Abstract
Neutral cholesterol ester hydrolase (NCEH) accounts for a large part of the nCEH activity in macrophage foam cells, a hallmark of atherosclerosis, but its subcellular localization and structure-function relationship are unknown. Here, we determined subcellular localization, glycosylation, and nCEH activity of a series of NCEH mutants expressed in macrophages. NCEH is a single-membrane-spanning type II membrane protein comprising three domains: N-terminal, catalytic, and lipid-binding domains. The N-terminal domain serves as a type II signal anchor sequence to recruit NCEH to the endoplasmic reticulum (ER) with its catalytic domain within the lumen. All of the putative N-linked glycosylation sites (Asn270, Asn367, and Asn389) of NCEH are glycosylated. Glycosylation at Asn270, which is located closest to the catalytic serine motif, is important for the enzymatic activity. Cholesterol loading by incubation with acetyl-LDL does not change the ER localization of NCEH. In conclusion, NCEH is targeted to the ER of macrophages, where it hydrolyzes CE to deliver cholesterol for efflux out of the cells.
Keywords: macrophage, atherosclerosis, lipid droplets, foam cells, glycosylation, cholesterol efflux, type II membrane protein, vesicular transport, KIAA1363, arylacetamide deacetylase like 1
The hydrolysis of intracellular cholesterol ester (CE) is the initial step of reverse cholesterol transport (1). Since this step is rate-limiting particularly in macrophage foam cells (2–5), a hallmark of atherosclerosis, it is important to clarify the mechanisms that mediate the hydrolysis of CE in foam cells (2–5).
As the hydrolysis of CE preceding reverse cholesterol transport takes place at neutral pH, the enzymes catalyzing it have been collectively called neutral CE hydrolases (nCEHs) to distinguish them from lysosomal acid lipase, the CE hydrolase in lysosome whose optimal pH is acidic. Since carboxyl ester lipase (also called cholesterol esterase or bile salt-dependent lipase) is a secreted protein, it is not involved in the hydrolysis of intracellular CE (6).
To date, three enzymes have been proposed to serve as nCEHs in macrophages: hormone-sensitive lipase (HSL) (7, 8); cholesterol ester hydrolase (CEH) (9), which is identical to a human liver carboxylesterase 1 (hCE-1) (10) or macrophage serine esterase1 (11), also known as a human ortholog of porcine triacylglycerol hydrolase (TGH) (12); and neutral cholesterol ester hydrolase (NCEH) (13), which is also known as KIAA1363 or arylacetamide deacetylase-like 1 (14). However, it is not clear which enzyme is of primary importance in the hydrolysis of CE in macrophages. In mice, the ablation of HSL did not significantly reduce nCEH activity in murine peritoneal macrophages (MPMs) (15, 16). Hence, HSL is not the major enzyme mediating nCEH activity in MPMs. Ghosh reported CEH as a promising candidate for an nCEH because its macrophage-specific overexpression under the promoter of scavenger receptor protected against diet-induced atherosclerosis in LDL receptor knockout mice (17). However, the effects of loss of function of CEH on nCEH activity in macrophages have not been reported. Furthermore, a mouse ortholog of CEH, TGH, was expressed at a low level in MPMs (13) and possessed a negligible nCEH activity compared with its hydrolase activity toward p-nitrophenyl butyrate (PNPB), triolein, and monoolein (18). In contrast, NCEH is robustly expressed in MPMs as well as in macrophage foam cells in atherosclerotic lesions, and the knockdown of NCEH by short interference RNA significantly reduced nCEH activity in MPMs (18). Therefore, NCEH is more likely to be involved in the hydrolysis of CE in MPMs.
Recently, it has become clear that NCEH is a mouse ortholog of KIAA1363. KIAA1363 was initially identified as a membrane-bound serine hydrolase whose expression is upregulated in invasive cancer cells (19). Subsequent studies in the labs of Cravatt and Casida have shown that KIAA1363 is chlorpyrifos oxon-binding protein (14), which has 2-acetyl monoalkylglycerol hydrolase activity and serves as a central node in an ether lipid signaling network that bridges platelet-activating factor and lysophosphatidic acid (20). These findings regarding the novel function of KIAA1363 do not negate the possibility that KIAA1363 also serves as NCEH because many lipid-metabolizing enzymes act on multiple substrates, thus being multifunctional.
Despite these intriguing functions of NCEH/KIAA1363, it is poorly understood where and how NCEH is associated with membranes and how posttranslational modifications influence its function. In this study, we show that NCEH is a single-membrane-spanning endoplasmic reticulum (ER) protein that anchors to the ER membrane via an N-terminal hydrophobic segment. Its catalytic and lipid-binding domains are located in the ER lumen. N-linked glycation at Asn270 is crucial for its enzymatic activity.
MATERIALS AND METHODS
Materials
Lecithin, BSA fraction V (BSA), adenosine 3′,5′-cyclic monophosphate sodium salt monohydrate, leupeptin, phenylmethylsulfonyl fluoride (PMSF), PNPB, and p-nitrophenol were purchased from Sigma-Aldrich (St. Louis, MO). Digitonin, saponin and tunicamycin were purchased from Wako Pure Chemicals (Osaka, Japan). Proteinase K was purchased from Roche. Adenosine-5′-triphosphate disodium salt trihydrate was purchased from MP Biomedicals. Protein G Sepharose was purchased from Applied Biosystems (GE Healthcare).
Preparation of lipoproteins
After an overnight fast, blood was collected from normolipidemic volunteers to isolate plasma. LDL (d 1.019–1.063 g/ml) and HDL (d 1.063–1.21 g/ml) were isolated from the plasma by sequential density ultracentrifugation (21). LDL was acetylated by repetitive additions of acetic anhydride (22).
Cells
HEK293, Hela, and RAW 264.7 cells were cultured in DMEM containing 10% (v/v) FBS with or without 1 mM sodium pyruvate. MPMs were harvested as described previously (18).
Mice
C57BL/6 was purchased from Clea (Tokyo, Japan). Mice were maintained and cared for according to the regulations of the Animal Care Committee of the University of Tokyo.
Vector and transfection
Mouse wild-type or mutant NCEH cDNA was subcloned into pcDNA3.1, pcDNA.3.1/myc-His (Invitrogen, Carlsbad, CA), or pCMV-Tag4 (Stratagene, La Jolla, CA) and used as NCEH or flag-tagged NCEH. Furthermore, NCEH cDNA or mutant NCEH cDNA was subcloned into pEGFP-N3 (Clontech, Palo Alto, CA) and used as NCEH-green fluorescent protein (GFP) fusion expression vector. Transfection was performed using Lipofectamin2000 (Invitrogen), Superfect (Qiagen, Hilden, Germany), or Fugene HD (Roche) as transfection reagents.
Site-directed mutagenesis and mutagenic oligonucleotides
Asn270, Asn367, and Asn389 were replaced by glutamine. Gly114 and Ser191 were replaced by alanine. Site-directed mutagenesis was performed using the megaprimer method (23). The oligonucleotides used to generate the mega primers are as follows: N270Q, 5′-gattgtgaacCAGcacacttcac-3′; N367Q, 5′-agtgcgggtgtgCAGgtgaccttgga-3′; N389Q, 5′-ctggccgaccCAGttctccgt-3′; G114A, 3′-cccagcctccGGCgtggatata-5′; and S191A, 3′-tcccaccagcGGCgtctccagaaat-5′.
Production of ΔN-NCEH, ΔN-NCEH-GFP, NCEH1-33-GFP, and NCEH1-89-GFP
A mutant NCEH that lacks the coding region for the N-terminal region of NCEH (1–33 amino acids) was generated by PCR using as a forward primer, 5′-cctggggacaatgctgctggacgccac-3′.
The full-length NCEH cDNA was used as a template. The PCR product was subcloned into the pcDNA3.1 or pEGFP-N3 vector, yielding ΔN-NCEH and ΔN-NCEH-GFP, respectively. In addition, the coding region for the N-terminal 1–33 and 1–89 amino acids of NCEH was generated by PCR and subcloned into the pEGFP-N3 vector, yielding NCEH1-33-GFP and NCEH1-89-GFP, respectively. The primers used were as follows: NCEH1-33-GFP, forward primer, 5′-ctggggacaatgaggtcgt-3′ and reverse primer, 5′-cagtttccaggggtcggac-3′; NCEH1-89-GFP, forward primer, 5′-ctggggacaatgaggtcgt-3′ and reverse primer,5′-cccatcgaagtcggtgtct-3′.
Adenovirus
We used the adenovirus overexpressing LacZ, NCEH, or HSL that had been used in the previous report (18).
Antibodies
Amino acid residues of human NCEH (amino acids 83–164) was expressed in bacteria as a glutathione S-transferase fusion protein, which was purified by glutathione affinity chromatography and used for immunization of rabbits according to the standard protocols as described previously (24). From serum samples with high antibody titers, IgG fractions were isolated using a protein G column (GE Healthcare). Serum of nonimmunized rabbits was used as a control. This rabbit polyclonal antibody of human NCEH recognized both human and mouse NCEH. Furthermore, we used rabbit polyclonal antibody of mouse NCEH, which recognizes amino acid residues containing catalytic core domain of mouse NCEH (amino acids 99–250). This antibody was already used in our previous report (18).
We used rabbit anti-HSL polyclonal antibody, which was used in the previous report (16). Mouse anti-ACAT1 monoclonal antibody was also used in the previous report (25). Mouse anti-protein disulfide isomerase (PDI) monoclonal antibody was from Stressgen, Assay Designs (Ann Arbor, MI). Mouse anti-FLAG monoclonal antibody was from Sigma-Aldrich. The mouse monoclonal 9E10 anti-myc antibody was from Abcam (Cambridge, UK). The rabbit anti-GFP polyclonal antibody was from MBL (Aichi Japan).
Western blot analyses
Cells were sonicated in buffer A (50 mmol/l Tris-HCl, pH 7.0, 250 mmol/l sucrose, 1 mmol/l EDTA, and 2 µg/ml leupeptin) and centrifuged at 15,000 g for 15 min at 4°C. Furthermore, the supernatant was used as whole-cell lysate. The supernatant was centrifuged at 100,000 g for 45 min at 4°C. The supernatant was centrifuged under the same conditions again and used as the S100 fraction. The precipitates of the first centrifugation were resuspended and centrifuged at 100,000 g for 45 min at 4°C again. The precipitates were resuspended again and used as a microsomal fraction. Alternatively, cells were suspended in buffer A containing 1% (v/v) Triton X-100 and were centrifuged at 15,000 g for 15 min at 4°C. The supernatant was used in Western blotting. Aliquots (10 µg) of proteins of various cellular fractions were subjected to 10% (w/v) SDS-PAGE and transferred to a nitrocellulose membrane. For detection of the proteins, the membranes were incubated with the appropriate antibody at a dilution of 1:1,000. Specifically, bound immunoglobulins were detected in a second reaction with a horseradish peroxidase-labeled IgG conjugate and visualized by enhanced chemiluminescence detection (ECL plus; GE Healthcare) on a Kodak image system.
Enzyme assays
Transfected HEK293 cells were sonicated in buffer A and centrifuged at 15,000 g for 15 min at 4°C. The supernatant was used to assay enzymatic activity. nCEH activity was measured essentially as described by Hajjar, Minick, and Fowler (26), using a reaction mixture containing 6.14 µM cholesterol [1-14C]oleate (48.8 µCi/µmol; 1 µCi = 37 kBq). PNPB-hydrolyzing activity was determined by measuring the rate of release of p-nitrophenol (absorbance at 405 nm at 37°C) by a modified procedure described previously (27).
Proteinase treatment
Permeabilized cells grown in monolayer were used. The method was based on the procedure described previously with minor modifications (28, 29). MPMs were cultured in 12-well plates to 80% confluence. Cells were rinsed with PBS and then permeabilized either with 1 µg/ml digitonin in PBS or with 2% (w/v) saponin in PBS, 1.0 ml/well, at room temperature for 10 min. Afterwards, 0.5 ml of buffer B (10 mM Hepes-KOH, pH 7.4, 100 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 5 mM sodium EDTA, and 250 mM sucrose) was added to each well. Protease digestion was started by adding the indicated final concentrations of proteinase K. The digestions proceeded at room temperature for 30 min. The reactions were stopped by addition of PMSF at a final concentration of 5 mM. Cells were suspended in buffer A containing 1% (v/v) Triton X-100 and were centrifuged at 15,000 g for 15 min at 4°C. The supernatant was used for Western blotting.
Immunocytochemistry
Cells were plated in four-chamber plates (Nunc Lab-Tek, Roskilde, Denmark). The culture media were aspirated and cells were fixed in 3.7% (w/v) PFA in PBS for 30 min at room temperature. Cells were permeabilized by exposure to 0.1% (v/v) Triton X-100, 1 µg/ml digitonin, or 2% (w/v) saponin in PBS for 10 min at room temperature, blocked with 2% (w/v) BSA in PBS for at least 30 min at room temperature and then incubated with primary antibody diluted 1:250 overnight at 4°C. BODIPY 493/503 was used to visualize neutral lipids. For detection, cells were subsequently incubated with either Alexa Fluor® 555 or Alexa Fluor 488-conjugated secondary antibodies (Invitrogen) diluted 1:200 in 2% (w/v) BSA in PBS for 1 h at room temperature, enclosed in Perma Fluor Aquaous Mounting Medium (Thermo Shandon) and observed with a Bio-Rad MRC 1024 microscope using a mercury lamp and a 60× objective or with Leica confocal microscopy (Leica) using an argon laser and a 100× objective. The images were processed with Photoshop (Adobe) software.
Deglycosylation
MPMs were suspended in buffer A containing 1% Triton X-100, and 20 µg was treated by PNGase and EndoH (New England Biolabs) according to the manufacturer's protocol.
Tunicamycin treatment
In 12-well plates, HEK293 cells were transfected with pcDNA3.1-based plasmids. After 12 h, media were replaced with DMEM containing 5 µg/ml tunicamycin. Cells were subsequently incubated for 24 h and were scraped into buffer A containing 1% Triton X-100. The prepared lysates were analyzed by Western blotting.
In vitro transcription/translation
pcDNA3.1-based plasmids were used for in vitro transcription/translation using the TNT™ Quick Coupled system (Promega) according to the manufacturer's protocol. The reaction products were analyzed by Western blotting.
Immunoprecipitation
Whole-cell lysates of HEK293 cells (0, 30 or 60 µg) transfected with FLAG-tagged NCEH or HSL were immunoprecipitated by 5 µg of mouse anti-FLAG monoclonal antibody (Sigma-Aidrich), which was bound to Protein G Sepharose. After SDS-PAGE, they were immunoblotted by mouse anti-FLAG monoclonal antibody. Mouse TrueBlot™ ULTRA HRP-Conjugated Anti-Mouse IgG (eBioscience) was used to detect the presence of anti-FLAG antibody.
Phospholylation
HEK293 cells were plated in 12-well plates and the next day transfected with 1 µg of pCMV-Tag4-based plasmids. After 24 h, the cells were preincubated in Krebs-Ringer buffer (25 mM HEPES, 129.4 mM NaCl, 5.2 mM KCl, 1.3 mM MgSO4, 50 mM KH2PO4, 2.5 mM CaCl2, 24.8 mM NaHCO3, and 2 mM glucose) for 6 h and subsequently incubated first in 1 ml of Krebs-Ringer buffer containing 0.5 mCi [32P] orthophosphate (GE Healthcare) for 1 h and then in the presence or absence of a protein kinase A (PKA) activator, forskolin (20 µM), for 30 min. Cells were suspended in buffer A containing 1% (v/v) Triton X-100 and centrifuged at 15,000 g for 15 min at 4°C. The concentration of protein in the supernatant was determined using a BCA Protein assay kit (Pierce). The supernatant was immunoprecipitated by 5 µg of mouse anti-FLAG monoclonal antibody. After SDS-PAGE, gels were dried, autoradiographed with a Phosphoimager Screen (FUJIFILM), and analyzed using BASTATION (FUJIFILM) software.
Statistical analyses
Results are presented as means ± SE. Student's t-test was employed to compare the means. All calculations were performed with STAT view version 5.0 for Macintosh (SAS Institute).
RESULTS
NCEH is a type II ER membrane integrated protein
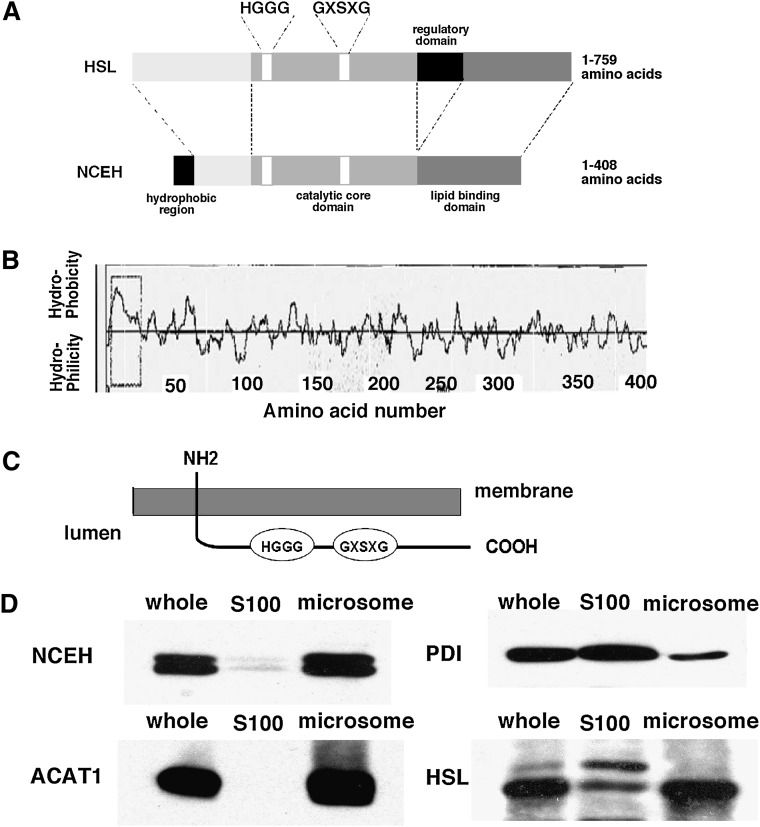
Figure 1A shows the structure of NCEH. It consists of three domains: i) an N-terminal domain, ii) a catalytic core domain, and iii) a lipid-binding domain, and lacks the regulatory domain present in HSL. The N-terminal region of NCEH contains a hydrophobic region. Figure 1B shows a hydropathy plot of the amino acid sequence of mouse NCEH. Based on this plot, NCEH is a type II membrane protein with a single transmembrane segment containing 23 amino acids. The predicted topology of NCEH is shown in Fig. 1C. In keeping with this prediction, duplets of NCEH proteins were predominantly distributed in the microsomal fraction when MPMs were homogenized by sonication (Fig. 1D). This pattern of subcellular distribution is similar to that of ACAT1, an integral ER protein that spans the membrane seven times (30, 31) but is distinct from that of either PDI, a soluble ER protein, or HSL, a predominantly cytosolic protein (32). PDI was distributed in both the S100 and microsomal fractions because PDI, a luminal soluble enzyme, leaked into the S100 fraction during sonication. HSL in macrophages was recognized as double bands as described previously (13). The upper band was distributed in the S100 fraction, while the lower band was predominantly distributed in the microsomal fraction.
Fig. 1.
Subcellular distribution of NCEH. A: Structure of HSL and NCEH. B: A hydropathy plot of NCEH produced with the SOSUI program. C: The membrane topology of NCEH predicted by the TMHMM program. D: MPMs fractionated as described in Materials and Methods. S100 and microsomal fractions were prepared by centrifugation of the sonicated MPMs at 100,000 g for 45 min at 4°C twice. Aliquots (10 µg) of whole-cell lysate, the S100, and microsomal fractions were analyzed by Western blotting with anti-NCEH, HSL, ACAT1, and PDI antibodies. Data are representative of three independent experiments.
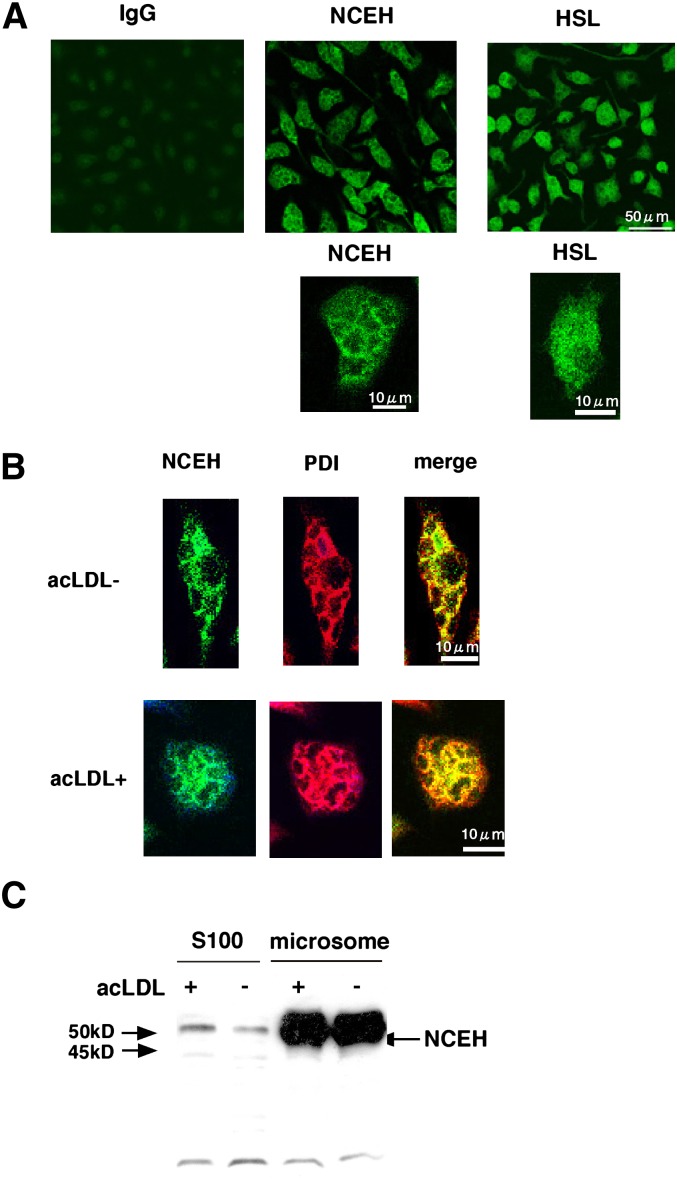
To confirm that NCEH is recruited to the ER, we performed immunostaining for endogenous NCEH in MPMs (Fig. 2A). Staining for NCEH showed a dense reticular network throughout the cells, a pattern characteristic of an ER-based localization. On the other hand, the staining for HSL was diffuse (Fig. 2A). We confirmed that the staining of NCEH occurred in the ER by using the ER marker protein PDI (Fig. 2B). Cholesterol loading by incubation with acLDL changed neither the pattern of staining (Fig. 2B) nor the amount of NCEH protein in either the S100 or microsomal fraction (Fig. 2C).
Fig. 2.
Subcellular localization of NCEH. A: Expression of NCEH and HSL in MPMs was determined by immunocytochemistry using affinity-purified anti-NCEH and HSL rabbit IgG. For detection, cells were incubated with Alexa Fluor® 488-conjugated secondary antibody. As a control, the staining of MPMs with preimmune rabbit IgG as the primary antibody did not produce any fluorescence. Data are representative of three independent experiments. B: The localization of NCEH was analyzed by combined immunocytochemical staining for NCEH (green) and PDI (red) in the presence or absence of 100 µg/ml acLDL. For detection, cells were incubated with either Alexa Fluor 488 or Alexa Fluor 555-conjugated secondary antibodies. Images were acquired with a Bio-Rad MRC 1024 microscope and a 60× objective. Data are representative of three independent experiments. C: S100 and microsomal fractions (10 µg each) of MPMs in the presence or absence of 100 µg/ml acLDL were analyzed by Westen blotting with anti-NCEH antibody. Data are representative of three independent experiments.
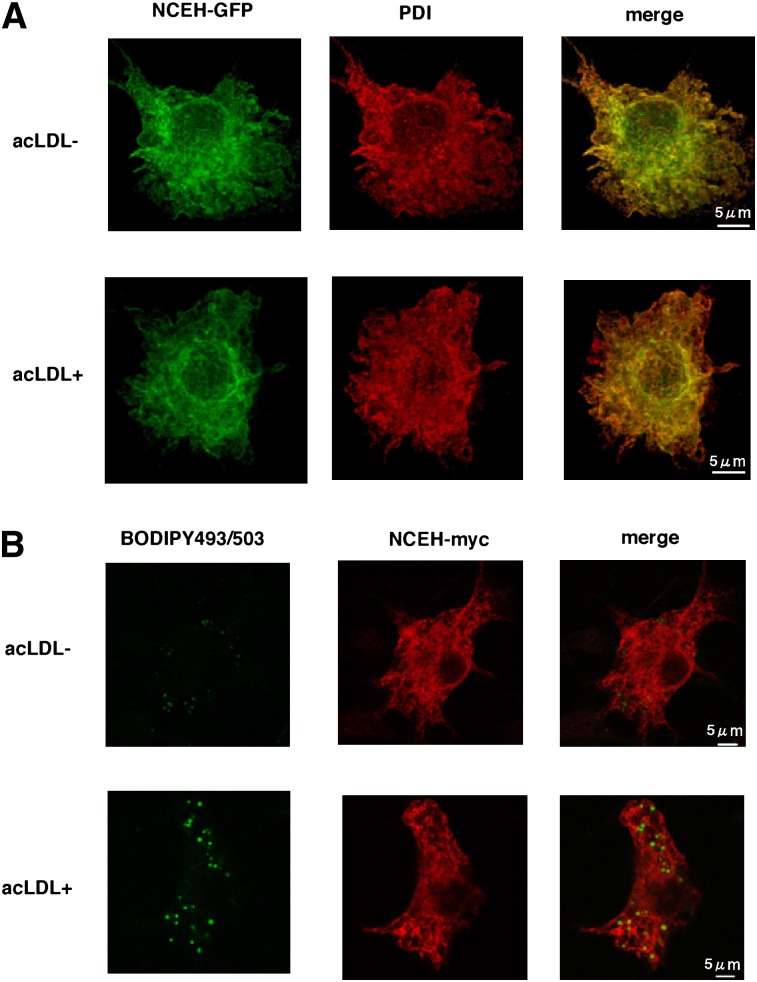
A similar distribution in the ER localization pattern was observed when GFP-tagged NCEH was overexpressed in RAW264.7 cells (Fig. 3). The fine reticular pattern of staining for GFP completely colocalized with the distribution of the ER marker PDI (Fig. 3A). Cholesterol loading by incubation with acLDL did not change the pattern of staining. To examine whether NCEH is present on lipid droplets (LDs), considered as a principal site for the hydrolysis of neutral lipids, we used BODIPY493/503 to visualize LDs (Fig. 3B). Amounts of myc-tagged NCEH localized around the ring of BODIPY493/503-positive LDs were negligible, and the vast majority of NCEH remained on the ER even after the cholesterol loading.
Fig. 3.
Positional relation between NCEH protein and lipid droplets in macrophages. A: NCEH-GFP was expressed in RAW264.7 macrophages with or without incubation with 100 µg/ml acLDL for 24 h. Cells were immunostained with mouse anti-PDI monoclonal antibody, followed by Alexa Fluor 555-conjugated secondary antibody. B: NCEH-myc was expressed in RAW264.7 macrophages with or without incubation with 100 µg/ml acLDL for 24 h. Cells were immunostained with mouse monoclonal 9E10 anti-myc antibody, followed by Alexa Fluor 555-conjugated secondary antibody. LDs were stained by BODIPY493/503 (green). Images were acquired with a confocal microscope (Leica) using an argon laser and a 100× objective. These data are representative of three independent experiments.
The catalytic domain is located in the ER lumen
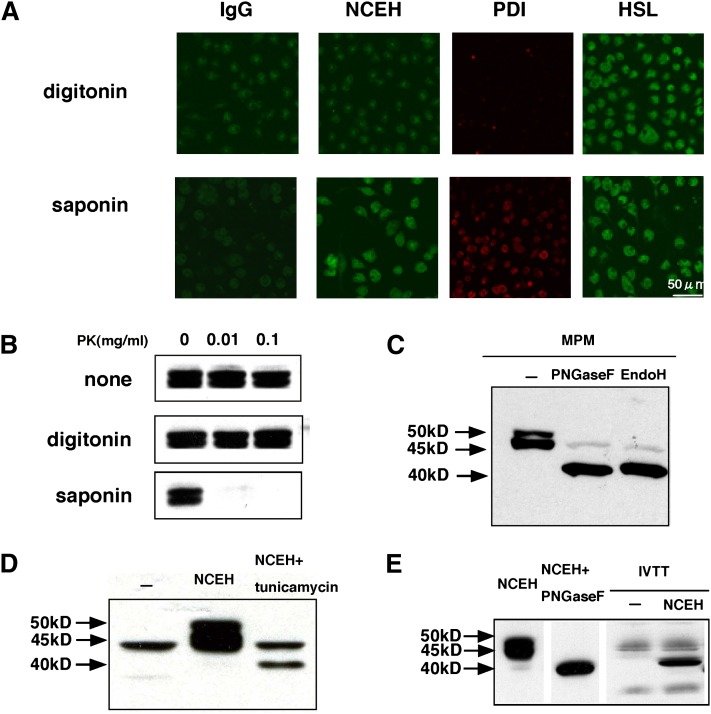
To confirm that most of NCEH including its catalytic domain and lipid-binding domain is in the ER lumen as predicted by the TMHMM program, we permeabilized MPMs with two different detergents, digitonin and saponin, and performed immunostaining using an antibody against the catalytic domain (99–250 amino acids) of NCEH and antibodies against PDI or HSL as controls. Treatment with digitonin exposed only cytosolic epitopes, whereas treatment with saponin exposed both cytosolic and ER lumenal epitopes (Fig. 4A). Treatment with digitonin visualized only HSL, while treatment with saponin visualized all three proteins, indicating that both NCEH and PDI are located in the ER lumen and that HSL is present in the cytosol.
Fig. 4.
Membrane topology of NCEH. A: MPMs permeabilized by exposure to 1 µg/ml digitonin or 2% saponin in PBS and then incubated with preimmune rabbit IgG (as a control), anti-NCEH, PDI, and HSL antibodies as primary antibodies. Anti-NCEH antibody recognized the catalytic core domain (99–250 amino acids). For detection, cells were subsequently incubated with either Alexa Fluor 555 or Alexa Fluor 488-conjugated secondary antibodies. Images were acquired with a Bio-Rad MRC 1024 microscope and a 60× objective. Data are representative of three independent experiments. B: MPMs cultured in 12-well plates were treated with PBS, 1 µg/ml digitonin in PBS, or with 2% (w/v) saponin in PBS. Protease digestion was started by adding the indicated final concentration of proteinase K. The reactions were stopped by addition of PMSF. Lysates from those cells were analyzed in Western blotting with anti-NCEH antibody. Data are representative of two independent experiments. C: Lysate (20 µg) prepared from MPMs was treated with PNGase F or Endo H and analyzed by Western blotting with NCEH antibody. Data are representative of four independent experiments. D: HEK293 cells transfected with NCEH were incubated in the presence or absence of 5 µg/ml tunicamycin. Lysate prepared from these cells was assessed by Western blotting. Data are representative of two independent experiments. E: pcDNA3.1-NCEH plasmid DNA (500 ng/reaction) translated using the TNT™ Quick Coupled system. The reaction products were analyzed by Western blotting. Data are representative of three independent experiments.
These results of immunostaining were supported by an experiment in which sensitivity to digestion by proteinase K was compared between different methods of permeabilization. When MPMs were treated with digitonin, NCEH was resistant to proteinase K. However, when treated with saponin, NCEH was sensitive to the proteinase (Fig. 4B).
To determine whether NCEH is a glycoprotein, we treated the membrane fraction with endoglycosidases (Fig. 4C). Treatment with either PNGase F or Endo H converted the double bands (50 and 45 kDa) of NCEH to a single band (40 kDa). Thus, the migration of NCEH as two distinct bands may result from heterogenous glycosylation. Because Endo H mainly catalyzes the deglycosylation of simple glycosylation formed in ER, NCEH's sensitivity to Endo H indicates that it is located in the ER lumen and does not reach the Golgi where N-linked carbohydrate chains become resistant to Endo H upon modification by mannosidase II in the Golgi. A similar shift in size was observed when HEK 293 cells transfected with the NCEH expression plasmid were incubated with 5 µg/ml tunicamycin, which inhibits N-linked glycosylation (Fig. 4D). NCEH does not receive other types of posttranslational modifications because the protein produced by the in vitro transcription/translation of NCEH without microsomes was almost identical in size to NCEH treated with PNGase F (Fig. 4E).
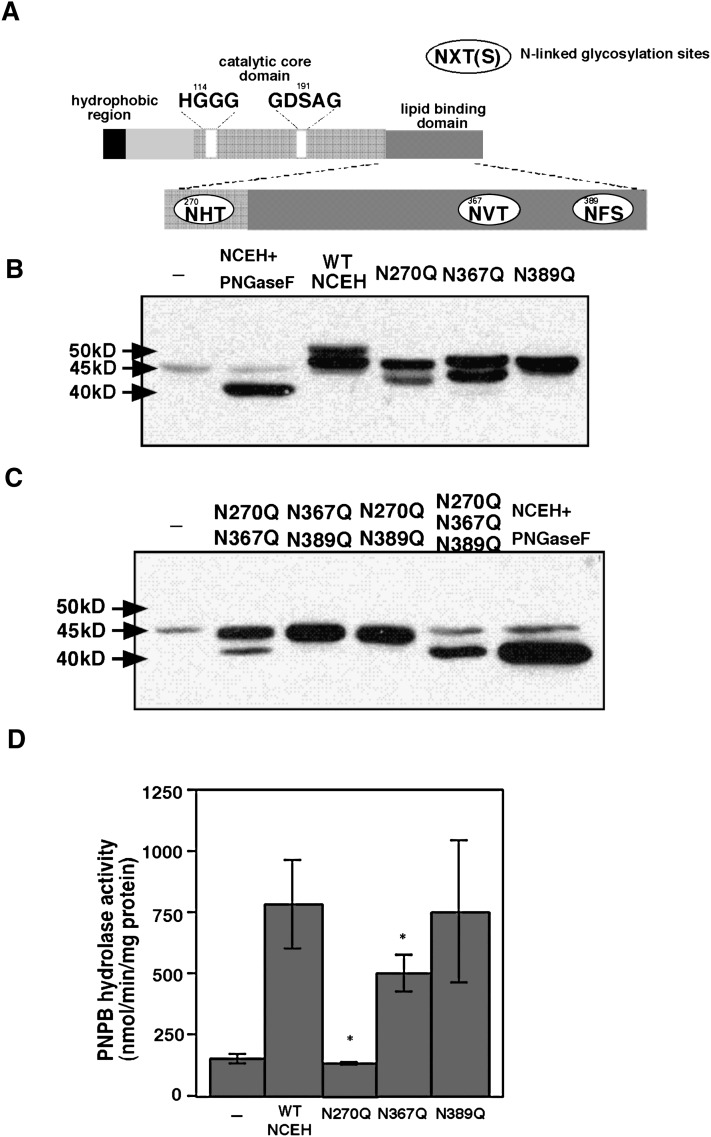
Role of N-linked glycosylation sites in the enzymatic activity and subcellular localization
NCEH contains three potential N-linked glycosylation sites (NXT or NXS): Asn270, Asn367, and Asn389 (Fig. 5A). To clarify whether those sites were glycosylated, each asparagine was mutated to glutamine by site-directed mutagenesis. The mutation of asparagine to glutamine is conservative, maintaining characteristics such as polarity or charge on this amino acid residue while preventing glycosylation. These mutations were named N270Q, N367Q, and N389Q. Mutant NCEH proteins containing one, two, or all of the mutations were transiently expressed in HEK293 cells (Fig. 5B, C).
Fig. 5.
Site-directed mutagenesis of N-linked glycosylation sites in NCEH. A: Three N-linked glycosylation sites: Asn270, Asn367, and Asn389. Whole-cell lysate was prepared from HEK293 cells transfected with wild-type or mutant NCEH containing one, two, or all of N270Q, N367Q, and N389Q. They were subjected to a Western blot analysis with anti-NCEH antibody (B and C) and measurements of enzymatic activity using PNPB as a substrate (D). Data in B and C are representative of two independent experiments. D: Data are expressed as means ± SE. Three independent measurements are representative of three independent experiments (*, P < 0.01, compared with wild-type NCEH).
A 50 kDa band, a high molecular weight form of wild-type NCEH, was not observed when mutants contained one of the following three substitutions: N270Q, N367Q, and N389Q (Fig. 5B), indicating that all three asparagine residues are glycosylated. Among the three mutants with a single substitution, only that having N389Q showed a single band (Fig. 5B). On the other hand, among the three with two mutations, only the mutant having N270Q and N367Q showed duplets (Fig. 5C). These results indicate that incomplete glycosylation at Asn389 causes the double band to form. The mutant NCEH containing all three substitutions had the same molecular weight as the wild-type NCEH digested with PNGase F (Fig. 5C), confirming that these three asparagines are only sites for N-linked glycosylation.
To determine which N-linked glycosylation is important for the enzymatic activity, we measured the PNPB hydrolase activity of mutant NCEH proteins (Fig. 5D). The N389Q mutant expressed almost as much PNPB hydrolase activity as the wild-type NCEH. On the other hand, nCEH activitiy levels of the N270Q and N367Q mutants were significantly reduced compared with wild-type NCEH. In particular, the PNPB hydrolase activity of the N270Q mutant was almost reduced to background levels. Hence, N-linked glycosylation in Asn270 might play an important role in catalysis. A contribution of Asn270, Asn367, and Asn389 to the subcellular distribution was not observed (see supplementary Fig. I).
To confirm that the His-Gly dipeptide motif or the GXSXG active serine motif of serine esterase is critical for enzymatic activity, we made mutants in which the catalytic Gly114 in HGGG or Ser191 in GXSXG was replaced by alanine: G114A and S191A, respectively. The PNPB hydrolase activity of each of these mutants was reduced to background levels (see supplementary Fig. II).
Role of the N-terminal region in the ER targeting
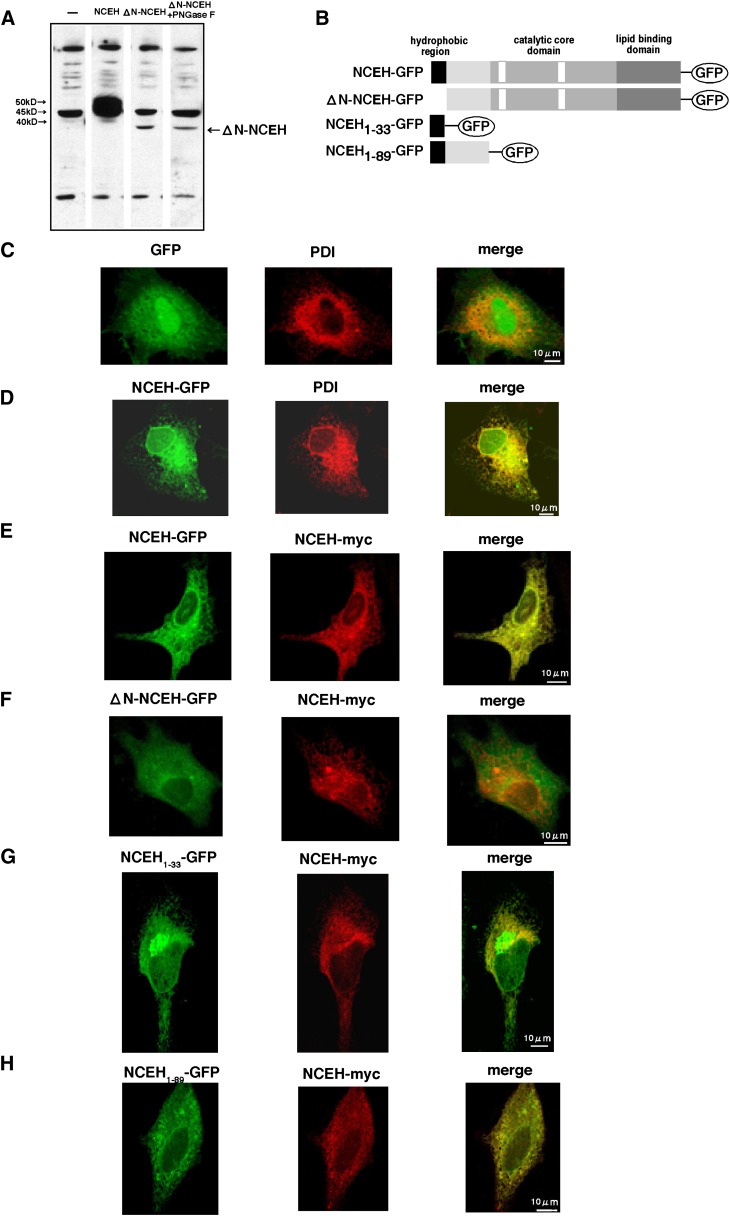
To determine the role of the N-terminal region of NCEH, we constructed a mutant NCEH that lacks the N-terminal region (1–33 amino acids) (ΔN-NCEH) and expressed it in HEK293 cells (Fig. 6). Amounts of the expressed ΔN-NCEH were significantly reduced compared with those of wild-type NCEH (Fig. 6A). Furthermore, the expressed ΔN-NCEH was a single band whose molecular weight was as small as that of wild-type NCEH treated with PNGase F or the mutant NCEH lacking all three potential N-linked glycosylation sites, suggesting that the ΔN-NCEH mutant did not undergo N-linked glycosylation. Consistent with this prediction, PNGase F treatment did not change the molecular weight of the ΔN-NCEH mutant (Fig. 6A). ΔN-NCEH was not detected in the media where the cells expressing ΔN-NCEH were incubated (data not shown). These results indicate that the N-terminal region of NCEH regulates the stability of the protein and that ΔN-NCEH is not glycosylated.
Fig. 6.
Role of the N-terminal region of NCEH. A: Mutant NCEH lacking N-terminal region (1–33 amino acids) (ΔN-NCEH) and wild-type NCEH were expressed in HEK293 cells. Lysate prepared from these cells was subjected to a Western blot analysis in the presence or absence of PNGase F. Data are representative of three independent experiments. GFP, NCEH-GFP, ΔN-NCEH-GFP, NCEH1-33-GFP, and NCEH1-89-GFP (B) were expressed as GFP fusion proteins in Hela cells. The cells were immunostained with mouse anti-PDI monoclonal antibody followed by Alexa Fluor 555-conjugated secondary antibody. In other sets of experiments, cells were cotransfected with pcDNA3.1-NCEH-myc and immunostained by mouse monoclonal 9E10 anti-myc antibody, followed by Alexa Fluor 555-conjugated secondary antibody. The intracellular distribution of these GFP fusion proteins was compared with that of PDI (C and D) of NCEH-myc (E–H). Images were acquired by confocal microscopy (Leica) using an argon laser and a 100× objective. Data are representative of three independent experiments.
Next, we took advantage of GFP-fused forms of several mutant proteins to determine the subcellular distribution of NCEH within Hela cells (Fig. 6B–H). NCEH-GFP colocalized with an ER marker protein, PDI (Fig. 6D). This is different from the diffuse cytoplasmic expression of GFP (Fig. 6C). NCEH-myc and NCEH-GFP colocalized when cotransfected (Fig. 6D), indicating that fusion of the GFP protein to the C-terminal end of NCEH did not influence the subcellular disnribution. On the other hand, ΔN-NCEH-GFP was distributed throughout the cell uniformly and did not colocalize with NCEH-myc (Fig. 6F). NCEH1-33-GFP, comprising the N-terminal region of NCEH (1–33 amino acids) fused directly with GFP, mostly colocalized with NCEH-myc (Fig. 6G). In addition to the ER, however, NCEH1-33-GFP was also found in the perinuclear region, where it was not localized with NCEH-myc. NCEH1-89-GFP, comprising the N-terminal amino acids (1–89 amino acids) of NCEH fused directly with GFP, entirely colocalized with NCEH-myc (Fig. 6H). These results indicate that the N-terminal region of NCEH serves as a type II signal anchor sequence which directs NCEH to the ER with its C-terminal side within the lumen (Ncyt/Cexo) (33).
Corroborating the results of immunostaining, Western blotting showed that the anti-GFP antibody detected both NCEH and NCEH1-33-GFP exclusively in the microsomal fraction, whereas it detected GFP itself almost exclusively in the S100 fraction (see supplementary Fig. III), supporting our premise that the N-terminal region of NCEH is a transmembrane domain.
NCEH is not a target of PKA
As deduced from the structural comparison with HSL, NCEH lacks a regulatory domain that contains many potential phospholylation sites for the regulation of enzymatic activities (Fig. 1A). To confirm that NCEH is not phospholylated, we overexpressed FLAG-tagged HSL or NCEH in HEK293 cells and cultured them in low phosphate media. After 6 h, the transfected cells were incubated in a medium containing H3PO4 labeled with 32P (see supplementary Fig. IV). The cell lysates in which HSL or NCEH tagged FLAG was overexpressed were immunoprecipitated by an antibody against the FLAG epitope tag and subjected to either Western blotting for HSL or NCEH (see supplementary Fig. IVA) or autoradiography (see supplementary Fig. IVB). Phospholylation of the 84 kDa HSL protein, which was further stimulated by treatment with forskolin, a PKA activator, was observed. In contrast, NCEH was not phospholylated even after the treatment with forskolin.
Next, we overexpressed HSL or NCEH in HEK 293 cells by adenovirus-mediated gene transfer and examined the effects of various concentrations of cAMP on the nCEH activity of the expressed enzymes (see supplementary Fig. IVC). Treatment with 1 μM cAMP significantly stimulated the nCEH activity of HSL by 20%, as reported previously (34, 35). However, even 10 μM cAMP did not increase the nCEH activity of NCEH, supporting that NCEH was not phospholylated at least under conditions where PKA is activated. Activation of the nCEH activity in the cell lysate from MPMs (see supplementary Fig. IVC) may result from the activation of HSL, not NCEH.
DISCUSSION
In this study, we showed that NCEH is tethered to the ER membrane with its catalytic domain residing in the ER lumen. The N-terminal amino acids are required for recruitment to the ER. NCEH contains three potential N-linked glycosylation sites that are all indeed glycosylated. In particular, the N-linked glycosylation at Asn270, which is located closest to the catalytic serine, is crucial for the enzymatic activity.
NCEH belongs to a family of ER proteins that are resident and catalytically active in the ER lumen. The best characterized proteins targeting this compartment are soluble and bear the C-terminal sequence KDEL (36, 37). NCEH belongs to another group of proteins that are membrane bound and lack the ER retrieval sequence. Most ER proteins involved in lipid biosynthesis have a polytopic topology: examples are HMG-CoA reductase, ACAT, diacylglycerol acyltransferase, and insig1. In contrast, NCEH has a bitopic topology: it is a type II membrane protein with its C-terminal projecting to the ER lumen. The N-terminal hydrophobic segment is the transmembrane domain that tethers NCEH to the ER membrane. In addition to anchoring the protein, the N-terminal segment (1–33 amino acids) containing the hydrophobic segment (4–26 amino acids) functions as a motif targeting the ER because NCEH deleted of this segment does not distribute in the ER, which is consistent with the loss of N-linked glycosylation of ΔN-NCEH. A longer stretch of the N terminus (1–89 amino acids) is required for the exact same distribution as wild-type NCEH. These features are consistent with the view that most ER proteins are recruited for insertion into membrane by a hydrophobic signal peptide at the N terminus (38). The specific topology of proteins is determined by factors like charge distribution, the length of the membrane segment and the charge of the segments following the membrane segment (39). Ozols and his colleagues proposed the existence of lumental targeting signal based on a comparison of the N-terminal transmembrane domains of two functionally distinct proteins: 11β-hydroxysteroid dehydrogenase, isozyme 1 (11β-HSD) and 50 kDa esterase (E3) (40). According to their model, an amino acid sequence consisting of short basic or neutral residues at the N terminus, followed by a specific array of hydrophobic residues terminating with acidic residues, is sufficient for lumental targeting of single-pass proteins. The amino acid sequence of the N terminus of NCEH is almost compatible with this model. Of note, region II of NCEH contains a cluster of tyrosyl residues (AYYXY) that are shared by 11β-SHD and E3.
NCEH is fully deglycosylated by Endo H (Fig. 4C), indicating that NCEH is localized to the ER and not transported to the Golgi body. Site-directed mutagenesis showed that all three putative N-glycosylation sites of NCEH (Asn270, Asn367, and Asn389) are glycosylated (Fig. 5B). In particular, N-linked glycosylation at Asn270 plays an important role in catalysis (Fig. 5D). The presence of N-oligosaccharides is required for the correct folding of many glycoproteins (41). N-linked glycosylation at Asn270 may play a more important role in the correct folding of NCEH. Asn389 was important for NCEH to form double bands (Fig. 5B, C). Asn389 of the 50 kDa band is glycosylated and Asn389 of the 45 kDa band is not. Because Asn389 is not important for the enzymatic activity of NCEH (Fig. 5D), the forms of NCEH differing in glycosylation at Asn389 may have similar enzymatic activity. A functional difference between these two is currently unknown.
Generally, the CE accumulated in macrophage foam cells is in LDs and its hydrolysis requires the association of the CE hydrolases with LDs. Indeed, HSL and CEH are largely cytosolic proteins. Egan et al. (32) reported that HSL translocates from the cytosol to LDs upon stimulation with catecholamines in adipocytes. Zhao et al. (42) reported that CEH translocates to LDs upon cholesterol loading in macrophages. Yet, hCE-1 and TGH, synonyms of CEH, have been reported to distribute to the ER via its C-terminal ER targeting signal, HIEL, in hepatocytes (43). It is unknown why subcellular localization of the same protein is different between different types of cells. In contrast to these two cytosolic enzymes, it is difficult to imagine how NCEH, an ER protein with its catalytic domain in the ER lumen, hydrolyses CE, which accumulates in LDs in foam cells primarily increasing the efflux of cholesterol from THP-1 cells (13). Recently, our understanding of the biogenesis of LDs in adipocytes has rapidly advanced (44). According to the current model, as lipid pools between the two leaflets of the ER enlarge, LDs are formed from the budding of cytoplasmic leaflets surrounding a lipid pool. Probably, CE synthesized by ACAT1, an ER polytopic protein with its catalytic site facing the cytoplasm (30, 31), accumulates in LDs in macrophages through the same pathway (45). Most LDs remain attached to the ER. Communication between LDs and ER may be more active than has been considered. For example, Rab 18 is present on LDs and its overexpression increases the association of LDs with the ER (46, 47). Therefore, it may not necessary to postulate a direct role for NCEH in the hydrolysis of CE in the LDs. CE in the LDs is continuously undergoing a futile cycle of hydrolysis, transport of the sterol to the ER for reesterification, and transport back to the LDs. NCEH may alter CE content in the LDs by disrupting this cycle. The apparently paradoxical hydrolysis of neutral lipids in LDs by ER luminal enzymes is also observed in adipocytes and possibly hepatocytes. TGH (TGH-1, hCE-1, and CEH) and TGH-2, which are mainly localized to the ER lumen, hydrolyzes triglyceride in LDs of hepatocytes and/or adipocytes (48).
It was reported that nCEH activity is increased by cAMP in WEHI, P388D1, or J774 macrophages (49–51). In this study, the nCEH activity of lysates prepared from MPMs was increased 20% by cAMP, but the difference was not significant. Previously, we reported that NCEH plays a major role in the nCEH activity of MPMs (13). On the other hand, it is thought that HSL plays a greater role than NCEH in the nCEH activity of macrophage immortal cell lines, such as RAW264.7 and J774(13). It depends on the ratio of expression of NCEH and HSL whether the nCEH activity of macrophage cell lysates is significantly increased by cAMP.
In summary, NCEH is a single-membrane-spanning ER protein that anchors to the ER membrane with an N-terminal hydrophobic segment. Its catalytic and lipid-binding domains are located in the ER lumen. The N-terminal region serves as a signal sequence that recruits NCEH to the ER lumen. N-linked glycosylation at Asn270 is crucial for its enzymatic activity. Elucidation of the mechanisms by which NCEH removes cholesterol from macrophage foam cells would pave the way for the development of novel therapies for the prevention of atherosclerosis.
Note added in proof
NCEH1 has been registered as an official name of NCEH, KIAA1363, or AADACL1 in HUGO. After acceptance of this paper, we reported the generation and phenotypic analysis of mice lacking Nceh1 (Cell Metab. 2009. 10:219–228).
Supplementary Material
Footnotes
Abbreviations:
- CE
- cholesterol ester
- CEH
- cholesterol ester hydrolase
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- HSL
- hormone-sensitive lipase
- LD
- lipid droplet
- MPM
- murine peritoneal macrophage
- nCEH
- neutral cholesterol ester hydrolase
- NCEH
- neutral cholesterol ester hydrolase
- PDI
- protein disulfide isomerase
- PKA
- protein kinase A
- PNPB
- p-nitrophenyl butyrate
- TGH
- triacylglycerol hydrolase
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture, the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, and the Takeda Science Foundation.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.Brown M. S., Goldstein J. L., Krieger M., Ho Y. K., Anderson R. G. 1979. Reversible accumulation of cholesteryl esters in macrophages incubated with acetylated lipoproteins. J. Cell Biol. 82: 597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kritharides L., Christian A., Stoudt G., Morel D., Rothblat G. H. 1998. Cholesterol metabolism and efflux in human THP-1 macrophages. Arterioscler. Thromb. Vasc. Biol. 18: 1589–1599. [DOI] [PubMed] [Google Scholar]
- 3.Graham A., Angell A. D., Jepson C. A., Yeaman S. J., Hassall D. G. 1996. Impaired mobilisation of cholesterol from stored cholesteryl esters in human (THP-1) macrophages. Atherosclerosis. 120: 135–145. [DOI] [PubMed] [Google Scholar]
- 4.Ishii I., Oka M., Katto N., Shirai K., Saito Y., Hirose S. 1992. Beta-VLDL-induced cholesterol ester deposition in macrophages may be regulated by neutral cholesterol esterase activity. Arterioscler. Thromb. 12: 1139–1145. [DOI] [PubMed] [Google Scholar]
- 5.Rothblat G. H., de la Llera-Moya M., Favari E., Yancey P. G., Kellner-Weibel G. 2002. Cellular cholesterol flux studies: methodological considerations. Atherosclerosis. 163: 1–8. [DOI] [PubMed] [Google Scholar]
- 6.Li F., Hui D. Y. 1997. Modified low density lipoprotein enhances the secretion of bile salt-stimulated cholesterol esterase by human monocyte-macrophages. Species-specific difference in macrophage cholesteryl ester hydrolase. J. Biol. Chem. 272: 28666–28671. [DOI] [PubMed] [Google Scholar]
- 7.Holm C. 2003. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem. Soc. Trans. 31: 1120–1124. [DOI] [PubMed] [Google Scholar]
- 8.Yeaman S. J. 2004. Hormone-sensitive lipase–new roles for an old enzyme. Biochem. J. 379: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S. 2000. Cholesteryl ester hydrolase in human monocyte/macrophage: cloning, sequencing, and expression of full-length cDNA. Physiol. Genomics. 2: 1–8. [DOI] [PubMed] [Google Scholar]
- 10.Long R. M., Calabrese M. R., Martin B. M., Pohl L. R. 1991. Cloning and sequencing of a human liver carboxylesterase isoenzyme. Life Sci. 48: PL43–PL49. [DOI] [PubMed] [Google Scholar]
- 11.Munger J. S., Shi G. P., Mark E. A., Chin D. T., Gerard C., Chapman H. A. 1991. A serine esterase released by human alveolar macrophages is closely related to liver microsomal carboxylesterases. J. Biol. Chem. 266: 18832–18838. [PubMed] [Google Scholar]
- 12.Lehner R., Vance D. E. 1999. Cloning and expression of a cDNA encoding a hepatic microsomal lipase that mobilizes stored triacylglycerol. Biochem. J. 343: 1–10. [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki H., Igarashi M., Nishi M., Sekiya M., Tajima M., Takase S., Takanashi M., Ohta K., Tamura Y., Okazaki S., et al. 2008. Identification of neutral cholesterol ester hydrolase, a key enzyme removing cholesterol from macrophages. J. Biol. Chem. 283: 33357–33364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nomura D. K., Leung D., Chiang K. P., Quistad G. B., Cravatt B. F., Casida J. E. 2005. A brain detoxifying enzyme for organophosphorus nerve poisons. Proc. Natl. Acad. Sci. USA. 102: 6195–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contreras J. A. 2002. Hormone-sensitive lipase is not required for cholesteryl ester hydrolysis in macrophages. Biochem. Biophys. Res. Commun. 292: 900–903. [DOI] [PubMed] [Google Scholar]
- 16.Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoiri F., Yahagi N., Kraemer F. B., Tsutsumi O., et al. 2000. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. USA. 97: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B., Song J., Chow W. N., St Clair R. W., Rudel L. L., Ghosh S. 2007. Macrophage-specific transgenic expression of cholesteryl ester hydrolase significantly reduces atherosclerosis and lesion necrosis in Ldlr mice. J. Clin. Invest. 117: 2983–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okazaki H., Igarashi M., Nishi M., Tajima M., Sekiya M., Okazaki S., Yahagi N., Ohashi K., Tsukamoto K., Amemiya-Kudo M., et al. 2006. Identification of a novel member of the carboxylesterase family that hydrolyzes triacylglycerol: a potential role in adipocyte lipolysis. Diabetes. 55: 2091–2097. [DOI] [PubMed] [Google Scholar]
- 19.Jessani N., Liu Y., Humphrey M., Cravatt B. F. 2002. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc. Natl. Acad. Sci. USA. 99: 10335–10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang K. P., Niessen S., Saghatelian A., Cravatt B. F. 2006. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 13: 1041–1050. [DOI] [PubMed] [Google Scholar]
- 21.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein J. L., Basu S. K., Brown M. S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260. [DOI] [PubMed] [Google Scholar]
- 23.Ling M. M., Robinson B. H. 1997. Approaches to DNA mutagenesis: an overview. Anal. Biochem. 254: 157–178. [DOI] [PubMed] [Google Scholar]
- 24.Okazaki H., Osuga J., Tamura Y., Yahagi N., Tomita S., Shionoiri F., Iizuka Y., Ohashi K., Harada K., Kimura S., et al. 2002. Lipolysis in the absence of hormone-sensitive lipase: evidence for a common mechanism regulating distinct lipases. Diabetes. 51: 3368–3375. [DOI] [PubMed] [Google Scholar]
- 25.Yagyu H., Kitamine T., Osuga J., Tozawa R., Chen Z., Kaji Y., Oka T., Perrey S., Tamura Y., Ohashi K., et al. 2000. Absence of ACAT-1 attenuates atherosclerosis but causes dry eye and cutaneous xanthomatosis in mice with congenital hyperlipidemia. J. Biol. Chem. 275: 21324–21330. [DOI] [PubMed] [Google Scholar]
- 26.Hajjar D. P., Minick C. R., Fowler S. 1983. Arterial neutral cholesteryl esterase. A hormone-sensitive enzyme distinct from lysosomal cholesteryl esterase. J. Biol. Chem. 258: 192–198. [PubMed] [Google Scholar]
- 27.Shirai K., Fitzharris T. J., Shinomiya M., Muntz H. G., Harmony J. A., Jackson R. L., Quinn D. M. 1983. Lipoprotein lipase-catalyzed hydrolysis of phosphatidylcholine of guinea pig very low density lipoproteins and discoidal complexes of phospholipid and apolipoprotein: effect of apolipoprotein C–II on the catalytic mechanism. J. Lipid Res. 24: 721–730. [PubMed] [Google Scholar]
- 28.Lin S., Lu X., Chang C. C., Chang T. Y. 2003. Human acyl-coenzyme A:cholesterol acyltransferase expressed in chinese hamster ovary cells: membrane topology and active site location. Mol. Biol. Cell. 14: 2447–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feramisco J. D., Goldstein J. L., Brown M. S. 2004. Membrane topology of human insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 279: 8487–8496. [DOI] [PubMed] [Google Scholar]
- 30.Joyce C. W., Shelness G. S., Davis M. A., Lee R. G., Skinner K., Anderson R. A., Rudel L. L. 2000. ACAT1 and ACAT2 membrane topology segregates a serine residue essential for activity to opposite sides of the endoplasmic reticulum membrane. Mol. Biol. Cell. 11: 3675–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S., Cheng D., Liu M. S., Chen J., Chang T. Y. 1999. Human acyl-CoA:cholesterol acyltransferase-1 in the endoplasmic reticulum contains seven transmembrane domains. J. Biol. Chem. 274: 23276–23285. [DOI] [PubMed] [Google Scholar]
- 32.Egan J. J., Greenberg A. S., Chang M. K., Wek S. A., Moos M. C., Jr., Londos C. 1992. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc. Natl. Acad. Sci. USA. 89: 8537–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakaguchi M., Tomiyoshi R., Kuroiwa T., Mihara K., Omura T. 1992. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc. Natl. Acad. Sci. USA. 89: 16–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook K. G., Colbran R. J., Snee J., Yeaman S. J. 1983. Cytosolic cholesterol ester hydrolase from bovine corpus luteum. Its purification, identification, and relationship to hormone-sensitive lipase. Biochim. Biophys. Acta. 752: 46–53. [DOI] [PubMed] [Google Scholar]
- 35.Escary J. L., Choy H. A., Reue K., Schotz M. C. 1998. Hormone-sensitive lipase overexpression increases cholesteryl ester hydrolysis in macrophage foam cells. Arterioscler. Thromb. Vasc. Biol. 18: 991–998. [DOI] [PubMed] [Google Scholar]
- 36.Pelham H. R. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15: 483–486. [DOI] [PubMed] [Google Scholar]
- 37.Munro S., Pelham H. R. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 48: 899–907. [DOI] [PubMed] [Google Scholar]
- 38.Blobel G. 1980. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA. 77: 1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gafvelin G., Sakaguchi M., Andersson H., von Heijne G. 1997. Topological rules for membrane protein assembly in eukaryotic cells. J. Biol. Chem. 272: 6119–6127. [DOI] [PubMed] [Google Scholar]
- 40.Mziaut H., Korza G., Hand A. R., Gerard C., Ozols J. 1999. Targeting proteins to the lumen of endoplasmic reticulum using N-terminal domains of 11beta-hydroxysteroid dehydrogenase and the 50-kDa esterase. J. Biol. Chem. 274: 14122–14129. [DOI] [PubMed] [Google Scholar]
- 41.Ronnett G. V., Knutson V. P., Kohanski R. A., Simpson T. L., Lane M. D. 1984. Role of glycosylation in the processing of newly translated insulin proreceptor in 3T3–L1 adipocytes. J. Biol. Chem. 259: 4566–4575. [PubMed] [Google Scholar]
- 42.Zhao B., Fisher B. J., St Clair R. W., Rudel L. L., Ghosh S. 2005. Redistribution of macrophage cholesteryl ester hydrolase from cytoplasm to lipid droplets upon lipid loading. J. Lipid Res. 46: 2114–2121. [DOI] [PubMed] [Google Scholar]
- 43.Gilham D., Alam M., Gao W., Vance D. E., Lehner R. 2005. Triacylglycerol hydrolase is localized to the endoplasmic reticulum by an unusual retrieval sequence where it participates in VLDL assembly without utilizing VLDL lipids as substrates. Mol. Biol. Cell. 16: 984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodman J. M. 2008. The gregarious lipid droplet. J. Biol. Chem. 283: 28005–28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang T. Y., Chang C. C., Ohgami N., Yamauchi Y. 2006. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22: 129–157. [DOI] [PubMed] [Google Scholar]
- 46.Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118: 2601–2611. [DOI] [PubMed] [Google Scholar]
- 47.Martin S., Driessen K., Nixon S. J., Zerial M., Parton R. G. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280: 42325–42335. [DOI] [PubMed] [Google Scholar]
- 48.Soni K. G., Lehner R., Metalnikov P., O'Donnell P., Semache M., Gao W., Ashman K., Pshezhetsky A. V., Mitchell G. A. 2004. Carboxylesterase 3 (EC 3.1.1.1) is a major adipocyte lipase. J. Biol. Chem. 279: 40683–40689. [DOI] [PubMed] [Google Scholar]
- 49.Khoo J. C., Mahoney E. M., Steinberg D. 1981. Neutral cholesterol esterase activity in macrophages and its enhancement by cAMP-dependent protein kinase. J. Biol. Chem. 256: 12659–12661. [PubMed] [Google Scholar]
- 50.Goldberg D. I., Khoo J. C. 1990. Stimulation of a neutral cholesteryl ester hydrolase by cAMP system in P388D1 macrophages. Biochim. Biophys. Acta. 1042: 132–137. [DOI] [PubMed] [Google Scholar]
- 51.Small C. A., Rogers M. P., Goodacre J. A., Yeaman S. J. 1991. Phosphorylation and activation of hormone-sensitive lipase in isolated macrophages. FEBS Lett. 279: 323–326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.