur in some birds and lizards (see below). Second, they may be trace primary bile acids, as have been described in the germ free rabbit (28). Finally, they may be formed by the intestinal bacteria and absorbed together with other secondary bile acids (29). The A/B ring juncture is not altered during transport through the hepatocyte.
Nuclear hydroxylation
Nuclear hydroxylation is the result of CYP-mediated hydroxylation (2, 3). If one accepts the concept of a default steroid nucleus with hydroxy groups at C-3 and C-7, then further modifications can be considered as additions to the default structure. In C27 bile alcohols and C27 bile acids, one additional nuclear hydroxylation occurs. The dominant site of additional hydroxylation is at C-12. Other sites of additional hydroxylation in C27 bile acids are at C-1, C-2, C-15, and C-16 (1).
In C24 bile acids, dominant sites of additional nuclear hydroxylation are at C-6, C-12, and C-16. Hydroxylation in a small number of species occurs at C-1, C-4, C-5, C-15, or C-19. In contrast to bile alcohols and C27 bile acids, two additional hydroxylations in the nucleus of C24 bile acids may occur. For example, some snakes have tetrahydroxy bile acids (C-3, C-7, C-12, and C-16) (30).
Side chain hydroxylation
The default structure of the C27 bile alcohol side chain has a hydroxy group at C-27. (In some nomenclature recommendations, substituents on the terminal carbon atom are designated as C-26 if the side chain has no additional hydroxylation). Additional hydroxylation to the default structure with a hydroxy group at C-27 may occur at C-24, C-25, or C-26. Some C27 bile alcohols have two additional hydroxylations on the side chain. As the nucleus commonly contains three hydroxy groups, such compounds are hexols; an example is the primary bile alcohol of Elasmobranchii (sharks, skates, and rays) called 5β-scymnol (3α,7α,12α,24,26,27-hexahydroxy-5β-cholestane) (31). C27 bile alcohols that are pentols, tetrols, and triols have also been identified (21, 22).
Some C27 bile acids have an unsubstituted side chain. Others undergo hydroxylation on the side chain at only one site. This may be at C-26 (amphibians and turtles), C-24 (amphibians and lizards), or C-22 (turtles). C24 bile acids hydroxylated at C-23 are common in birds and marine mammals. A C24 bile acid hydroxylated at C-22 (haemulcholic acid) is present in some fish (32).
In this review, we summarize available information on biliary bile alcohol and bile acid structure in fish, reptiles, birds, and mammals based on analyses of bile from 677 vertebrate species performed over the past three decades. Bile salts of amphibians are discussed only briefly because of their complexity and because existing analyses have generally not used state of the art techniques. In addition, compared with fish and reptiles, surveys of amphibian bile salts have been much more limited in covering the phylogenetic diversity of amphibian species.
Bile alcohols and bile acids are the result of complex biochemical pathways and they may be considered phenotypic traits. As such, they should provide information on phylogenetic relationships. We tabulate bile salt composition in different species and then compare information provided by bile salt structure with that provided by gene analyses or more traditional morphological evidence. A driving force for evolution should be increased survival value. Therefore, we discuss physiological function even though we have very little information on physicochemical or physiological properties of bile alcohol sulfates and C27 bile acids.
Our work builds on the work of many others, in particular that of G. A. D. Haslewood (33), now deceased, and T. Hoshita of Hiroshima University (21, 22). We note the synthetic efforts of the laboratory of G. Salen (34) and T. Iida (35, 36) who has developed new methods for synthesizing natural bile acids. We also acknowledge the extensive work on bile acid metabolism by Swedish workers in the school of bile acids launched by the late Sune Bergstrom (37) and continued by his pupils such as Sjövall (38), Norman, Lindstedt, and Danielsson (39) and in turn by their pupils such as Axelson, Gustafson(s), Björkhem, Wikwall, Einarsson, Angelin, Marschall, and many others who cannot be mentioned here.
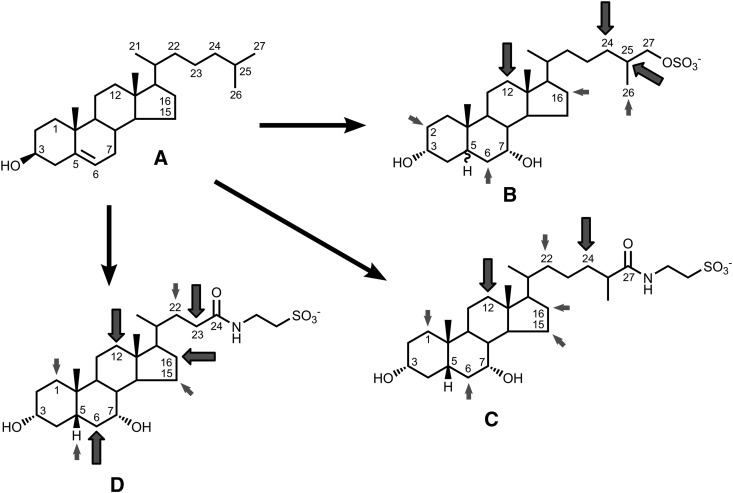
Table 1 summarizes the chemical structure of natural C27 bile alcohols. In the species tables, individual bile alcohols are denoted by Roman numerals. Table 2 summarizes C27 bile acid structure; in the species tables, C27 bile alcohols are denoted by capital letters. Table 3 summarizes C24 bile acid structure; in the species tables, Arabic numbers are used to indicate individual C24 bile acids. A detailed tabulation for C27 bile salts has been authored by Une and Hoshita (22) and their numbering scheme has also been included in Tables 1 and 2. Table 4 and supplementary Tables V–X tabulate bile salt composition for individual species using the descriptors of Tables 1–3. Table 4 gives bile salt composition in turtles and crocodilians and provides an example of the format used in supplementary Tables V–X. Supplementary Table V tabulates bile salt composition for nonperciform fish, supplementary Table VI for perciform fish, supplementary Table VII for squamates, supplementary Table VIII for snakes, supplementary Table IX for birds, and supplementary Table X for mammals. Species are denoted by their common English name. Systematic names of the species reviewed are easily found on the world wide web. Figure 1 shows the structure of the three classes of bile salts and their common and most of the uncommon sites of hydroxylation based on our own studies and those of many previous workers in the bile salt field.
TABLE 1.
Biliary C27 bile alcoholsa
| U & Hb | Ring A | Ring B | Ring C | Ring D | C-24 | C-25 | C-26b | C-27c | Trivial Name | |
|---|---|---|---|---|---|---|---|---|---|---|
| A. C27 triols | ||||||||||
| I | 0418 | 3αOH | 7αOH | CH2OH | CH3 | |||||
| II | 0419 | 3βOH | 7αOH | CH2OH | CH3 | 16-Deoxymyxinol | ||||
| B. C27 tetrols and a C24 tetrol | ||||||||||
| III | 0610 | 3αOH | 7αOH | 12αOH | CH2OH | (C24 tetrol) | Petromyzonol | |||
| IV | 0309 | 3αOH | 7αOH | 12αOH | CH2OH | CH | CH3 | CH3 | ||
| V | 0316 | 3αOH | 7αOH | 12αOH | CH2OH | CH3 | CH3 | |||
| VI | 0323 | 3αOH | 7αOH | 12αOH | CH2OH | CH3 | ||||
| VII | 0331 | 3αOH | 7αOH | 16αOH | CH2OH | CH3 | ||||
| VIII | 0332 | 3βOH | 7αOH | 16αOH | CH2OH | CH3 | Myxinol | |||
| IX | 0335 | 3αOH | 7αOH | OH | CH2OH | CH3 | ||||
| X | 0338 | 3αOH | 7αOH | OH | CH2OH | |||||
| XI | 0301 | 3αOH | 6βOH, 7αOH | CH2OH | CH3 | |||||
| XII | 3-oxo | 7αOH | 12αOH | CH2OH | CH3 | |||||
| C. C27 pentols | ||||||||||
| XIII | 0217 | 3αOH | 7αOH | 12αOH | OH | CH2OH | CH3 | Chimaerol | ||
| XIV | 0220 | 3αOH | 7αOH | 12αOH | OH | CH2OH | CH3 | Bufol | ||
| XV | 0219 | 3βOH | 7αOH | 12αOH | OH | CH2OH | CH3 | |||
| XVI | 0222 | 3βOH | 7αOH | 12αOH | OH | CH2OH | Latimerol | |||
| XVII | 0223 | 3αOH | 7αOH | 12αOH | OH | CH2OH | Cyprinol | |||
| XVIII | 3-oxo | 7αOH | 12αOH | |||||||
| XIX | 0201 | 2βOH,3αOH | 7αOH | 12αOH | CH2OH | CH3 | ||||
| XX | 0202 | 3αOH | 6αOH, 7βOH | OH | CH2OH | CH3 | ω-Trichechol | |||
| XXI | 0203 | 3αOH | 6βOH, 7αOH | OH | CH2OH | CH3 | α-Trichechol | |||
| XXII | 0204 | 3αOH | 6βOH, 7βOH | OH | CH2OH | CH3 | β-Trichechol | |||
| D. C27 hexols | ||||||||||
| XXIII | 0104 | 3αOH | 7αOH | 12αOH | ROH | OH | CH2OH | Scymnol | ||
| XXIV | 0106 | 3αOH | 7αOH | 12αOH | OH | OH | CH2OH | Dermophol | ||
| XXV | 0101 | 2βOH,3αOH | 7αOH | 12αOH | OH | CH2OH | Arapaimol-B | |||
The table also includes petromyzonol, the unique C24 bile alcohol that is present in the lamprey
Numbers are those given by Une and Hoshita in their thorough tabulation of bile alcohols (22). Data in that review also give structural information for bile alcohols with chain lengths other than C24 and C27. In this tabulation, we have not distinguished 5α or 5β isomers, as any alcohol may exist as either geometric isomer. We have also not distinguished the two diastereoisomers that can occur at C-25.
The IUPAC nomenclature recommendation is that when there is only a single substituent at C-26 or C-27, the terminal carbon is designated as C-26. Therefore cholesterol 27-hydroxylase generates a bile alcohol with a primary alcoholic group at C-26.
TABLE 2.
Biliary C27 bile acids
| I. 5α bile acids (A/B trans) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U & Ha | Ring A | Ring B | Ring C | Ring D | C-25 Diast. | C-22 | C-23 | C-24 | C-25 | C-26 | C-27 | |
| A | 3αOH | 7αOH | R | COOH | ||||||||
| B | 3αOH | 7αOH | S | COOH | ||||||||
| C | 3αOH | 7αOH | R | OH | COOH | |||||||
| D | 3αOH | 7αOH | SOH | COOH | ||||||||
| E | 0901 | 3αOH | 7αOH | 12αOH | COOH | |||||||
| F | 0808 | 3αOH | 7αOH | 12αOH | OH | COOH | ||||||
| G | 0811 | 3αOH | 7αOH | 12αOH | OH | COOH | ||||||
| H | 0906 | 3αOH | 7αOH | 12αOH | Δ23 | COOH | ||||||
| I | 0911 | 3-oxo | 7αOH | 12αOH | COOH | |||||||
| J | 3αOH | 12αOH | OH | COOH | ||||||||
| II. 5β bile acids (A/B cis) | ||||||||||||
| K | 1002 | 3αOH | 7αOH | COOH | ||||||||
| L | 1009 | 3αOH | 7βOH | COOH | ||||||||
| M | 0917 | 3αOH | 7αOH | OH | COOH | |||||||
| N | 1003 | 3αOH | 7αOH | Δ23 | COOH | |||||||
| O | 1004 | 3αOH | 7αOH | Δ24 | COOH | |||||||
| P | 1βOH,3αOH | 7αOH | R | COOH | ||||||||
| Q | 0801 | 1βOH,3αOH | 7αOH | 12αOH | R | COOH | ||||||
| R | 0802 | 2βOH,3αOH | 7αOH | 12αOH | S | COOH | ||||||
| S | 0902 | 3αOH | 7αOH | 12αOH | R | COOH | ||||||
| T | 0902 | 3αOH | 7αOH | 12αOH | S | COOH | ||||||
| U | 0806 | 3αOH | 7αOH | 12αOH | SOH | COOH | ||||||
| V | 0809a | 3αOH | 7αOH | 12αOH | R | ROH | COOH | |||||
| W | 0809b | 3αOH | 7αOH | 12αOH | R | SOH | COOH | |||||
| X | 0809c | 3αOH | 7αOH | 12αOH | S | ROH | COOH | |||||
| Y | 0809d | 3αOH | 7αOH | 12αOH | S | SOH | COOH | |||||
| Z | 0812 | 3αOH | 7αOH | 12αOH | OH | COOH | ||||||
| AA | 0907 | 3αOH | 7αOH | 12αOH | Δ23 | COOH | ||||||
| AB | 0813 | 3αOH | 7αOH | 12αOH | Δ23 | OH | COOH | |||||
| AC | 0908a | 3αOH | 7αOH | 12αOH | Δ24 E | COOH | ||||||
| AD | 0908b | 3αOH | 7αOH | 12αOH | Δ24 Z | COOH | ||||||
| AE | 0904 | 3βOH | 7αOH | 12αOH | COOH | |||||||
| AF | 0905 | 3αOH | 7βOH | 12αOH | COOH | |||||||
| AG | 0912 | 3-oxo | 7αOH | 12αOH | COOH | |||||||
| AH | 0914 | 3αOH | 7-oxo | 12αOH | COOH | |||||||
| AI | 1013 | 3αOH | 12αOH | COOH | ||||||||
| AJ | 3αOH | 12αOH | SOH | COOH | ||||||||
| AK | 1015 | 3αOH | 12αOH | R | ROH | COOH | ||||||
| AL | 3βOH | 12αOH | COOH | |||||||||
| AM | 3αOH | 7αOH | 15αOH | COOH | ||||||||
| AN | 3αOH | 7αOH | 15αOH | SOH | COOH | |||||||
| AO | 3αOH | 7αOH | 16αOH | COOH | ||||||||
| AP | 3αOH | 7αOH | 16αOH | SOH | COOH | |||||||
| AQ | 3αOH | 7αOH | 16αOH | ROH | COOH | |||||||
| AR | 3αOH | 7αOH | 12αOH | 16αOH | COOH | |||||||
Diast., diastereomeric configuration.
Numbers are those given by Une and Hoshita (22) in their thorough tabulation which gives original references. Their review also tabulates C25, C26, C28. and C29 bile acids. These are mostly found in frogs.
TABLE 3.
Biliary C24 bile acids
| I. 5β (A/B cis) Bile Acids | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trivial Name | Steroid Nucleus (C19) | Side Chain | Type | Occurrence | ||||||
| Ring A | Ring B | Ring C | Ring D | C-22 | C-23 | C-24 | ||||
| 1 | Chenodeoxycholic | 3αOH | 7αOH | COOH | P | Many species | ||||
| 2 | (none proposed) | 3αOH | 7oxo | COOH | P | Cavimorphs | ||||
| 3 | (none proposed) | 3αOH | 7αOH | Δ22 | COOH | ? | Caviomorphs | |||
| 4 | (none proposed) | 3αOH | 7βOH | Δ22 | COOH | ? | Agouti | |||
| 5 | Phocaecholic | 3αOH | 7αOH | ROH | COOH | P | Sea mammals, birds | |||
| 6 | Haemulcholic | 3αOH | 7αOH | SOH | COOH | P | Fish | |||
| 7 | Lithocholic | 3αOH | COOH | S | Many species | |||||
| 8 | (none proposed) | 3αOH | 15αOH | COOH | S | Wombat | ||||
| 9 | Cholic | 3αOH | 7αOH | 12αOH | COOH | P | Many species | |||
| 10 | (none proposed) | 3αOH | 7oxo | 12αOH | COOH | ? | Sloth | |||
| 11 | (none proposed) | 3αOH | 7αOH | 12oxo | COOH | ? | Birds | |||
| 12 | (none proposed) | 3αOH | 7αOH | 12αOH | Δ22 | COOH | P,S | Snakes | ||
| 13 | (none proposed | 3αOH | 7αOH | 12αOH | ROH | COOH | S | Snakes | ||
| 14 | Deoxycholic | 3αOH | 12αOH | COOH | S | Many species | ||||
| 15 | Bitocholic | 3αOH | 12αOH | ROH | COOH | S | Snakes | |||
| 16 | Lagodeoxycholic | 3αOH | 12βOH | COOH | S | Rabbit | ||||
| 17 | Ursodeoxychoic | 3αOH | 7βOH | COOH | P,S | Nutria, bears | ||||
| 18 | Ursocholic | 3αOH | 7βOH | COOH | P,S | Humans | ||||
| 19 | α-muricholic | 3αOH | 6βOH,7αOH | COOH | S | Rodents | ||||
| 20 | β-muricholic | 3αOH | 6βOH,7βOH | COOH | P | Rodents | ||||
| 21 | (none proposed) | 3αOH | 6βOH,7βOH | Δ22 | COOH | P,S | Rodents | |||
| 22 | ω-muricholic | 3αOH | 6αOH,7βαOH | COOH | P,S | Rodents | ||||
| 23 | Murideoxycholic | 3αOH | 6βOH | COOH | S | Rodents | ||||
| 24 | Hyocholic | 3αOH | 6αOH,7αOH | COOH | P | Pigs | ||||
| 25 | Hyodeoxycholic | 3αOH | 6αOH | COOH | S | Pigs | ||||
| 26 | (none proposed) | 3αOH | 7αOH,19βOH | COOH | P | Infants | ||||
| 27 | Vulpecholic | 1αOH,3αOH | 7αOH | COOH | P | Marsupials | ||||
| 28 | (none proposed) | 1βOH,3αOH | 7αOH | COOH | P | Pigeons | ||||
| 29 | (none proposed) | 3αOH,4βOH | 7αOH | COOH | P | Pheasants | ||||
| 30 | (none proposed) | 3αOH | 5βOH,7αOH | COOH | P | Pheasants | ||||
| 31 | Cygnocholic | 3αOH | 7αOH | 15αOH | COOH | P | Swans | |||
| 32 | Avicholic | 3αOH | 7αOH | 16αOH | COOH | P | Birds | |||
| 33 | Avideoxycholic | 3αOH | 16αOH | COOH | S | Birds | ||||
| 34 | (none proposed) | 3αOH | 7αOH | 12αOH | 16αOH | COOH | P | Snakes | ||
| 35 | (none proposed) | 3αOH | 12αOH | 16αOH | COOH | ?P,S | Snakes | |||
| 36 | Norchenodeoxycholic | 3αOH | 7αOH | COOH | P | Pinnipeds | ||||
| II. 5α (A/B trans) bile acids | ||||||||||
| 37A | Allochenodeoxycholic | 3αOH | 7αOH | COOH | P | Reptiles | ||||
| 38A | Allocholic | 3αOH | 7αOH | 12αOH | COOH | P | Reptiles | |||
| 39A | Allodeoxycholic | 3αOH | 12αOH | COOH | S | Reptiles | ||||
| 40A | Alloavicholic | 3αOH | 7αOH | 16αOH | COOH | S | Birds | |||
P, primary bile acid (formed in hepatocyte); S, secondary, formed in intestine by bacteria. Default substitutents are indicated by bold font.
TABLE 4.
Bile salts of turtles and crocodilians
| Phylogeny | SN | Species | Bile salt class(es) | C27 bile alcohols | C27 bile acids |
|---|---|---|---|---|---|
| Reptilia Subclass: Archosauria Order: Testudines (Turtles) | |||||
| Geomydidae | R1 | Reeves turtle | C27 5β-acids | D,U | |
| Testudinae | R2 | Leopard tortoise | C27 5β-acids | S,U,AJ | |
| R3 | African spurred tortoise | C27 5β-acids | A,B,D,S,AJ | ||
| R4 | Asian brown tortoise | C27 5β-acids | S,U, AJ | ||
| R5 | California desert tortoise | C27 5β-acids, C27 alcohols | NA | NA | |
| R6 | Madagascar spider tortoise | C27 5β-acids | NA | ||
| Emydidae | R7 | Wood turtle | C27 5β-acids | U | |
| R8 | Chinese 3-stripe box turtle | C27 5β-acids | D,U | ||
| R9 | Diamondback terrapin | C27 5β-acids | U | ||
| Chelydridae | R10 | Alligator snapping turtle | C27 5β-acids | U,AJ | |
| Cheloniidae | R11 | Green turtle | C27 5β-acids | U,AJ | |
| R12 | Loggerhead turtle | C27 5β-acids | U,AJ | ||
| R13 | Hawksbill turtle | C27 5β-acids | U,AJ | ||
| R14 | Olive Ridley turtle | C27 5β-acids | U,AJ, others | ||
| Dermochelyidae | R15 | Leatherback turtle | C27 5β-acids | D, other | |
| Trionychidae | R16 | Chinese soft shell turtle | C27 5β-acids | U | |
| Chelidae | R17 | Siebenrock's snake-neck turtle | C27 5β-acids,C27 alcohols | NA | NA |
| Reptilia: Crocodilia | |||||
| Eusuchia | |||||
| Crocodilidae | R18 | American crocodile | C27-5β acids; C24-5α- acids | S,T | 38A |
| R19 | Orinoco crocodile | C27-5β acids; | S,T | ||
| R20 | Nile crocodile | C27-5β acids; | A, S,T | ||
| R21 | Mugger | C27-5β acids; C24-5β acids; | S,T | 9 | |
| Gavilidae | R22 | Indian gavial | C27-5β acids | S,T | |
Fig. 1.
Chemical structure of cholesterol (A) and the three main classes of bile salts: C27 bile alcohols (B), C27 bile acids (C), and C24 bile acids (D). Bile salts are shown as their natural conjugates: C27 bile alcohols are shown esterified with sulfate; C27 bile acids are shown as their N-acyl amidate conjugates with taurine. Glycine conjugation is not shown. Also not shown is modification of the 7α-hydroxy group; in some caviomorphs, it is oxidized to a 7-oxo group, and in some rodents and bears, it is epimerized to a 7β-hydroxy group. C27 bile alcohols are shown as either 5α (trans A/B ring juncture) or 5β (cis A/B ring juncture). C27 and C24 bile acids are shown only in the 5β configuration, although 5α-C27 bile acids are likely to occur, and 5α-C24 bile acids (allo bile acids) are common in nature. The large arrows indicate sites of hydroxylation on the nucleus or side chain that occur in many species. The small arrows indicate sites of hydroxylation that occur in only a few species. For C24 bile acids, some sites of hydroxylation that occur in only a few species and constitute <10% of biliary bile acids are not shown, for example, hydroxylation at C-4. Additional sites of hydroxylation are likely to be discovered in the future.
METHODS
Bile samples were obtained from a great many sources (23). These included zoos, aquaria, biologists, veterinarians, and a commercial fish farm. Samples were collected by aspiration from the gallbladder or from the common duct in animals not possessing gallbladders. Bile was then dispersed in at least three volumes of reagent grade isopropanol in brown glass bottles. The isopropanol solution was then shipped by airmail to the University of California, San Diego. Samples were stored at 4°C.
Samples were first analyzed qualitatively by TLC to detect the possible presence of unconjugated bile acids using a developing system that separates unconjugated from conjugated bile acids (40). Bile acids (C27 and C24) were then analyzed by reverse-phase HPLC using a C18 column and the methanol-phosphate buffer solvent system described by Rossi, Converse, and Hofmann (41). In this method, conjugated bile acids are quantified by the absorbance of their amide bond at 205 nm. Unconjugated bile acids and bile alcohol sulfates are not detected. Bile acid classes were analyzed by ESI-MS-MS as described by Chatman et al. (42). The mode of bile salt conjugation was determined using the Q3 cell. This analytical technique provides information on compounds differing in their m/z values, and in principle, detects all bile salts. For determination of individual C24 bile acids and bile alcohols, samples were analyzed by GC-MS using a Series II model HP 5890/5970 MS and a 30m SPB-35 capillary column as described (43). For GC-MS, C24 bile acid samples underwent deconjugation using either sodium hydroxide (2 M NaOH, 4 h, 130°C, Parr bombs) or the enzyme cholylglycine hydrolase (Sigma Aldrich, St. Louis, MO). C27 bile acids were not analyzed by GC-MS as they are not hydrolyzed by cholylglycine hydrolase and are degraded by the conditions of alkaline deconjugation; therefore, identification of C27 bile acids was based solely on retention time using HPLC as well as ESI-MS-MS. Bile alcohol sulfates were deconjugated enzymatically using sulfatase [aryl sulfatase from Helix Pomatia (Sigma Aldrich)] or by solvolysis (44, 45). After deconjugation, C24 bile acids were esterified with methanol. Bile acids and bile alcohols were then converted to their per-acetates or per-TMS derivatives. To establish the location and orientation of hydroxyl groups in novel bile acids, proton and 13C nuclear magnetic resonance analyses were used in a continuing collaboration with T. Iida of Nihon University (35, 36, 45, 46).
For most species, only a single bile sample was available. However, in seven of 103 fish species, values were the mean of two or more fishes. In squamates, 13 of 103 species had samples from two or more animals. In birds, 22 of 271 samples came from two or more animals. In mammals, more than one-third of samples came from multiple animals. Proportions of individual bile acids from different animals in the same species were usually in fair agreement. In the tables, only bile acids composing more than 10% of bile salts are listed and all analyses tabulated here were performed at the University of California, San Diego. Some samples were analyzed by ESI-MS-MS but not by GC-MS. For these samples, only bile salt classes are known; only classes that were >10% of biliary bile salt composition are listed.
Samples were from adult animals so far as is known and no information was available on gender. However, gender effects on biliary bile salt composition are thought to be small, at least in mammals (47–49).
RESULTS AND DISCUSSION
Bile salts of fish
Overview of bile salt composition in fish species.
Supplementary Tables V and VI give bile salt composition for over 100 fish species. There are some 30,000 fish species in Fish Base (http://www.fishbase.org); thus our database contains information on bile salt composition in about 0.3 percent of living fish species. Older literature on fish bile salts was summarized by Tammar (50).
The earliest evolving fish are believed to be the jawless fish, also known as Agnatha, which are currently limited to hagfishes and lampreys (51, 52). Jawless fish had predominantly C27 bile alcohols. The sea lamprey is unique in also having a C24 bile alcohol (5α-petromyzonol; F2), a type of bile salt not found in any other animal species characterized to date (20, 33). Its mechanism of formation is unknown. Hagfish are distinctive in having their major bile alcohol 5α-myxinol (3β,7α,16α,27-tetrahydroxy-5α-cholestane) present as a disulfate (C-3 and C-27) (53). We have found disulfated bile alcohols in only one other fish species, the sea lamprey, for which a minor fraction (∼10%) of the bile alcohols is disulfated.
Disulfated bile alcohols have not been studied with regard to physicochemical properties but are unlikely to have favorable properties for solubilizing dietary lipids. This raises the possibility that hagfish use their bile salts for functions other than lipid digestion, such as communication, similar to the use of bile salts as olfactory markers or pheremones by the sea lamprey and other fish species (54–58).
5α-Myxinol is a bile salt unique to hagfish. The bile salt is unusual in retaining the 3β-hydroxy group of cholesterol. All other known primary bile salts have the 3-hydroxy group in the α configuration, an exception being the pheromonal 3-oxo bile alcohols of the sea lamprey (55). Although 3β-hydroxy bile acids are common fecal bile acids, at least in humans (59), they are never present in bile in appreciable proportions because they are epimerized to 3α-hydroxy bile acids during hepatocyte transport (11, 12).
In contrast to the unique bile salts of jawless fish, the major bile salt of all species of Elasmobranchii (sharks, skates, and rays; subgroup of Chondrichthyes or cartilaginous fish) was a C27 5β bile alcohol known as 5β-scymnol (3α,7α,12α,24,26,27-hexahydroxy-5β-cholestane) (31). The default C27 bile alcohol has hydroxyl groups at C-3, C-7, and C-27, meaning that scymnol has undergone three additional hydroxylations (one on the nucleus at C-12, two on the side chain at C-24 and C-26). Outside of Elasmobranchii, hexahydroxy bile alcohols were present in biliary bile salts in many other species but only in trace proportions. Other than Elasmobranchii, the other fish within Chondrichthyes are the Chimaerae. The major bile salt of Chimaerae was 5β-chimaerol (3α,7α,12α,24,27-pentahydroxy-5β-cholestane), a bile alcohol that differs from 5β-scymnol by lacking C-26 hydroxylation (60).
Ray-finned fish (Actinopterygii) currently contain the largest number of fish species (52). In species characterized to date, the dominant bile salts of ray-finned fish were the common C24 bile acids cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid). Several early evolving ray-finned fish orders (Polypteriformes, Acipenseriformes, Amiiformes), including species such as the sturgeon (F15) and paddlefish (F16), showed varying proportions of C24 bile acids and C27 bile alcohols in their bile. Another example is the bowfin (F18), for which C27 bile alcohols account for more than 20% of the biliary bile salt pool. C27 bile alcohols were also present in the European eel (F22), Kroyer's deep sea angler fish (F38), bulbous dreamer (F39), coralfish (F73, F74), butterfly fish (F75, F76), moon fish (F94), and Pacific barracuda (F102). The bile salts of Cypriniformes (F28–F33 in supplementary Table V) were unusual in being 5α C27 bile alcohols (13, 61). C27 bile acids were rare in the ray-finned fish species examined to date but were found in the Japanese medaka (F46) and arapaima (F21).
In ray-finned fish, all bile acids were found to be conjugated to taurine with a few exceptions. In the sea bream (F101), C24 bile acids were conjugated with cysteinolic acid, a taurine congener that is thought to be of dietary origin, specifically in ingested algae (62, 63). In some species of angel fish (F84–F93) and the bulbous dreamer (F39), bile acids were conjugated with N-methyltaurine. N-methyltaurine conjugates are resistant to bacterial deconjugation in mammals (64, 65) and the same is likely to be true for angelfish. In all species except the soda cichlid (F77) and the Mozambique tilapia (F80), bile acids were present largely in conjugated form. In these two species, a major fraction of bile acids was present in unconjugated form. The occurrence of a major fraction of biliary bile acids in unconjugated form is very rare in vertebrates, having been observed previously only in the Australian opossum (66). Some fish species [hagfish (F1), the South American lungfish (F12), the arapaima (F21)] contained very complex mixtures of C27 compounds in bile.
Bile alcohols from early evolving fish [hagfish (F1), lamprey (F2), coelacanth (F11), lungfish (F12), mudfish (F13)] were 5α (A/B cis), as was 5α-cyprinol, the dominant bile salt of Cypriniformes (F28–F31). C27 bile acids and C24 bile acids were predominantly 5β, but occasional species had moderate proportions of 5α C24 bile acids [European conger, (F24)]. The structure of the side chain of C27 bile salts occurring in fish is not known.
Nuclear hydroxylation occurred at C-16 in the hagfish (F1) but in no other fish species. The only site of nuclear hydroxylation other than those of the default structure at C-3 and C-7 was at C-12 in many fish (forming CA in the case of C24 bile acids) and at C-2 in the arapaima (F21) (67). No evidence for any secondary bile acids was observed except in six of 10 species of angelfish (F84–F85, F87–F88, F91, F93) in whom deoxycholic acid (DCA; 3α,12α-dihydroxy-5β-cholan-24-oic acid) was present. The presence of DCA indicates a resident intestinal anaerobic flora capable of dehydroxylating CA at C-7. In two species of angelfish (F90, F91), the 12β-epimer of DCA was present; this bile acid is presumably also of bacterial origin.
Side chain hydroxylation of C27 alcohols (in addition to the default hydroxylation at C-27) was at C-24, C-25, or C-26. Side chain hydroxylation of C24 bile acids was not observed in fish, with the exception of the chicken grunt (F81) in which a novel bile acid with a hydroxy group at C-22 was present (32). The functional significance of this side chain hydroxylation is not known. Confirmation of the assigned structure of this bile acid has been shown by direct synthesis (68).
Evolutionary implications for fish species.
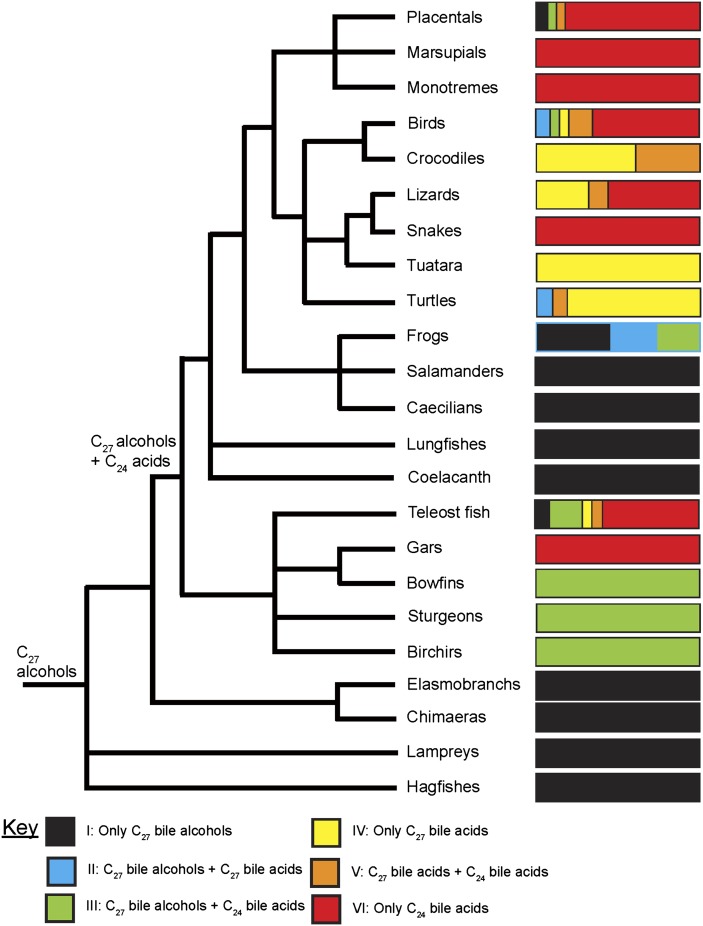
The variation of bile salts in fish is mostly consistent with current theories on evolution of fish and other vertebrates (52, 69). Figure 2 shows a generic vertebrate phylogenetic tree (69) with bile salt variation overlaid in color code. Each of the colored bars indicates the proportion of species with certain bile salt profiles ranging from type I (>95% C27 bile alcohols, “ancestral phenotype”) to type VI (>95% C24 bile acids, “derived phenotype”). The base of the fish tree contains the jawless fish (lampreys, hagfish) and cartilaginous fish (chimaerae, rays, sharks, skates), all of which use C27 bile alcohols (or C24 bile alcohols for sea lamprey) as their dominant bile salts. Some of the basal bony fish (birchirs, bowfins, sturgeons) have a mixture of C27 bile alcohols (ancestral phenotype) and C24 bile acids (derived phenotype) (i.e., a type III profile). These fish demonstrate the ability to synthesize both C27 bile alcohols and C24 bile acids without the formation of C27 bile acids. Obviously, it cannot be predicted whether the basal bony fish are on their way to eventually having a solely derived bile salt pattern (as seen in the majority of ray-finned fish in our sample) or if the mixture of ancestral and derived forms of bile salts is advantageous in these fish. The majority of teleost fish surveyed so far have biliary bile salt profiles consisting of mostly C24 bile acids with the exception of cypriniform fish such as carp and zebrafish. As seems to be true throughout all vertebrates, structural variation of bile salts in fish appears to have occurred on a relatively slow timescale, with significant variation seen between fish orders but little variation between the more narrow evolutionary units of families, genera, and species.
Fig. 2.
Variation of bile salt structures across vertebrate species. The phylogeny is a general one for vertebrates (69) with debated evolutionary relationships (e.g., frogs, salamanders, caecilians) depicted as polyotomies. Each bar chart shows the proportion of animals in each vertebrate group (using data from animal species analyzed so far) that are classified into one of six bile salt profiles based on the one or two bile salt classes (C27 bile alcohols, C27 bile acids, C24 bile acids) that account for 10% or more of the total biliary bile salt pool (see key). There are a small number of fish and amphibian species that have all three major classes of bile salts present in bile, each at 10% or more of the total bile salt pool. For simplicity, these species are still classified based on the two bile salt types that account for the greatest percentage of the bile salt pool. The inferred bile salt profiles for the last common ancestor to all living vertebrates and the last common ancestor to bony fish and land vertebrates (basal gnathostome) are indicated.
The lobe-finned fish (coelacanths, lungfishes) are currently positioned at the base of the tree that also has amphibians and land animals (e.g., mammals, reptiles) (69). Similar to jawless fish, lobe-finned fish have 5α-C27 bile alcohols in their bile, suggesting that 5α-C27 bile alcohols are the ancestral trait. Thus, there appears to be two broad transitions in vertebrate evolution from C27 5α bile alcohols to C24 5β bile acids: 1) from jawless fish to bony fish and 2) from lobe-finned fish to land tetrapods.
In terms of fish evolution, there is a striking contrast between the major bile salts of hagfish and lampreys, which are quite different from one another in both side-chain length (C8 for hagfish, C5 for lampreys) and hydroxylation patterns (3β,7α,16α,27 for hagfish and 3α,7α,12α,24 for lamprey). There is debate on how closely related lampreys and hagfish are to one another. Mitochondrial DNA evidence has been used to argue that lampreys and hagfish are closely related (70), whereas morphology and developmental comparisons suggest that they are only very distantly related (71). Our finding of marked differences between lamprey and hagfish bile salts are consistent with proposals that these two groups of jawless fish are not very closely related to one another and have evolved independently for a long time.
In analyzing the bile salt variation patterns across fish, there appears to be at least two main pathways in the evolutionary transition from C27 bile alcohols to C24 bile acids: a ‘direct’ pathway (a) and an ‘indirect’ pathway (b) that uses C27 bile acids as an ‘intermediate’ step. This concept was originally proposed by Haslewood (33) and has now been confirmed by our more extensive sampling of vertebrate species. Pathway (a) appears to be the more common one in ray-finned fish, with many examples of species with C27 bile alcohols and C24 bile acids in their bile but not appreciable amounts of C27 bile acids. This pattern in fish differs from that in amphibians and reptiles, which use pathway (b) where C27 bile acids are common (21, 22).
It is tempting to infer what the ancestral bile salt profile was at some of the major nodes on the tree in Fig. 2. As discussed above, the earliest ancestral vertebrates are predicted to have produced only C27 bile alcohols as their major bile salts. The more difficult question is, when did the ability to synthesize C24 bile acids first appear. There are two main possibilities here. First, the last common ancestor to fish and mammals (basal gnathostome) already had the ability to synthesize C24 bile acids. In this case, the observation of extant gnathostomes (e.g., lobe-finned fish, salamanders, some frogs) that secrete only C27 bile alcohols would reflect lineage-specific loss of the ability to synthesize C24 bile acids during evolution. Alternatively, the basal gnathostome had only C27 bile alcohols and the ability to synthesize C24 bile acids has occurred multiple times in vertebrate evolution (i.e., convergent evolution).
Bile salts of amphibians
Overview of bile salt composition in amphibians.
The class Amphibia includes three extant orders: Anura (frogs and toads), Caudata (salamanders and newts), and Gymnophiona (caecilians). There is estimated to be over 5,000 anuran species and at least 500 species within Caudata. Most studies of amphibian bile salts have been on species within Anura (22). Only a handful of studies have examined the bile salts within Caudata. We have so far analyzed the bile of only one caecilian species. Consequently, with respect to the species diversity of amphibians, the diversity of bile salts has barely been probed.
Amphibian species have some of the most complex biliary bile salt profiles found within vertebrates, with some species having significant amounts of C27, C26, C28, or other side chain length bile alcohols as well as C27 bile acids and C24 bile acids (9, 22, 72–74). The complexity of anuran bile salt profiles makes it difficult to summarize patterns of variation. The major bile salt of salamander species examined so far is dermophol (3α,7α,12α,25,26,27-tetrahydroxy-cholestane), found in both 5α and 5β orientation in various species (75). The bile of the only caecilian species analyzed so far was found to contain tetrahydroxy- and pentahydroxy-C27 bile alcohols (L. R. Hagey, unpublished observations).
Bile salts of reptiles
Overview of bile salt composition in reptilian species.
The bile salts of turtles and crocodilians (alligators, crocodiles, and gavials) are summarized in Table 4. In turtles, biliary bile salts generally consisted of primary and secondary C27 bile acids. In two species, the California desert tortoise (R5) and Siebenrock's snake-neck turtle (R17), C27 bile alcohols were also present and comprised more than 10% of the total biliary bile salt pool. In Crocodilia, there was a mixed picture. The Orinoco crocodile (R19) and Nile crocodile (R20) had predominantly C27 acids. The American crocodile (R18), the mugger (R21), and the Indian gavial (R22) had mixtures of C27 and C24 acids.
In squamates (lizards and snakes), bile acids predominated (supplementary Tables VII, VIII). The majority of lizards (21 of 34 species) had solely C24 bile acids; a minority (10 of 34 species) had solely C27 bile acids. Three species had both C24 and C27 bile acids. In contrast, all snakes analyzed had exclusively C24 bile acids.
Bile salts were entirely 5β in turtles and snakes. In the American crocodile (F18), the C27 bile acids were 5β whereas the C24 bile acids were largely 5α. In lizards, about one-fourth of the species examined had C24 bile acids that were entirely 5α (mainly allo-CDCA or allo-CA). The reason for this finding, unique among vertebrates, is not known. In humans and rodents, 5β-reduction of bile acid intermediates is catalyzed by aldo-keto reductase 1D1 (AKR1D1) (2, 3). In lizards whose bile salt pool consists mainly of allo-bile acids, perhaps AKR1D1 (if present) catalyzes 5α- and not 5β- reduction of bile acid intermediates or, alternatively, another enzyme catalyzes reduction to the 5α orientation. At any rate, the finding indicates that 5α bile acids (as taurine conjugates) appear to function adequately in the digestive process. The finding also suggests that such 5α bile acids are substrates for the hepatocyte and ileal enterocyte transport systems that mediate the enterohepatic circulation of bile salts.
Additional nuclear hydroxylation of the default structure occurs at C-12 in crocodilians, lizards, snakes, and turtles. Bile acids hydroxylated at C-15 occurred in some turtle species. In pythons and boas, hydroxylation occurs at C-16 in addition to C-12. What was remarkable was the identification of some snake species in whom all bile acids lacked a 7-hydroxy group at C-7. The dominant bile acid was a 3α,12α,16α-trihydroxy bile acid, a bile acid considered by Haslewood (30) to be formed by 16α-hydroxylation of DCA, based on earlier studies by Bergström et al. (76) in the python. Usually, 7-deoxy bile acid are considered to be secondary bile acids, formed by bacterial 7-dehydroxylation of primary bile acids (1). Cholesterol 7α-hydroxylase (CYP7A1) is considered the rate limiting step in bile acid biosynthesis, and a pathway not involving hydroxylation at C-7 has never been considered to occur (2, 3). The simplest explanation for the apparent absence of primary bile acids is that bile acid synthesis diminishes almost completely between meals.
Three species of snakes (R58, R59, and R61) had tetrahydroxy bile acids with hydroxylation at C-12 and C-16 in addition to the default hydroxylation at C-3 and C-7. To date, this is the only known instance of tetra-hydroxy C24 bile acids being the dominant primary bile salts of any vertebrate species in health.
Hydroxylation on the side chain occurred at C-22 in turtles. So far, C27 bile acids with 22-hydroxylation have only been observed in turtles. The functional significance of this novel site of hydroxylation is not known. In lizards with C27 bile acids, hydroxylation at C-24 occurred. Again, the effect of such hydroxylation on physicochemical and physiological properties is not known.
In snakes whose bile salts are entirely C24 bile acids, hydroxylation at C-23 was present. In adders and vipers, α-oxidation of such compounds gives rise to 24-nor derivatives (C23 bile acids) (77) and norchenodeoxycholic acid (as its taurine conjugate) was identified in the bile of the Horned sand viper (R93). Snake bile acids also contained a few percent of Δ22 bile acids, presumably formed as the first step in continuing β-oxidation.
Evolutionary implications for reptilian species.
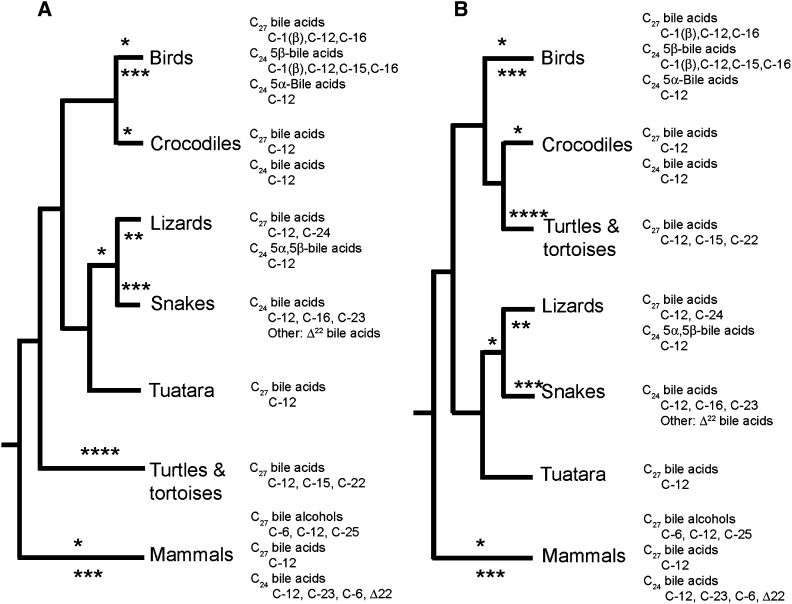
The phylogeny of reptiles is still an active area of debate, especially with regard to the inter-relationships within squamates and of the relationship of Testudines (turtles and tortoises) to other reptile groups (69, 78). Figure 3 shows bile salt structural variation overlaid on two phylogenetic trees of the major reptile groups (crocodiles, lizards, snakes, tuatara, turtles/tortoises), birds (technically part of reptiles as well), and mammals. Tree A in Fig. 3 is the more traditional hypothesis based on morphologic and paleontological data whereas tree B is probably best supported by current molecular data (69). In either tree, one could speculate that the “ancestral” bile salt profile of the last common ancestor to living reptiles mainly contained a C27 bile acid (perhaps with a 3α,7α,12α-trihydroxy pattern). With respect to the evolution of bile salt structural diversity in reptiles, four major “innovations” are noted: 1) 24R-hydroxylation as first seen in bile acids of varanid lizards and the tuatara (* in Fig. 3); 2) synthesis of 5α (allo) C24 bile acids in agamid lizards (** in Fig. 3); 3) multiple changes unique to snake bile salts as compared with other reptiles (7-dehydroxylation, 16α-hydroxylation, 23R-hydroxylation, and unsaturation of the side-chain; *** in Fig. 3); and 4) 15- and 22-hydroxylation of C27 bile acids unique to turtles/tortoises (**** in Fig. 3).
Fig. 3.
Variation of bile salt structures across reptile groups in comparison with birds and mammals. The major bile salts of each group are indicated with hydroxylation patterns in addition to the default hydroxylation for each bile salt class noted. The evolutionary relationships of living reptile groups are still debated, especially with regard to the placement of Testudines (turtles and tortoises) (69, 78). The traditional hypothesis, based on morphological and paleontological data, placed Testudines as the most basal group (A). More recent molecular phylogenies challenge this hypothesis and place Testudines as a sister-group to crocodiles (B) (69). In either arrangement, one may hypothesize that the common ancestor to all living reptile groups possessed a bile salt profile consisting mainly of C27 bile acids (possibly with C27 bile alcohols as well). In this case, four main “innovations” (indicated by, *, **, ***, and ****) with regard to evolution of bile salt structures in reptiles are postulated: *, ability to synthesize C24 bile acids; **, formation of 5α (allo) C24 bile acids in some lizard lineages; ***, novel additional modifications to nucleus and side-chain (e.g., C-16 and C-23 hydroxylation; introduction of double bond between C-22 and C-23) to C24 bile acids in snakes; ****, 15- and 22-hydroxylation in the C27 bile salts of turtles and tortoises.
There are similarities between the bile salts of reptiles and birds. Paleognath birds (cassowaries, emus, kiwis, ostriches, and tinamous), currently considered to be at the base of the avian evolutionary tree (69, 79, 80), have bile salt profiles consisting mainly of dihydroxy- and trihydroxy- C27 bile acids (supplementary Table IX ), a profile very similar to crocodilians (Table 4). 5α C24 bile acids are common in waterfowl, although no bird species yet analyzed has biliary bile salts consisting mostly of 5α bile acids as seen in agamid lizards. 16α- and 23R-hydroxylation, as seen in snakes, are also common in birds.
Bile salts of birds
Overview of bile salt composition in bird species.
Supplementary Table IX lists the bile salt composition of 271 avian species, about 3% of the total number of living bird species. Most birds have gallbladders but some do not. Absence of the gallbladder has been reported for pigeons (A122–A135), some psittacine species (A189–A196), species in the Cuculidae family, hummingbirds, pea fowls, the ostrich, and the rhea (81).
Bile acids were fully conjugated in all species and conjugation was predominantly with taurine. However, conjugation with glycine was observed in some fruit doves (A104–A118) and pigeons (A122–A135), as reported previously (82). The majority of avian bile salts were C24 bile acids. CDCA was the most common bile acid, being present in three fourths of the avian species.
C27 bile acids were present in 12% of the avian species analyzed. They were present in ratites (A1–A3) and in the Andean condor (A61), species considered to have evolved quite early (77, 78). C27 bile acids were also present in the order Coraciiformes [hornbills (A136, A137), rollers (A138–A141)], the Micronesian kingfisher (A142), bee-eaters (A143, A144), and in the order Musophagiformes [turacos and plantain eaters (A157–A161)]. In the order Piciformes, C27 bile acids were present in two barbets (A172, A173), two toucans (A180, A181), one aracari (A182), and one of two woodpecker species (A186). In the order Passiformes, C27 bile acids were present in the family Corvidae [Hawaiian crow (A216) and the spotted nutcracker (A218)]. In the family Contingidae, C27 bile acids were present in the Capuchin bird (A219), bare-throated bellbird (A220), the Guianan cock of the rock (A221) and the pompadour cotinga (A222). They were also present in the family Eurylamidae [Northern lesser green broadbill (A230)]. In the family Icteridae, they were present in the long-crested helmet shrike (A236). In the family Pipridae, they were present in two of three manakins (A247, A248). In the family Plocidae, they were present in the Northern white-headed buffalo weaver (A249).
C27 bile alcohols were present in 3% of avian species. These included tinamous (A2, A3) and in three of five species in the order Musophagiformes, [the great blue and Livingston's turaco (A157, A160) and the Western gray plantain eater (A158)]. C27 bile alcohols were also present in the black-spotted barbet (A172), a trogon (A211), and two passeriform birds, the northern nutcracker (A218) and the Guianan cock of the rock (A221).
The A/B ring juncture of C27 and C24 bile acids was 5β in most bird species. Only 3% of species with C24 bile acids had allo bile acids exceeding 10% of total biliary bile acids. The Southern screamer (A4) had a bile salt pool consisting of over 60% allo-CDCA. Many species had a small percent (<5%) of allo bile acids.
In C27 bile acids, additional sites of hydroxylation were at C-1 in the red-winged tinamou (A3) (44) and at C-16 in the hornbills (A136, A137) and the Andean condor (A61).
In bird species with C24 bile acids, the dominant bile acid was CDCA, occurring in three-fourths of the species analyzed. Additional major sites of hydroxylation were at C-12 (CA) or C-16 (avicholic acid). Thus, hydroxylation at C-16 is a common pathway in birds and occurs as frequently as hydroxylation at C-12 in our samples. Far less common sites of hydroxylation were at C-1 (β-hydroxy) (82) and at C-15 (α-hydroxy) (83). Very low proportions of bile acids hydroxylated at C-4 [β-hydroxy (A51, A52, A54)] or C-5 [β-hydroxy; (A45, A52, A54)] were noted in some pheasants and tragopans.
Only three species analyzed had 7-deoxy bile acids; these are presumed to have been formed by bacterial dehydroxylation. The lumen of the avian colon in most avian species has a small volume, likely explaining the lack of 7-deoxy bile acids (14). Species containing 7-deoxy bile acids were the chinstrap penguin (A201), the Australian stone curlew (A76), and the snowy sheathbill (A80).
Side chain hydroxylation of C27 bile acids was not detected but there are many as yet unidentified C27 bile acids. In C24 bile acids, hydroxylation was only at C-23 and was present in 8% of species.
Evolutionary implications for bird species.
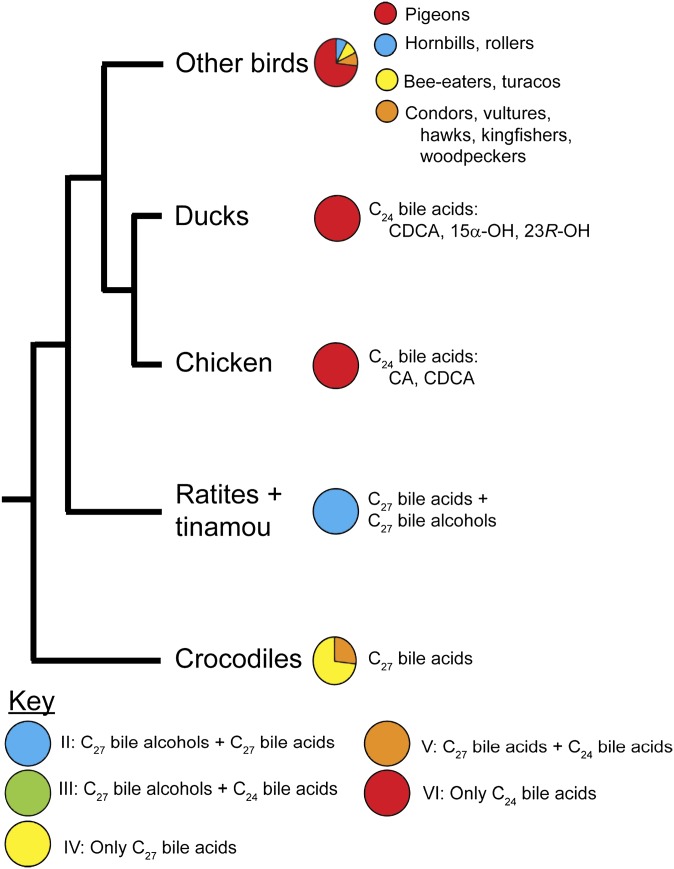
The phylogeny of birds has been actively debated for decades with both agreement and disagreement between morphology-based and genome-based phylogenies (69). Two recently published studies attempt to classify living birds based on comparisons of morphological characteristics (80) or DNA variation (79) across a large number of species from phylogenetically diverse bird families. Both studies place paleognath birds at the base of the avian phylogenetic tree. Figure 4 shows bile salt structural variation overlaid on a simplified phylogeny for birds with crocodilians as the most closely related outgroup. The bile salts of paleognath birds and crocodilians share the similarity of having trihydroxy (mainly 3α,7α,12α-trihydroxy) C27 bile acids as the dominant bile salts. However, unlike crocodilians, paleognath birds also have more than 10% C27 bile alcohols as well. There has obviously been substantial evolution of bile salt structural variation in birds, whose bile salts show hydroxylation sites uncommon in other vertebrates such as C-1, C-4, C-5, C-15, and C-23.
Fig. 4.
Variation of bile salt structures across birds. The evolutionary relationships of living bird groups are controversial, especially within the large number of passeriform birds (69, 79, 80). Nevertheless, recent large-scale morphological/paleontological (80) and molecular phylogenies (79) share agreement in placing ratite birds (cassowaries, rheas, emus, kiwis, ostriches) and tinamous (collectively called paleognathic birds) at the base of the bird evolutionary tree. Paleognathic birds share with crocodiles (hypothesized to be the closest living reptile relatives to modern birds) the phenotype of having bile salt profiles consisting largely of C27 bile acids. Each pie chart shows the proportion of birds in each group (using data from species analyzed so far) that are classified into one of six bile salt profiles based on the one or two bile salt classes (C27 bile alcohols, C27 bile acids, C24 bile acids) that account for 10% or more of the total biliary bile salt pool (see key). No bird species analyzed so far have a type I (C27 bile alcohols only) profile, so this type is not included in the plots.
Bile salts of mammals
Overview of bile salt composition in mammals.
Supplementary Table X contains the biliary bile salt profile of 172 mammalian species, some 4% of extant mammalian species. Bile salt profiles of mammals differed from those of nonmammals in three principal ways. First, in nine species, the 7α-hydroxy group of CDCA, the default C24 bile acid, was altered to either a 7-oxo group or a 7β-hydroxy group. Second, conjugation with glycine rather than taurine was much more frequent in mammals compared with nonmammals. Outside of mammals, glycine conjugation has been identified only in fruit doves and pigeons (82). Third, a much greater proportion of species analyzed (about one-third) contained ≥10% DCA in their biliary bile salts, a secondary bile acid formed by anaerobic bacteria. In fish, only five of 102 species analyzed contained DCA (in reptiles, only three of 103 species; in birds, only three of 272 species).
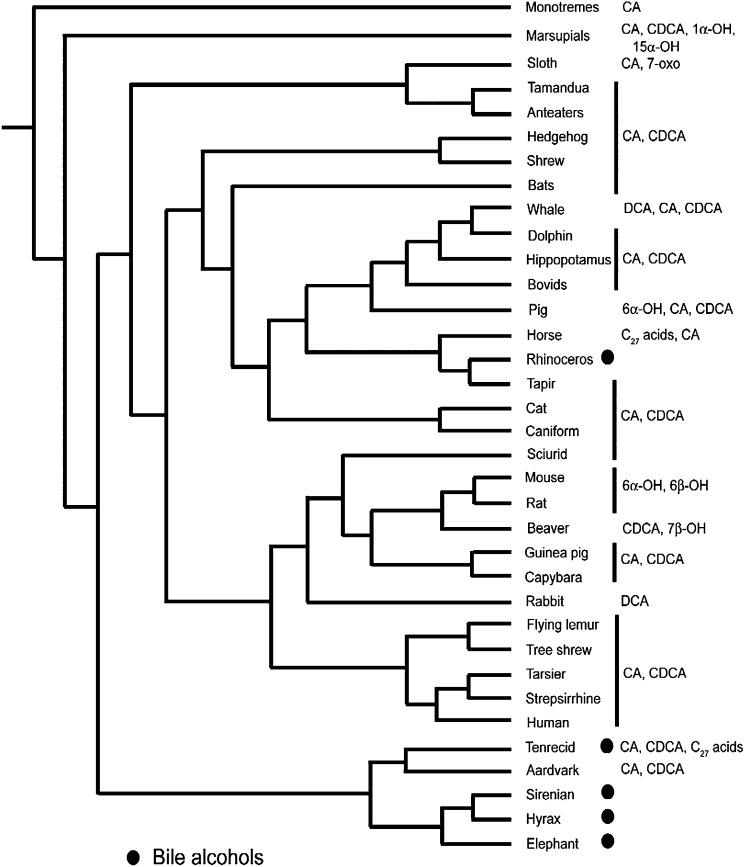
Most mammals had C24 bile acids. C27 bile alcohols were present in the related species of elephant (M19), manatee (M20–M22), and the rock hyrax (M23) (collectively referred to as Paenungulates) as well as the black rhinoceros (M50). C27 bile acids were present at ≥10% of total biliary bile salts in three species: the horse (M52) and two primates in the Loridae family [bushbaby (M171) and the Bengal slow loris (M172)].
The 7α-hydroxy group of the default C24 bile acid CDCA was oxidized to a 7-oxo group in the Queensland koala (M15, a marsupial) and two hutias (M40, M41), part of an infraorder of rodents known as caviomorphs that are found in South America. The 7α-hydroxy group of CDCA was epimerized to a 7β-hydroxy group (ursodeoxycholic acid, UDCA; 3α,7β-dihydroxy-5β-cholan-24-oic acid) in four caviomorphs (M41–M44), as well as in four bear species (M73–76) and in the North American beaver (M38).
Bile acids were generally present in conjugated form. Two exceptions were the Australian striped opossum (M13) and the South Eastern spotted cuscus (M14). The bile of these marsupials contained 1α,3α,7α-trihydroxy bile acid, the majority in unconjugated form (66). This bile acid, unique to marsupials, is much more hydrophilic than any other natural trihydroxy bile acid based on its HPLC retention time. Its hydrophilicity may explain its incomplete conjugation based on the observation that in rats, intravenously infused hydrophilic epimers of CA are secreted into bile in considerable part in unconjugated form (84).
Whether bile acids are conjugated with glycine or taurine, at least in mammals, depends on the taurine concentration in hepatocyte peroxisomes (85) and the taurine/glycine specificity of the bile acid amino transferase (86). Bile acids were conjugated mostly with taurine in monotremes (platypus and echidna; M1, M2), marsupials (M3–M16), insectivores (M26), and carnivores (M54–M94) including pinnipeds. Taurine conjugation predominated in rodents such as mice (M31–M33) and squirrels (M36, M37). In primates, both glycine and taurine conjugation was present in Hominidae [orangutans (M144) and humans (M146)], as well as in Cercopithecidae [guenons, macaques, and baboons (M146–M152)] and Atelidae [howlers (M158, M159) and the sifaka (M169)]. Among rodents, the golden hamster (M30), nutria (M34), and the beaver (M38) conjugated bile acids with both glycine and taurine. In bovids (M116–M143), both glycine and taurine conjugation was observed, as previously reported from this laboratory (48). Conjugation solely with glycine was observed for bile acids in lagomorphs (M45, M46), caviomorphs (M39–M44), as well as deer (M108–M110) and the Lesser Malay chevrotain (M107).
The presence of DCA as a major biliary bile acid indicates that DCA was formed in the colon, absorbed, and circulated enterohepatically with the primary bile acids. The absence of DCA in the biliary bile acids in species in which CA was a major bile acid may have several explanations. First, DCA may not be formed. Second, DCA may be formed and not absorbed; this is highly unlikely as DCA is membrane permeable and readily absorbed from the colon (87). Third, DCA may be absorbed but rehydroxylated during hepatocyte transport. Rehydroxylation is likely to be at C-7 in most species, generating CA, but can occur at other sites. Some species are known to efficiently rehydroxylate DCA, for example, the prairie dog (88). Others show limited rehydroxylation capacity, for example, the rat (89), the hamster (90), guinea pig (91), and probably the mouse (92). Still others, notably humans, are incapable of rehydroxylating DCA (93–95). Because DCA has a longer half life than that of its precursor CA, its proportion in bile can considerably exceed that CA. In supplementary Table XI, we have listed the species having >50% DCA in their biliary bile acids.
In C27 bile alcohols, hydroxylation at C-6 (β-OH) was observed in the manatee (M20–M22) and at C-12 in the rock hyrax (M23). The structure of the C27 bile acids occurring in the horse (M52) and the two primates in the Loridae family (M171, M172) has not been determined.
Additional hydroxylation to the default C24 bile acid (CDCA) was mainly at C-12 (generating CA). 6β-Hydroxylation is known to occur in mice (M32). 6α-Hydroxylation was observed in the Suidae (M94–97). Hydroxylation at C-1 (α-OH), as noted, was present in two marsupials (M13, M14).
The C27 bile alcohols of Paenungulates (M19–M23) were hydroxylated at C-25 in the side chain. In C24 bile acids, side chain hydroxylation was at C-23 and occurred only in pinnipeds (M68–M72).
The side chain of the C24 bile acids present in two agouti species (M41, M42) has a double bond at C-22. Such unsaturation of the side chain seems likely to occur in the hepatocyte during bile acid biosynthesis because of incomplete β-oxidation. In rats, the Δ22 derivative of β-muricholic acid also occurs and is not only formed in the hepatocyte but also in the intestine by bacterial enzymes (96, 97).
Evolutionary implications for mammals.
Although many aspects of mammalian phylogeny have been settled, there is still ongoing debate about which mammals form the base of the placental mammal tree (69, 98–100), with recent molecular studies defining a cohort Afrotheria that includes the Paenungulates (elephants, hyraxes, manatees) and also golden moles, elephant shrews, tenrecs, and aardvarks (101, 102). Bile salt structural variation in mammals is overlaid on a recent molecular-based phylogeny (100) in Fig. 5. It is striking that the bile salts of Paenungulates (100% C27 bile alcohols) are quite different from that of the tenrec (M25) and aardvark (M26), although the tenrec does have a minor fraction of C27 bile alcohols in its bile. The bile alcohols of Paenungulates also have uncommon hydroxylation at C-25, a trait also found in the bile alcohols of lobe-finned fish (103, 104), animals positioned at the base of the tetrapod evolutionary tree. The complete lack of bile acids in Paenungulates suggests that CYP27A1, a multi-functional enzyme that cannot only add a hydroxy group at C-27 but also convert the terminal methyl group to a carboxyl group, appears to have a more limited catalytic capability in Paenungulates.
Fig. 5.
Variation of bile salt structures across mammals. The evolutionary relationships of living mammals are still actively researched (69, 98). The phylogeny depicted is based on variation of 20 nuclear DNA sequences (100). The phylogeny is annotated with the major bile salts of each mammalian group. CA, CDCA, and DCA refer to cholic acid, chenodeoxycholic acid, and deoxycholic acid, respectively. C27 acids refers to the presence of C27 bile acids at more than 10% of the total bile salt pool. Modifications to the stem C24 bile acid including additional hydroxylation (1α-OH, 6α-OH, 6β-OH, 7β-OH, 15α-OH) and conversion of the 7α-hydroxyl group to a 7-oxo group are indicated.
The predominance of CA in mammals limits the phylogenetic inferences that can be made from bile salt structural variation. However, there are a number of mammals with relatively unique bile acids that confirm current phylogenetic models. First, the Paenungulates have C27 bile alcohols with hydroxylation at C-25 and form a recognized cluster of early evolving Afrotheres. Second, the entire groups of Pinnipeds, comprising the families Odobenidae (walruses), Otariidae (sea lions and fur seals), and Phocidae (earless seals), have C24 bile acids with hydroxylation at C-23, a phenotype not seen in other animals in the large order of Carnivora. Third, as mentioned above, bears share the phenotype of synthesizing bile acids with 7β-hydroxylation. Finally, marsupials show a number of unusual bile acid modifications, attesting to the geographic isolation of this group of mammals.
The evolutionary shift from C27 bile alcohols to C24 bile acids
If the last common ancestor to all extant vertebrates used C27 bile alcohols, then there has been a remarkable evolutionary shift to C24 5β- bile acids, commonly with one additional hydroxy group added to the nucleus at C-12. CA occurs in more species than any other bile acid in our sample of 650 vertebrate bile samples. We can speculate about the possible functional superiority of this C24 5β bile acid as compared with its C27 homolog or corresponding C27 alcohol.
For the elimination of cholesterol, the only structural requirement is water solubility and inefficient absorption from the biliary tract and intestine. The key parameter of aqueous solubility is adequate solubility of the calcium salt. The organ most likely to be the site of deposition of an insoluble calcium salt is the gallbladder because here, calcium activity is high (1 mM) and residence time is long (hours) (105). Indeed, in experimental animals, there are three known examples of the precipitation of insoluble calcium salts of bile acids when the circulating bile acids were enriched in an atypical bile acid. The first is precipitation of the calcium and sodium salts of the taurine conjugate of murideoxycholic acid (3α,6β-dihydroxy-5β-cholan-24-oic acid) in the gallbladder of prairie dogs to whom the unconjugated compound had been administered (106). The second is precipitation of the calcium salts of lithocholic acid (LCA, 3α-hydroxy-5β-cholan-24-oic acid) and its 6β-hydroxy metabolite, murideoxycholic acid, in taurine-depleted rats to whom LCA had been administered (107, 108). The third is precipitation of the calcium salt of the glycine conjugate of allo-DCA in rabbits that are fed 5α-cholestane-3β-ol (109). This 5α saturated derivative of cholesterol is absorbed and metabolized to allo-CA. Allo-CA undergoes bacterial 7-dehydroxylation to form allo-DCA which, in turn, is absorbed and conjugated with glycine. The calcium salt of the glycine conjugate of allo-DCA precipitates from solution in the gallbladder. Although this induced disease indicates that glycine- conjugated allo-DCA is unsatisfactory in its solubility properties, it is not relevant biologically for animals such as agamid lizards or other species whose biliary bile acids contain substantial proportions of 5α- bile acids because these species conjugate their bile acids with taurine, and the available information on 5β bile acids suggests that the calcium salts of taurine conjugates are quite water soluble (110).
Inefficient passive absorption in the biliary tract and intestine requires a molecule too large to pass through the paracellular junctions and too polar to flipflop across the lipid bilayer. Glycine and taurine conjugated bile acids and bile alcohol sulfates are fully ionized at the pH conditions prevailing in the small intestine. The negative charge on the amino acid or sulfate moiety precludes passive absorption across the lipid bilayer even for the most hydrophobic bile acids. Active absorption is mediated by the ileal bile acid transport system (111–113). The localization of the ileal bile acid transport system to the terminal ileum together with the membrane impermeant nature of conjugated bile salts results contributes to the high luminal concentration of bile salts throughout the majority of the small intestine.
Intestinal conservation of circulating bile acids results in the accumulation of a circulating bile salt pool, which is conveniently stored under sterile conditions in the gallbladder. The gallbladder is present in the ancient vertebrates (e.g., jawless fish) indicating that it evolved together with the enterohepatic circulation of bile salts.
A micellar phase that promotes rapid absorption of lipids should be useful for species that obtain most of their caloric needs from absorption of nutrients in the small intestine. For micellar solubilization of dietary lipids, amphipathic properties are required. So far as is known, all natural primary bile salts are amphipathic, capable of rapidly transforming lipid bilayers to mixed micelles at low concentrations (114). However, the addition of a nuclear hydroxy group increases the critical micellization concentration (CMC) value (115), suggesting that there must be other reasons for forming trihydroxy bile acids.
The amphipathic properties of bile salts arise from the hydroxy groups of both the default structure and the additional hydroxy group all being present on one side (the α face) of the bile acid molecule (116). No information is available on micelle formation of most of the natural bile alcohol sulfates or of C27 bile acids. The CMC of 5α-cyprinol sulfate and its lipid solubilizing properties were nearly identical to that of taurocholate (13). The CMC of C27 bile acids with unsubstituted side chains should be lower than those of their corresponding C24 homologs (116). The natural bile acid with the highest CMC is the 3-hydroxy-7-oxo- compound occurring in caviomorphs. Guinea pigs have bile that is extremely dilute (117) and it remains possible that a micellar phase is not present in the small intestinal content of caviomorphs.
To promote lipid solubilization in the small intestine, conjugated bile salts must remain intact. Pancreatic juice is lacking in sulfatases, precluding hydrolysis of the ester bond linking the sulfate group in bile alcohol sulfates. C24 bile acids conjugated with amino acids other than glycine or taurine are rapidly hydrolyzed by pancreatic carboxypeptidases, but glycine and taurine conjugated bile acids are completely resistant (118).
In some fish, reptiles, birds, and mammals, bile acids encounter intestinal bacterial enzymes during their enterohepatic cycling. So far as is known, both bile alcohol sulfates and conjugated bile acids undergo deconjugation in species with a distal intestinal bacterial flora. Probably, most liberated bile alcohols are too polar to be absorbed passively. A gallstone consisting mostly of unsulfated bile alcohols has been reported to occur in the elephant (119). In contrast to unconjugated bile alcohols, unconjugated bile acids can be absorbed passively from the distal intestine (87). Therefore, a bile acid is superior to a bile alcohol for retention in the enterohepatic circulation.
Hind gut fermenters such as horses rely on their cecal bacteria to convert nonabsorbed polysaccharides into short-chain fatty acids that are absorbed and provide an important caloric source. Bile acids have antimicrobial properties (120–123), and a high cecal concentration of bile acids might jeopardize efficient conversion of unabsorbed polysaccharides to short-chain fatty acids. The aqueous concentration of bile acids in the colon is decreased by deconjugation and 7-dehydroxylation, as well as by the acidic pH of cecal contents, as unconjugated bile acids are poorly soluble at cecal pH. In humans, cecal content has a bile acid concentration averaging 0.4 mM (124) in contrast to a concentration twenty times higher in the proximal small intestine.
CDCA was a primary bile acid in more than half of the mammalian species and there is potential toxicity arising from its bacterial metabolite LCA. LCA is an extremely toxic bile acid when fed to animals (13, 125, 126) and is never present in bile above 4% of biliary bile acids. One biological solution to the LCA toxicity problem is to add an additional hydroxy group to the default structure during primary bile acid biosynthesis. When trihydroxy bile acids undergo 7-dehydroxylation, the result is a dihydroxy bile acid having a 3,X-dihydroxy nuclear structure, where X is the additional hydroxy group. Detoxification of LCA is also achieved by efficient sulfation of its C-3 hydroxy group. Sulfation of LCA is the mode of LCA detoxification in humans (127, 128). Sulfated lithocholyl amidates are secreted into bile. They are not substrates for the ileal bile acid transport system and are rapidly excreted.
There are striking species differences in the toxicity of orally administered CDCA and these differences can be explained by success in LCA detoxification. In humans, CDCA is rather safe when given at a dose of 10 mg/kg-day (129), but in rabbits (130), rhesus monkeys (131, 132), and baboons (133), species that cannot detoxify LCA, CDCA is highly toxic. Another striking example of species-specific toxicity of bile acids is the guinea pig. Administration of the taurine conjugate of CA to the guinea pig results in the accumulation of DCA and its 3-oxo derivative, followed by death for unknown reasons (L. R. Hagey and A. F. Hofmann, unpublished observations).
As noted, the addition of a hydroxy group to the default structure (CDCA) precludes the formation of LCA. However, hydroxylation at C-12 is present in bile alcohol sulfates in ancient fish and as noted bile alcohols if formed by bacterial esterases are unlikely to be absorbed. Therefore, hydroxylation at C-12 is likely to have another function such as enhancing aqueous solubility.
In snakes and pinniped mammals, bile acids with a hydroxy group at C-23 are formed. The taurine conjugates of these acids are more resistant to bacterial deconjugation than the taurine conjugate of the corresponding bile acid with an unsubstituted side chain (77).
Coevolution of bile salts and bile salt receptors
The variation of bile salt structures across species suggests that receptors for bile salts would have cross-species variations in their ligand selectivity. Indeed, investigations in this area have revealed interesting model systems for the study of ligand-receptor coevolution. In humans and rodents, three nuclear hormone receptors are involved in regulation of bile acid synthesis and/or excretion: farnesoid X receptor (FXR, NR1H4) (5–7), pregnane X receptor (PXR, NR1I2) (134), and vitamin D receptor (VDR, NR1I1) (135, 136). FXR serves as the major transcriptional regulator of bile salt synthesis, in part by controlling the expression of CYP7A1, the rate-limiting enzyme in bile acid and presumably bile alcohol synthesis (137, 138). Human FXR is strongly activated by the primary bile acid CDCA (5–7, 137). VDR and PXR both regulate the expression of enzymes (e.g., CYP3A, sulfotransferases) in enterocytes and hepatocytes that can hydroxylate or sulfate LCA (135, 136, 139).
Studies of nonmammalian FXRs have shown significant differences in protein sequence and ligand specificity. The African clawed frog (Xenopus laevis) expressed an unusual FXR (also termed FOR, FXR-like orphan receptor) that has a 33 amino acid insert (relative to mammalian FXRs) in helix-7 of the ligand-binding domain. Recombinant Xenopus laevis FXR was not activated by C24 bile acids such as CDCA but was activated by a partially purified extract of frog bile (140). Further study showed that Xenopus laevis FXR could be activated by C27 bile alcohols, which are the dominant primary bile salts of this amphibian (9). The ligand specificities of recombinant sea lamprey (Petromyzon marinus) and zebrafish (Danio rerio) FXRs were also studied in a reporter assay system. These two animals synthesize 5α bile alcohols as their primary bile salts. Lamprey and zebrafish FXRs were each found to be activated strongly by 5α bile alcohols but not 5β bile acids or alcohols (9). Homology modeling of the structure of these FXRs, using the high-resolution crystal structure of rat FXR as the template (141), predicted a ligand-binding pocket with a shape ideal for accommodating planar 5α bile alcohols but not for the bent structure of 5β bile salts (9). This contrasts with the structure of rat FXR, which has a curved ligand-binding pocket suitable for binding 5β bile acids (141). Thus, these studies demonstrate an interesting coevolution of FXR and bile salt structures with changes in the architecture of the FXR ligand-binding pocket between species.
The first comprehensive study of multiple nonmammalian PXRs showed no activation by C24 bile acids (142). Later studies, however, found that zebrafish PXR was strongly activated by C27 5α bile alcohols that are the primary bile salts of zebrafish (143). Homology modeling of zebrafish PXR, using a high-resolution crystal structure of human PXR as a template (144), predicted a planar binding pocket that could accommodate 5α but not 5β bile salts, a finding similar to that observed for lamprey and zebrafish FXRs (9).
Human and mice VDRs are activated by a small number of bile acids, mainly LCA and its derivatives (135, 136). It has been proposed that activation of VDR by LCA serves as a protective response against this potentially toxic secondary bile acid (136), although the in vivo importance of this has yet to be ascertained. Interestingly, three nonmammalian VDRs (lamprey, zebrafish, Xenopus laevis) studied so far were found to be insensitive to all bile acids and alcohols (143, 145, 146). This raises the possibility that the ability of VDRs to bind bile acids is a more recent evolutionary development, possibly as an adaptation to changes in intestinal anatomy and microbial colonization that allow for generation of toxic secondary bile acids such as LCA.
Areas of ignorance and future studies
The extensive structural variation of bile salts across vertebrates highlights a number of questions. The most basic is what evolutionary factors drive the variation in bile salts. We have speculated as to the possible superiority of C24 5β bile acids. With the exception of 5α-cyprinol sulfate (main bile salt of cyprinid fish) (14), the physicochemical properties of C27 bile acids and C27 bile alcohols have not been studied in detail. Beyond known functions such as lipid solubilization and induction of bile flow, what other functions do bile salts perform? Signaling properties of bile acids are now being identified (147) as well as hormonal actions (148). Bile acids have also been proposed to be osmosensors, regulating colonic secretion (149).
There are many unanswered questions with regard to how bile composition is regulated and how the bile salt synthetic pathways vary in animals using bile salts other than typical C24 bile acids such as CA or CDCA. For instance, why do the Paenungulates produce only C27 bile alcohols whereas bile salts of the closely related aardvark are nearly all C24 bile acids? For animals that synthesize mainly C27 bile acids, how is side-chain cleavage avoided, given that the peroxisomal enzymes involved in bile salt side-chain cleavage are also involved in fatty acid oxidation and thus are likely conserved across most vertebrates (2, 3)? The pathways by which 5α-bile salts are synthesized have also not been defined in any vertebrate species. Ongoing or future genome sequencing efforts in animals with unusual bile salt profiles (e.g., elephant, hyrax, anole lizard, zebrafish, coelacanth, sea lamprey) will likely be helpful in defining differences and similarities between the bile salt enzymes of these species as compared with humans and rodents.
Note added in proof
After this manuscript had been submitted, Bi et al (Chem. Pharm Bull 2009;57:528-31) reported the presence in the bile of the sun bear (Selenaretos thibetanus Cuvier) trace proportions of a previously undescribed bile acid, 3α,7α,9α-trihydroxy-5β-cholan-24-oic acid.
Supplementary Material
Acknowledgments
We acknowledge with gratitude the receipt of bile samples from our colleagues throughout the world.
Footnotes
Abbreviations:
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- CMC
- critical micellization concentration
- DCA
- deoxycholic acid
- FXR
- farnesoid X receptor
- LCA
- lithocholic acid
- PXR
- pregnane X receptor
- UDCA
- ursodeoxycholic acid
- VDR
- vitamin D receptor
M.D.K. is supported by K08-GM074238 from the National Institutes of Health. A.F.H. and L.R.H. have been supported in the past by grants from the National Institutes of Health as well as grants-in-aid from the Falk Foundation e.V., Freiburg, Germany and the University of California, San Diego. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six tables. These tables give biliary bile acid composition for individual vertebrate species.
REFERENCES
- 1.Hofmann A. F., Hagey L. R. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norlin M., Wikvall K. 2007. Enzymes in the conversion of cholesterol into bile acids. Curr. Mol. Med. 7: 199–218. [DOI] [PubMed] [Google Scholar]
- 3.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 4.Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., Noonan D. J., Burka L. T., McMorris T., Lamph W. W., et al. 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 81: 687–693. [DOI] [PubMed] [Google Scholar]
- 5.Makishima M., Okamato A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365. [DOI] [PubMed] [Google Scholar]
- 6.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 7.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. 1999. Endogenous bile acids are ligand for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553. [DOI] [PubMed] [Google Scholar]
- 8.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 9.Reschly E. J., Ai N., Ekins S., Welsh W. J., Hagey L. R., Hofmann A. F., Krasowski M. D. 2008. Evolution of the bile salt nuclear receptor FXR in vertebrates. J. Lipid Res. 49: 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridlon J. M., Kang D. J., Hylemon P. B. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 11.Shefer S., Salen G., Hauser S., Dayal B., Batta A. K. 1982. Metabolism of iso-bile acids in the rat. J. Biol. Chem. 257: 1401–1406. [PubMed] [Google Scholar]
- 12.Marschall H. U., Broome U., Einarsson C., Alvelius G., Thomas H. G., Matern S. 2001. Isoursodeoxycholic acid: metabolism and therapeutic effects in primary biliary cirrhosis. J. Lipid Res. 42: 735–742. [PubMed] [Google Scholar]
- 13.Goto T., Holzinger F., Hagey L. R., Cerrè C., Ton-Nu H-T., Schteingart C. D., Steinbach J. H., Shneider B. L., Hofmann A. F. 2003. Physicochemical and physiological properties of 5a-cyprinol sulfate, the toxic bile salt of cyprinid fish. J. Lipid Res. 44: 1643–1651. [DOI] [PubMed] [Google Scholar]
- 14.Stevens C. E. 1988. Comparative Physiology of the Vertebrate Digestive System. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 15.Okuda K. I. 1994. Liver mitochondrial P450 involved in cholesterol catabolism and vitamin D activation. J. Lipid Res. 35: 361–372. [PubMed] [Google Scholar]
- 16.Hagey L. R., Krisans S. K. 1982. Degradation of cholesterol to propionic acid by rat liver peroxisomes. Biochem. Biophys. Res. Commun. 107: 834–841. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen J. I., Gustafsson J. 1980. Conversion of 3α,7α,12α-trihydroxy-5β-cholestanoic acid into cholic acid by rat liver peroxisomes. FEBS Lett. 121: 345–348. [DOI] [PubMed] [Google Scholar]
- 18.Björkhem I. 1992. Mechanism of degradation of the steroid side chain in the formation of bile acids. J. Lipid Res. 33: 455–471. [PubMed] [Google Scholar]
- 19.Ferdinandusse S., Denis S., Faust P. L., Wanders R. J. 2009. Bile acids: role of peroxisomes. J. Lipid Res. 50:2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haslewood G. A. D., Tökés L. 1969. Comparative studies of bile salts: bile salts of the lamprey Petromyzon marinus L. Biochem. J. 114: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshita T. 1985. Bile alcohols and primitive bile acids. Sterols and Bile Acids. Danielsson H., Sjövall J., Elsevier Science Publishers, Amsterdam: 279–302. [Google Scholar]
- 22.Une M., Hoshita T. 1994. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima J. Med. Sci. 43: 37–67. [PubMed] [Google Scholar]
- 23.Moschetta A., Xu F., Hagey L. R., van Berge Henegouwen G. P., van Erpecum K. J., Brouwers J. F., Cohen J. C., Bierman M., Hobbs H. H., Steinbach J. H., et al. 2005. A phylogenetic survey of biliary lipids in vertebrates. J. Lipid Res. 46: 2221–2232. [DOI] [PubMed] [Google Scholar]
- 24.Kuramoto T., Moriwaki S., Kawamoto K., Hoshita T. 1987. Intestinal absorption and metabolism of homoursodeoxycholic acid in rats. J. Pharmacobiodyn. 10: 309–316. [DOI] [PubMed] [Google Scholar]
- 25.Katona B. W., Cummins C. L., Ferguson A. D., Li T., Schmidt D. R., Mangelsdorf D. J., Covey D. F. 2007. Synthesis, characterization, and receptor interaction profiles of enantiomeric bile acids. J. Med. Chem. 50: 6048–6058. [DOI] [PubMed] [Google Scholar]
- 26.Une M., Kuramoto T., Hoshita T. 1983. The minor bile acids of the toad, Bufo vulgaris formosus. J. Lipid Res. 24: 1468–1474. [PubMed] [Google Scholar]
- 27.Mui M. M., Kamat S. Y., Elliott W. H. 1974. Bile acids XLII. Preparation of 5a-cholestan-26-oic acids from kryptogenin. Steroids. 24: 239–250. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann A. F., Mosbach E. H., Sweeley C. C. 1969. Bile acid composition of bile from germ-free rabbits. Biochim. Biophys. Acta. 176: 204–207. [DOI] [PubMed] [Google Scholar]
- 29.Kallner A. 1967. On the biosynthesis and metabolism of allodeoxycholic acid in the rat. Acta Chem. Scand. 21: 315–321. [DOI] [PubMed] [Google Scholar]
- 30.Haslewood G. A. D., Wootton V. M. 1951. Comparative studies of ‘bile salts’. 2. Pythocholic acid. Biochem. J. 49: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridgwater R. J., Briggs T., Haslewood G. A. D. 1962. Comparative studies of ‘bile salts’. 14. Isolation from shark bile and partial synthesis of scymnol. Biochem. J. 82: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshita T., Hirofuji S., Sasaki T., Kazuno T. 1967. Stero-bile acids and bile alcohols. LXXXVII. Isolation of a new bile acid, haemulcholic acid from the bile of Parapristipoma trilineatum. J. Biochem. 61: 136–141. [DOI] [PubMed] [Google Scholar]
- 33.Haslewood G. A. D. 1967. Bile salt evolution. J. Lipid Res. 8: 535–550. [PubMed] [Google Scholar]
- 34.Dayal B., Tint G. S., Batta A. K., Shefer S., Salen G., Bose A. K., Pramanik B. N. 1983. Synthesis of 3α,7α,12α,25-tetrahyroxy-5β-cholestan-24-one, an intermediate in the 25-hydroxylation pathway of cholic acid biosynthesis from cholesterol. J. Lipid Res. 24: 208–219. [PubMed] [Google Scholar]
- 35.Iida T., Hikosaka M., Kakiyama G., Shiraishi K., Schteingart C. D., Hagey L. R., Ton-Nu H. T., Hofmann A. F., Mano N., Goto J., et al. 2002. Potential bile acid metbolites. 25. Synthesis and chemical properties of stereoisomeric 3α,7α,16- and 3α,7α,15-trihydroxy-5β-cholan-24-oic acids. Chem. Pharm. Bull. (Tokyo). 50: 1327–1334. [DOI] [PubMed] [Google Scholar]
- 36.Kakiyama G., Tamegai H., Iida T., Mitamura K., Ikegawa S., Goto T., Mano N., Goto J., Holz P., Hagey L. R., et al. 2007. Isolation and chemical synthesis of a major, novel biliary bile acid in the common wombat (Vombatus Ursinus): 15α-hydroxylithocholic acid. J. Lipid Res. 48: 2682–2692. [DOI] [PubMed] [Google Scholar]
- 37.Bergström S., Danielsson H. 1963. Factors influencing formation and excretion of bile acids. Biochem. Soc. Symp. 24: 63–73. [PubMed] [Google Scholar]
- 38.Sjövall J. 2004. Fifty years with bile acids and steroids in health and disease. Lipids. 39: 703–722. [DOI] [PubMed] [Google Scholar]
- 39.Danielsson H., Sjövall J. 1975. Bile acid metabolism. Annu. Rev. Biochem. 44: 233–253. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann A. F. 1962. Thin layer adsorption chromatography of free and conjugated bile acids on silicic acid. J. Lipid Res. 3: 127–128. [Google Scholar]
- 41.Rossi S. S., Converse J. L., Hofmann A. F. 1987. High pressure liquid chromatography analysis of conjugated bile acids in human bile: simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugated bile acids. J. Lipid Res. 28: 589–595. [PubMed] [Google Scholar]
- 42.Chatman K., Hollenbeck T., Hagey L. R., Vallee M., Purdy R., Weiss F., Suizdak G. 1999. Nanoelectrospray mass spectrometry and precursor ion monitoring for quantitative steroid analysis and attomole sensitivity. Anal. Chem. 71: 2358–2363. [DOI] [PubMed] [Google Scholar]
- 43.Hagey L. R., Schteingart C. D., Ton-Nu H. T., Hofmann A. F. 2002. A novel primary bile acid in the Shoebill stork and herons and its phylogenetic significance. J. Lipid Res. 43: 685–690. [PubMed] [Google Scholar]
- 44.Cohen B. I., Budai K., Javitt N. B. 1981. Solvolysis of chenodeoxycholic acid sulfates. Steroids. 37: 621–626. [DOI] [PubMed] [Google Scholar]
- 45.Kuroki S., Schteingart C. D., Hagey L. R., Cohen B. I., Mosbach E. H., Rossi S. S., Hofmann A. F., Matoba N., Une M., Hoshita T., et al. 1988. Bile salts of the West Indian manatee, Trichechus manatus latirostris: novel bile alcohol sulfates and absence of bile acids. J. Lipid Res. 29: 509–552. [PubMed] [Google Scholar]
- 46.Hagey L. R., Kakiyama G., Muto A., Iida T., Mushiake K., Goto T., Mano N., Goto J., Oliveira C. A., Hofmann A. F. 2009. A new, major C27 biliary bile acid in the Red-winged tinamou (Rhynchotus rufescens): (25R)-1β,3α,7α-trihydroxy-5β-cholestan-27-oic acid. J. Lipid Res. 50: 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hofmann A. F., Grundy S. M., Lachin J. M., Lan S. P., Baum R. A., Hanson R. F., Hersh T., Hightower N. C., Jr., Marks J. W., Mekhjian H., et al. 1982. Pretreatment biliary lipid composition in white patients with radiolucent gallstones in the National Cooperative Gallstone Study. Gastroenterology. 83: 738–752. [PubMed] [Google Scholar]
- 48.Hagey L. R., Gavrilkina M. A., Hofmann A. F. 1997. Age-related changes in the biliary bile acid composition of bovids. Can. J. Zool. 75: 1193–1201. [Google Scholar]
- 49.Hellou J., King A., Ni I. H. 1988. Bile acids from the harp seals, Phoca groenlandica. Comp. Biochem. Physiol. Comp. Physiol. 89: 211–214. [DOI] [PubMed] [Google Scholar]
- 50.Tammar A. R. 1974. Bile salts in fishes. Chem. Zool. 8: 595–612. [Google Scholar]
- 51.Forey P., Janvier P. 1993. Agnathans and the origin of jawed vertebrates. Nature. 361: 129–134. [Google Scholar]
- 52.Nelson J. S. 2006. Fishes of the World. 4th ed John Wiley and Sons, Inc., Hoboken, NJ. [Google Scholar]
- 53.Haslewood G. A. D. 1966. Comparative studies of bile salts. Myxinol disulphate, the principal bile salt of hagfish (Myxinidae). Biochem. J. 100: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huertas M., Hubbard P. C., Canário A. M., Cerdà J. 2007. Olfactory sensitivity to conspecific bile fluid and skin mucus in the European eel Anguilla anguilla (L). J. Fish Biol. 70: 1907–1920. [Google Scholar]
- 55.Li W., Scott A. P., Siefkes M. J., Yan H., Liu Q., Yun S. S., Gage D. A. 2002. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 296: 138–141. [DOI] [PubMed] [Google Scholar]
- 56.Sorensen P. W., Fine J. M., Dvornikovs V., Jeffrey C. S., Shao F., Wang J., Vrieze L. A., Anderson K. R., Hoye T. R. 2005. Mixture of new sulfated steroids functions as a migratory pheromone in the sea lamprey. Nat. Chem. Biol. 1: 324–328. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C., Brown S. B., Hara T. J. 2001. Biochemical and physiological evidence that bile acids produced and released by lake char (Salvelinus namaycush) function as chemical signals. J. Comp. Physiol. [B]. 171: 161–171. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C., Hara T. J. 2009. Lake char (Salvelinus namaycush) olfactory neurons are highly sensitive and specific to bile acids. J. Comp. Physiol. A. Neuroethol. Sens. Neural Behav. Physiol. 195: 203–215. [DOI] [PubMed] [Google Scholar]
- 59.Hofmann A. F., Loening-Baucke V., Lavine J. E., Hagey L. R., Steinbach J. H., Packard C. A., Griffin T. L., Chatfield D. A. 2008. Altered bile acid metabolism in childhood functional constipation: inactivation of secretory bile acids by sulfation in a subset of patients. J. Pediatr. Gastroenterol. Nutr. 47: 598–606. [DOI] [PubMed] [Google Scholar]
- 60.Bridgwater R. J., Haslewood G. A. D., Watt J. R. 1963. Comparative studies of ‘bile salts’. 17. A bile alcohol from Chimaera monstrosa. Biochem. J. 87: 28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haslewood G. A. 1954. A bile alcohol from Cyprinidae. Biochem. J. 59: xi. [PubMed] [Google Scholar]
- 62.Goto T., Ui T., Une M., Kuramoto T., Kihira K., Hoshita T. 1996. Bile salt composition and distribution of the D-cysteinolic acid conjugated bile salts in fish. Fish. Sci. 62: 606–609. [Google Scholar]
- 63.Une M., Goto T., Kihira K., Kuramoto T., Hagiwara K., Nakajima T., Hoshita T. 1991. Isolation and identification of bile salts conjugated with cysteinolic acid from bile of the red seabream, Pagrosomus major. J. Lipid Res. 32: 1619–1623. [PubMed] [Google Scholar]
- 64.Schmassmann A., Fehr H. F., Locher J., Lillienau J., Schteingart C. D., Rossi S. S., Hofmann A. F. 1993. Cholylsarcosine, a new bile acid analogue: metabolism and effect on biliary secretion in humans. Gastroenterology. 104: 1171–1181. [DOI] [PubMed] [Google Scholar]
- 65.Schmassmann A., Hofmann A. F., Angellotti M. A., Ton-Nu H. T., Schteingart C. D., Clerici C., Rossi S. S., Rothschild M. A., Cohen B. I., Stenger R. J., et al. 1990. Prevention of ursodeoxycholate hepatotoxicity in the rabbit by conjugation with N-methyl amino acids. Hepatology. 11: 989–996. [DOI] [PubMed] [Google Scholar]
- 66.Lee S. P., Lester R., Pyrek J. S. 1987. Vulpecholic acid (1α, 3α,7α-trihydroxy-5β-cholan-24-oic acid): a novel bile acid of a marsupial, Trichosurus vulpecula (Lesson). J. Lipid Res. 28: 19–31. [PubMed] [Google Scholar]
- 67.Haslewood G. A. D., Tökés L. 1972. Comparative studies of bile salts. A new type of bile salt from Arapaima gigas (Cuvier) (family Osteoglossidae). Biochem. J. 126: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roda A., Grigolo B., Minutello A., Pellicciari R., Natalini B. 1990. Physicochemical and biological properties of natural and synthetic C-22 and C-23 hydroxylated bile acids. J. Lipid Res. 31: 289–298. [PubMed] [Google Scholar]
- 69.Meyer A., Zardoya R. 2003. Recent advances in the (molecular) phylogeny of vertebrates. Annu. Rev. Ecol. Evol. Syst. 34: 311–338. [Google Scholar]
- 70.Delarbre C., Gallut C., Barriel V., Janvier P., Gachelin G. 2002. Complete mitochondrial DNA of the hagfish, Eptatretus burgeri: the comparative analysis of mitochondrial DNA sequences strongly supports the cyclostome monophyly. Mol. Phylogenet. Evol. 22: 184–192. [DOI] [PubMed] [Google Scholar]
- 71.Janvier P. 2007. Evolutionary biology: born-again hagfishes. Nature. 446: 622–623. [DOI] [PubMed] [Google Scholar]
- 72.Anderson I. G., Briggs T., Haslewood G. A. D., Oldham R. S., Scharen H., Tökés L. 1979. Comparison of the bile salts of frogs with those of their tadpoles. Bile-salt changes during the metamorphosis of Rana Catesbeiana Shaw. Biochem. J. 183: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson I. G., Haslewood G. A. D., Oldham R. S., Amos B., Tökés L. 1974. A more detailed study of bile salt evolution, including techniques for small-scale identification and their application to amphibian biles. Biochem. J. 141: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reschly E. J., Ai N., Welsh W. J., Ekins S., Hagey L. R., Krasowski M. D. 2008. Ligand specificity and evolution of liver X receptors. J. Steroid Biochem. Mol. Biol. 110: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kihira K., Yasuhara M., Kuramoto T., Hoshita T. 1977. New bile alcohols. 5α- and 5β-dermophols from amphibians. Tetrahedron Lett. 8: 687–690. [Google Scholar]
- 76.Bergström S., Danielsson H., Kazuno T. 1960. Bile acids and steroids 98. The metabolism of bile acids in python and constrictor snakes. J. Biol. Chem. 235: 983–988. [PubMed] [Google Scholar]
- 77.Merrill J. R., Schteingart C. D., Hagey L. R., Peng Y., Ton-Nu H. T., Frick E., Jirsa M., Hofmann A. F. 1996. Hepatic biotransformation in rodents and physicochemical properties of 23(R)-hydroxychenodeoxycholic acid, a natural α-hydroxy bile acid. J. Lipid Res. 37: 98–112. [PubMed] [Google Scholar]
- 78.Vidal N., Hedges S. B. 2009. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C. R. Biol. 332: 129–139. [DOI] [PubMed] [Google Scholar]
- 79.Hackett S. J., Kimball R. T., Reddy S., Bowie R. C. K., Braun E. L., Braun M. J., Chojnowski J. L., Cox W. A., Han K-L., Harshman J., et al. 2008. A phylogenomic study of birds reveals their evolutionary history. Science. 320: 1763–1768. [DOI] [PubMed] [Google Scholar]
- 80.Livezey B. C., Zusi R. L. 2007. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zool. J. Linn. Soc. 149: 1–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devine J. 2008. A note on the morphology of the gallbladder. ANZ J. Surg. 11: 264–267. [Google Scholar]
- 82.Hagey L. R., Schteingart C. D., Ton-Nu H-T., Hofmann A. F. 1994. Biliary bile acids of fruit pigeons and doves (Columbiformes): presence of 1β-hydroxy-chenodeoxycholic acid and conjugation with glycine as well as taurine. J. Lipid Res. 35: 2041–2048. [PubMed] [Google Scholar]
- 83.Kakiyama G., Iida T., Goto T., Mano N., Goto J., Nambara T., Hagey L. R., Schteingart C. D., Hofmann A. F. 2006. Identification of a novel bile acid in swans, tree ducks, and geese: 3α,7α,15α-trihydroxy-5β-cholan-24-oic acid. J. Lipid Res. 47: 1551–1558. [DOI] [PubMed] [Google Scholar]
- 84.Borgström B., Barrowman J., Krabisch L., Lindstrom M., Lillienau J. 1986. Effects of cholic acid, 7β-hydroxy- and 12β-hydroxy-isocholic acid on bile flow, lipid secretion and bile acid synthesis in the rat. Scand. J. Clin. Lab. Invest. 46: 167–175. [DOI] [PubMed] [Google Scholar]
- 85.Hardison W. G. 1978. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology. 75: 71–75. [PubMed] [Google Scholar]
- 86.He D., Barnes S., Falany C. N. 2003. Rat liver bile acid CoA:amino acid N-acyltransferase: expression, characterization, and peroxisomal localization. J. Lipid Res. 44: 2242–2249. [DOI] [PubMed] [Google Scholar]
- 87.Mekhjian H. S., Phillips S. F., Hofmann A. F. 1979. Colonic absorption of unconjugated bile acids: perfusion studies in man. Dig. Dis. Sci. 24: 545–550. [DOI] [PubMed] [Google Scholar]
- 88.Cohen B. I., Singhal A. K., Mongelli J., Rothschild M. A., McSherry C. K., Mosbach E. H. 1983. Hydroxylation of secondary bile acids in the perfused prairie dog liver. Lipids. 18: 909–912. [DOI] [PubMed] [Google Scholar]
- 89.Clayton L. M., Gurantz D., Hofmann A. F., Hagey L. R., Schteingart C. D. 1989. Role of bile acid conjugation in hepatic transport of dihydroxy bile acids. J. Pharmacol. Exp. Ther. 248: 1130–1137. [PubMed] [Google Scholar]
- 90.Kuroki S., Mosbach E. H., Stenger R. J., Cohen B. I., McSherry C. K. 1987. Comparative effects of deoxycholate and 7-methyl-deoxycholate in the hamster. Hepatology. 7: 229–234. [DOI] [PubMed] [Google Scholar]
- 91.Cantafora A., Alvaro D., Attili A. F., Di Biase A., Anza M., Mantovani A., Angelico M. 1986. Hepatic 3α-dehydrogenation and 7α-hydroxylation of deoxycholic acid in the guinea-pig. Comp. Biochem. Physiol. B. 85: 805–810. [DOI] [PubMed] [Google Scholar]
- 92.Wang D. Q., Tazuma S., Cohen D. E., Carey M. C. 2003. Feeding natural hydrophilic bile acids inhibits intestinal cholesterol absorption: studies in the gallstone-susceptible mouse. Am. J. Physiol. Gastrointest. Liver Physiol. 285: G494–G502. [DOI] [PubMed] [Google Scholar]
- 93.Hanson R. F., Williams G. 1971. Metabolism of deoxycholic acid in bile fistula patients. J. Lipid Res. 12: 688–691. [PubMed] [Google Scholar]
- 94.LaRusso N. F., Szczepanik P. A., Hofmann A. F. 1977. Effect of deoxycholic acid ingestion on bile acid metabolism and biliary lipid secretion in normal subjects. Gastroenterology. 72: 132–140. [PubMed] [Google Scholar]
- 95.Matern S., Sjövall J., Pomare E. W., Heaton K. W., Low-Beer T. S. 1975. Metabolism of deoxycholic acid in man. Med. Biol. 53: 107–113. [PubMed] [Google Scholar]
- 96.Kakiyama G., Iida T., Yoshimoto A., Goto T., Mano N., Goto J., Nambara T., Hagey L. R., Hofmann A. F. 2004. Chemical synthesis of (22E)-3α,6β,7β-trihydroxy-5β-chol-22-en-24-oic acid and its taurine and glycine conjugates: a major bile acid in the rat. J. Lipid Res. 45: 567–573. [DOI] [PubMed] [Google Scholar]
- 97.Rodrigues C. M., Kren B. T., Steer C. J., Setchell K. D. 1996. Formation of delta 22-bile acids in rats is not gender specific and occurs in the peroxisome. J. Lipid Res. 37: 540–550. [PubMed] [Google Scholar]
- 98.Delgado S., Vidal N., Veron G., Sire J. Y. 2008. Amelogenin, the major protein of tooth enamel: a new phylogenetic marker for ordinal mammal relationships. Mol. Phylogenet. Evol. 47: 865–869. [DOI] [PubMed] [Google Scholar]
- 99.Prasad A. B., Allard M. W., Program N. C. S., Green E. D. 2008. Confirming the phylogeny of mammals by use of large comparative sequence datasets. Mol. Biol. Evol. 25: 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Springer M. S., Burk-Herrick A., Meredith R., Eizirik E., Teeling E., O'Brien S. J., Murphy W. J. 2007. The adequacy of morphology for reconstructing the early history of placental mammals. Syst. Biol. 56: 673–684. [DOI] [PubMed] [Google Scholar]
- 101.Murphy W. J., Eizirik E., Johnson W. E., Zhang Y. P., Ryder O. A., O'Brien S. J. 2001. Molecular phylogenetics and the origins of placental mammals. Nature. 409: 614–618. [DOI] [PubMed] [Google Scholar]
- 102.Murphy W. J., Eizirik E., O'Brien S. J., Madsen O., Scally M., Douady C. J., Teeling E., Ryder O. A., Stanhope M. J., de Jong W. W., et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 294: 2348–2351. [DOI] [PubMed] [Google Scholar]
- 103.Amos B., Anderson I. G., Haslewood G. A. D., Tökés L. 1977. Bile salts of the lungfishes Lepidosiren, Neoceratodus and Protopterus and those of the coelacanth Latimeria chalumnae Smith. Biochem. J. 161: 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anderson I. G., Haslewood G. A. D. 1964. Comparative studies of ‘bile salts’. 20. Bile salts of the coelacanth, Latimeria chalumnae Smith. Biochem. J. 93: 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hofmann A. F., Mysels K. J. 1992. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J. Lipid Res. 33: 617–626. [PubMed] [Google Scholar]
- 106.Cohen B. I., Ayyad N., Mosbach E. H., McSherry C. K., Matoba N., Hofmann A. F., Ton-Nu H. T., Peng Y., Schteingart C. D., Stenger R. J. 1991. Replacement of cholesterol gallstones by murideoxycholyl taurine gallstones in prairie dogs fed murideoxycholic acid. Hepatology. 14: 158–168. [DOI] [PubMed] [Google Scholar]
- 107.Palmer R. H., Hruban Z. 1966. Production of bile duct hyperplasia and gallstones by lithocholic acid. J. Clin. Invest. 45: 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zaki F. G., Carey J. B., Jr., Hoffbauer F. W., Nwokolo C. 1967. Biliary reaction and choledocholithiasis induced in the rat by lithocholic acid. J. Lab. Clin. Med. 69: 737–748. [PubMed] [Google Scholar]
- 109.Hofmann A. F., Mosbach E. H. 1964. Identification of allodeoxycholic acid as the major component of gallstones induced in the rabbit by 5α-cholestan-3β-ol. J. Biol. Chem. 239: 2813–2821. [PubMed] [Google Scholar]
- 110.Gu J. J., Hofmann A. F., Ton-Nu H. T., Schteingart C. D., Mysels K. J. 1992. Solubility of calcium salts of unconjugated and conjugated natural bile acids. J. Lipid Res. 33: 635–646. [PubMed] [Google Scholar]
- 111.Alrefai W. A., Gill R. K. 2007. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm. Res. 24: 1803–1823. [DOI] [PubMed] [Google Scholar]
- 112.Kosters A., Karpen S. J. 2008. Bile acid transporters in health and disease. Xenobiotica. 38: 1043–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pellicoro A., Faber K. N. 2007. Review article: The function and regulation of proteins involved in bile salt biosynthesis and transport. Aliment. Pharmacol. Ther. 26(Suppl 2): 149–160. [DOI] [PubMed] [Google Scholar]
- 114.Hofmann A. F., Mysels K. J. 1988. Bile salts as biological surfactants. Colloids Surf. 30: 145–173. [Google Scholar]
- 115.Hofmann A. F., Roda A. 1984. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J. Lipid Res. 25: 1477–1489. [PubMed] [Google Scholar]
- 116.Roda A., Hofmann A. F., Mysels K. J. 1983. The influence of bile salt structure on self-association in aqueous solutions. J. Biol. Chem. 258: 6362–6370. [PubMed] [Google Scholar]
- 117.Guertin F., Loranger A., Lepage G., Roy C. C., Yousef I. M., Domingo N., Chanussot F., Lafont H., Tuchweber B. 1995. Bile formation and hepatic plasma membrane composition in guinea-pigs and rats. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 111: 523–531. [DOI] [PubMed] [Google Scholar]
- 118.Huijghebaert S. M., Hofmann A. F. 1986. Pancreatic carboxypeptidase hydrolysis of bile acid-amino conjugates: selective resistance of glycine and taurine amidates. Gastroenterology. 90: 306–315. [DOI] [PubMed] [Google Scholar]
- 119.Agnew D. W., Hagey L. R., Shoshani J. 2005. The elephants of Zoba Gash Barka, Eritrea: part 4. Cholelithiasis in a wild African elephant (Loxodonta africana). J. Zoo Wildl. Med. 36: 677–683. [DOI] [PubMed] [Google Scholar]
- 120.D'Aldebert E., Biyeyeme Bi Mve M. J., Mergey M., Wendum D., Firrincieli D., Coilly A., Fouassier L., Corpechot C., Poupon R., Housset C., et al. 2009. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology. 136: 1435–1443. [DOI] [PubMed] [Google Scholar]
- 121.Lorenzo-Zuniga V., Bartoli R., Planas R., Hofmann A. F., Vinado B., Hagey L. R., Hernandez J. M., Mahe J., Alvarez M. A., Ausina V., et al. 2003. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 37: 551–557. [DOI] [PubMed] [Google Scholar]
- 122.Inagaki T., Moschetta A., Lee Y. K., Pena L., Zhao G., Downes M., Yu R. T., Shelton J. M., Richardson J. A., Repa J. J., et al. 2006. Regulation of antibacterial defenses in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA. 103: 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hofmann A. F., Eckmann L. 2006. How bile acids confer gut mucosal protection against bacteria. Proc. Natl. Acad. Sci. USA. 103: 4333–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hamilton J. P., Xie G., Raufman J. P., Hogan S., Griffin T. L., Packard C. A., Chatfield D. A., Hagey L. R., Steinbach J. H., Hofmann A. F. 2007. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. 293: G256–G263. [DOI] [PubMed] [Google Scholar]
- 125.Hofmann A. F. 2004. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab. Rev. 36: 703–722. [DOI] [PubMed] [Google Scholar]
- 126.Fickert P., Fuchsbichler A., Marschall H. U., Wagner M., Zollner G., Krause R., Zatloukal K., Jaeschke H., Denk H., Trauner M. 2006. Lithocholic acid feeding induces segmental bile duct obstruction and destructive cholangitis in mice. Am. J. Pathol. 168: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Allan R. N., Thistle J. L., Hofmann A. F. 1976. Lithocholate metabolism during chemotherapy for gallstone dissolution. 2. Absorption and sulphation. Gut. 17: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fisher R. L., Hofmann A. F., Converse J. L., Rossi S. S., Lan S. P. 1991. The lack of relationship between hepatotoxicity and lithocholic-acid sulfation in biliary bile acids during chenodiol therapy in the National Cooperative Gallstone Study. Hepatology. 14: 454–463. [PubMed] [Google Scholar]
- 129.Schoenfield L. J., Lachin J. M. 1981. Chenodiol (chenodeoxycholic acid) for dissolution of gallstones: the National Cooperative Gallstone Study. A controlled trial of efficacy and safety. Ann. Intern. Med. 95: 257–282. [DOI] [PubMed] [Google Scholar]
- 130.Fischer C. D., Cooper N. S., Rothschild M. A., Mosbach E. H. 1974. Effect of dietary chenodeoxycholic acid and lithocholic acid in the rabbit. Am. J. Dig. Dis. 19: 877–886. [DOI] [PubMed] [Google Scholar]
- 131.Dyrszka H., Salen G., Zaki F. G., Chen T., Mosbach E. H. 1976. Hepatic toxicity in the rhesus monkey treated with chenodeoxycholic acid for 6 months: biochemical and ultrastructural studies. Gastroenterology. 70: 93–104. [PubMed] [Google Scholar]
- 132.Webster K. H., Lancaster M. C., Hofmann A. F., Wease D. F., Baggenstoss A. H. 1975. Influence of primary bile acid feeding on cholesterol metabolism and hepatic function in the rhesus monkey. Mayo Clin. Proc. 50: 134–138. [PubMed] [Google Scholar]
- 133.Morrissey K. P., McSherry C. K., Swarm R. L., Nieman W. H., Deitrick J. E. 1975. Toxicity of chenodeoxycholic acid in the nonhuman primate. Surgery. 77: 851–860. [PubMed] [Google Scholar]
- 134.Kliewer S. A., Willson T. M. 2002. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J. Lipid Res. 43: 359–364. [PubMed] [Google Scholar]
- 135.Adachi R., Shulman A. I., Yamamoto K., Shimomura I., Yamada S., Mangelsdorf D. J., Makishima M. 2004. Structural determinants for vitamin D responses to endocrine and xenobiotic signals. Mol. Endocrinol. 18: 43–52. [DOI] [PubMed] [Google Scholar]
- 136.Makishima M., Lu T. T., Xie W., Whitfield G. K., Domoto H., Evans R. M., Haussler M. R., Mangelsdorf D. J. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science. 296: 1313–1316. [DOI] [PubMed] [Google Scholar]
- 137.Kalaany N. Y., Mangelsdorf D. J. 2006. LXRs and FXR: the Yin and Yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68: 159–191. [DOI] [PubMed] [Google Scholar]
- 138.Downes M., Verdecia M. A., Roecker A. J., Hughes R., Hogenesch J. B., Kast-Woelbern H. R., Bowman M. E., Ferrer J-L., Anisfeld A. M., Edwards P. A., et al. 2003. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell. 11: 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sonoda J., Xie W., Rosenfeld J. M., Barwick J. L., Guzelian P. S., Evans R. M. 2002. Regulation of a xenobiotics sulfonation cascade by nuclear pregnane X receptor (PXR). Proc. Natl. Acad. Sci. USA. 99: 13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Seo Y-W., Sanyal S., Kim H-J., Won D. H., An J-Y., Amano T., Zavacki A. M., Kwon H-B., Shi Y-B., Kim W-S., et al. 2002. FOR, a novel orphan nuclear receptor related to farnesoid X receptor. J. Biol. Chem. 277: 17836–17844. [DOI] [PubMed] [Google Scholar]
- 141.Mi L-Z., Devarakonda S., Harp J. M., Han Q., Pellicciari R., Willson T. M., Khorasanizadeh S., Rastinejad F. 2003. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell. 11: 1093–1100. [DOI] [PubMed] [Google Scholar]
- 142.Moore L. B., Maglich J. M., McKee D. D., Wisely B., Willson T. M., Kliewer S. A., Lambert M. H., Moore J. T. 2002. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 16: 977–986. [DOI] [PubMed] [Google Scholar]
- 143.Krasowski M. D., Yasuda K., Hagey L. R., Schuetz E. G. 2005. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol. Endocrinol. 19: 1720–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Watkins R. E., Wisely G. B., Moore L. B., Collins J. L., Lambert M. H., Williams S. P., Willson T. M., Kliewer S. A., Redinbo M. R. 2001. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 292: 2329–2333. [DOI] [PubMed] [Google Scholar]
- 145.Krasowski M. D., Yasuda K., Hagey L. R., Schuetz E. G. 2005. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors). Nucl. Recept. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Reschly E. J., Bainy A. C. D., Mattos J. J., Hagey L. R., Bahary N., Mada S. R., Ou J., Venkataramanan R., Krasowski M. D. 2007. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol. Biol. 7: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. 2009. Bile acids as regulatory molecules. J. Lipid Res. 50: 1509–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191. [DOI] [PubMed] [Google Scholar]
- 149.Keely S. J., Scharl M. M., Bertelsen L. B., Hagey L. R., Barrett K. R., Hofmann A. F. 2007. Bile acid-induced secretion in polarized monolayers of T84 colonic epithelial cells: structure-activity relationships. Am. J. Physiol. Gastrointest. Liver Physiol. 292: G290–G297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.